Abstract
Disassembly of RecA protein subunits from a RecA filament has long been known to occur during DNA strand exchange, although its importance to this process has been controversial. An Escherichia coli RecA E38K/ΔC17 double mutant protein displays a unique and pH-dependent mutational separation of DNA pairing and extended DNA strand exchange. Single strand DNA-dependent ATP hydrolysis is catalyzed by this mutant protein nearly normally from pH 6 to 8.5. It will also form filaments on DNA and promote DNA pairing. However, below pH 7.3, ATP hydrolysis is completely uncoupled from extended DNA strand exchange. The products of extended DNA strand exchange do not form. At the lower pH values, disassembly of RecA E38K/ΔC17 filaments is strongly suppressed, even when homologous DNAs are paired and available for extended DNA strand exchange. Disassembly of RecA E38K/ΔC17 filaments improves at pH 8.5, whereas complete DNA strand exchange is also restored. Under these sets of conditions, a tight correlation between filament disassembly and completion of DNA strand exchange is observed. This correlation provides evidence that RecA filament disassembly plays a major role in, and may be required for, DNA strand exchange. A requirement for RecA filament disassembly in DNA strand exchange has a variety of ramifications for the current models linking ATP hydrolysis to DNA strand exchange.
Keywords: DNA, DNA/Recombination, DNA/Repair, Nucleic Acid/Enzymology, Protein/Nucleic Acid Interaction, RecA
Introduction
The RecA protein of Escherichia coli catalyzes homologous DNA pairing and strand exchange reactions that are central to recombination and recombinational DNA repair (1–4). RecA protein is found in virtually all bacteria. RecA homologs, such as RadA (5, 6) and Rad51 (7–10), are ubiquitous in archaea and eukaryotes, respectively.
The DNA strand exchange reaction promoted by RecA protein occurs in several distinct phases. The first phase is binding of RecA protein to single-stranded DNA (ssDNA)2 to form a helical nucleoprotein filament. Filament formation occurs in at least two steps, with a slow nucleation step preceding a rapid 5′ to 3′ extension of the filament to encompass the available DNA (11–13). During the second phase of DNA strand exchange, RecA aligns homologous DNA strands and catalyzes strand exchange over a short distance producing a length of hybrid DNA that typically consists of up to 1000 bp (2, 14). In the third phase of DNA strand exchange, RecA catalyzes extension of the hybrid DNA region via facilitated branch migration that occurs 5′ to 3′ with respect to the bound ssDNA (15).
RecA protein subunits within a nucleoprotein filament hydrolyze ATP in a DNA-dependent manner. ATP is hydrolyzed uniformly throughout the RecA nucleoprotein filament (16) with a kcat of about 30 min−1 when RecA is bound to ssDNA. The kcat is reduced to 20 min−1 when RecA binds directly to double-stranded (ds) DNA (3) or when a RecA filament is catalyzing DNA strand exchange (17, 18). Homologous pairing and formation of short stretches of exchanged DNA require the binding of ATP but not ATP hydrolysis (19–22). However, the efficient extension of this newly formed heteroduplex DNA via RecA-promoted branch migration requires ATP hydrolysis (21, 22). When ATP is hydrolyzed, the rates of branch migration and ATP hydrolysis are coupled (17), and the reaction proceeds unidirectionally (15, 22). ATP hydrolysis also enables RecA to bypass a variety of structural barriers during DNA strand exchange, such as heterologous insertions (22, 23), and to carry out exchange between two homologous duplex DNA molecules (22, 24).
Efficient RecA-mediated DNA strand exchange is facilitated by additional proteins, in particular the single strand DNA-binding protein (SSB). SSB melts the secondary structure in ssDNA that can block filament extension (25, 26) and sequesters the single strand of DNA that is displaced from the duplex substrate during exchange (27). However, if SSB is bound to ssDNA before RecA, the nucleation step of filament formation is strongly inhibited (12, 28–31). Thus, SSB is included in most RecA reactions only after RecA nucleation has already occurred. In vivo, SSB provides a barrier to RecA filament nucleation that is overcome with the aid of recombination mediator proteins (32). In E. coli, the proteins most clearly implicated in facilitating RecA nucleation onto SSB-bound DNA are the RecF, RecO, and RecR proteins (12, 30, 33–35) and the RecBCD helicase/nuclease (36–38).
Certain RecA mutants show an enhanced ability to nucleate onto SSB-coated DNA in vitro without the aid of recombination mediator proteins. Two of these mutants include RecA730, a single point mutation of residue glutamate 38 to lysine (E38K) (39, 40), and RecA ΔC17, a truncation of the last 17 C-terminal residues of the protein (29, 41, 42).
In the crystal structures of RecA (43–46), Glu-38 is positioned on the outer surface of the RecA filament. The crystal structure of RecA protein bound to DNA shows that Glu-38 makes no contact with the DNA sequestered inside the filament (46). It is not clear how this mutation speeds RecA nucleation onto ssDNA bound by SSB.
The last 25 C-terminal amino acid residues of RecA protein, here referred to as the C terminus (only a part of the C-terminal domain of RecA), include 7 residues that are negatively charged. All of these negatively charged residues are removed in the ΔC17 truncation mutant protein. The RecA C terminus modulates the optimal reaction conditions for RecA reactions (41, 42, 47, 48), the entry of linear double-stranded (lds) DNA into a RecA-ssDNA filament during DNA strand exchange (41), and nucleation of RecA filaments on SSB-coated ssDNA (29).
The RecA E38K/ΔC17 double mutant further enhances the capacity of RecA to nucleate onto SSB-coated ssDNA when compared with each individual mutant (29). Because the last 24 residues of the C terminus are not ordered or are omitted in most RecA crystal structures (44–46), we do not know if there is a functional interaction between the C terminus and residue Glu-38.
In this study, we find that the combination of the two mutations also affects the coupling of ATP hydrolysis to DNA strand exchange. Although we know that hydrolysis is necessary to complete exchange of long DNA substrates, it is still not known how these processes are coupled. There are basically three extant models to link DNA strand exchange to RecA-catalyzed ATP hydrolysis.
The facilitated DNA rotation model (reviewed in Ref. 49) proposes that the energy from ATP hydrolysis is used to rotate DNA strands around the outside of a RecA protein filament. Rotation of one DNA molecule around another could create the torsional stress needed to unwind regions of heterology during DNA strand exchange, such that they could be bypassed. Rotation of one DNA strand around the outside of a RecA filament could also explain how RecA filaments catalyze exchange between two duplex DNA molecules when only three DNA strands can be accommodated inside a filament (reviewed in Ref. 50). However, rotation of DNA around RecA filaments has never been observed directly, and the DNA-binding sites that would be needed on the exterior of the filament to effect this rotation have never been identified.
In the RecA redistribution model (21, 51, 52), disassembly of RecA monomers from a filament is the only ATP hydrolysis-dependent step. According to this model, DNA strand exchange is limited by discontinuities in RecA filaments. These discontinuities can be filled if RecA filaments dissociate and the monomers are redistributed. This model also allows for bypass of heterologous inserts during DNA strand exchange (21, 51, 52). However, no proposal has been made for how the RecA redistribution model might couple ATP hydrolysis to exchange involving two duplex DNA substrates.
A third model can be articulated, based in large measure on an early proposal of Howard-Flanders et al. (53) for RecA-mediated DNA strand exchange. Instead of focusing on discontinuities in the RecA filament, this model posits that branch migration is directly coupled to dissociation of RecA monomers and a concurrent conformational change that allows for exchange of the DNA strands (49). It is not clear how disassembly of RecA filament subunits would allow for exchange between two duplex DNA molecules. Because ATP hydrolysis is only linked to DNA strand exchange through disassembly of RecA subunits from the 5′-proximal end of the filament, neither this model nor the RecA redistribution model explains why the entire RecA filament hydrolyzes ATP.
The RecA filament redistribution and RecA disassembly models both rely on the dissociation of filament subunits. Such dissociation is known to occur during DNA strand exchange (54). However, this dissociation could be incidental, and arguments have been advanced both for and against a direct link between this dissociation and DNA strand exchange (21, 22, 49, 55, 56). Conditions or RecA filament alterations that permit ATP hydrolysis, but block subunit dissociation, would allow a much more complete correlation and exploration of the role of disassembly in strand exchange.
Here, we find that there is a pH-dependent separation of RecA-mediated homologous pairing and RecA-mediated branch migration in the RecA E38K/ΔC17 mutant. At lower pH values, ATP hydrolysis and extended DNA strand exchange are completely uncoupled. We exploit this characteristic of the RecA double mutant to further investigate the role of RecA filament disassembly in DNA strand exchange. This study reveals that disassembly of mutant RecA filaments during DNA strand exchange is also uncoupled from ATP hydrolysis at pH values that are nonpermissive for branch migration. Altering the pH to restore filament disassembly also restores branch migration. Formation of products during DNA strand exchange does not occur when disassembly is inhibited, providing the first experimental evidence that disassembly not only occurs during DNA strand exchange but may be necessary for the process to be completed.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Potassium phosphate, potassium chloride, magnesium acetate, EDTA, and glycerol were purchased from Fisher. Ammonium sulfate was purchased from MP Biomedicals. Dithiothreitol (DTT) was purchased from Research Organics. Tris base was purchased from RPI. Restriction endonucleases and the 2-log DNA molecular weight marker were purchased from New England Biolabs. Chromatography resins were purchased from GE Healthcare except for the ceramic hydroxyapatite purchased from Bio-Rad. All other chemicals and reagents were purchased from Sigma unless otherwise noted.
Proteins
The wild type E. coli RecA protein was purified as described (31). Its concentration was determined using the extinction coefficient ϵ280 = 2.23 × 104 m−1 cm−1 (57). The SSB protein was purified as described (58); its concentration was determined using the extinction coefficient ϵ280 = 2.38 × 104 m−1 cm−1. The RecA E38K/ΔC17 protein was purified using a modified purification protocol for the RecA ΔC17 protein (42). All purification steps were carried out at 4 °C. Cell paste containing RecA E38K/ΔC17 protein was flash-frozen with liquid N2 and then thawed overnight on ice in a solution of 250 mm Tris-HCl (80% cation, pH 7.8) and 25% (w/v) sucrose. The resuspended cells were adjusted to 20% (w/v). The cells were lysed for 45 min with a lysozyme solution (2.5 mg/ml final concentration lysozyme in 250 mm Tris-HCl (80% cation, pH 7.8)), followed by the addition of 0.4 ml 25 mm EDTA per ml of lysis solution. The lysate was sonicated and then centrifuged to remove cell debris. RecA E38K/ΔC17 protein was precipitated from the lysate supernatant with 0.111 ml of 5% (w/v) polyethyleneimine per ml of lysate. The pellet was washed with R-buffer (20 mm Tris-HCl (80% cation, pH 7.8), 10% glycerol, 0.1 mm EDTA, 1 mm DTT), and RecA E38K/ΔC17 was then extracted from the pellet twice with R-buffer + 150 mm ammonium sulfate. The extracted RecA protein was precipitated with 0.31 g/ml ammonium sulfate and centrifuged. The resulting pellet was washed twice with R-buffer + 0.31 g/ml ammonium sulfate. The pellet was resuspended in R-buffer + 1 m ammonium sulfate and then loaded on a phenyl-Sepharose 6 fast flow (low sub) column. Most of the RecA E38K/ΔC17 protein was not retained by the column and was collected in the flow-through fraction. This fraction was dialyzed into R-buffer and applied to a Source 15-Q column. RecA E38K/ΔC17 protein was eluted from the Source 15-Q column with a 10 column volume linear gradient of R-buffer to R-buffer + 1 m KCl. The eluted RecA mutant protein was dialyzed into P-buffer (20 mm potassium phosphate, 10% glycerol, 0.1 mm EDTA, and 1 mm DTT) and applied to a ceramic hydroxyapatite column. RecA E38K/ΔC17 protein was eluted from the column with a linear gradient from P-buffer to 1 m P-buffer (same as P-buffer, except with 1 m potassium phosphate). Peak fractions were analyzed by SDS-PAGE. Fractions containing the mutant RecA protein were pooled and dialyzed into R-buffer for storage. The concentration was determined using the wild type RecA protein extinction coefficient. The purified protein was free of detectable nuclease activity.
The RecA K72R protein was purified as described (22) with the following changes. The RecA K72R protein precipitated by ammonium sulfate was resuspended in R-buffer + 1 m ammonium sulfate and loaded onto a phenyl-Sepharose 6 fast flow (low sub) column. The RecA K72R protein eluted in the flow-through from the phenyl-Sepharose column. The flow-through fractions were pooled, dialyzed into R-buffer, and applied to a Source 15 Q column. The RecA K72R protein was eluted with a linear 10-column volume gradient of R-buffer to R-buffer + 1 m KCl. The fractions containing RecA K72R were dialyzed into P-buffer and applied to a ceramic hydroxyapatite and eluted with a 10 column volume gradient of P-buffer to 0.5 m P-buffer (same as P-buffer except 0.5 m potassium phosphate). Fractions containing RecA K72R protein were identified via SDS-PAGE, and the RecA K72R protein was concentrated by precipitation with 0.35 mg/ml ammonium sulfate. The pellet was resuspended in R-buffer and dialyzed against it for storage. The RecA K72R protein was free of detectable nuclease activity. The concentration of the purified protein was determined via absorbance at 280 nm using the wild type RecA protein extinction coefficient.
DNA Substrates
Supercoiled DNA and cssDNA substrates were derived from the bacteriophages M13mp7 and M13mp18 and were purified by CsCl banding as described (59–61). The ldsDNA used in the strand exchange reactions was generated by complete digestion of M13mp7 or M13mp18 supercoiled circular DNA with BsmBI according to the manufacturer's instructions. The nicked circular double strand DNA used in the ATP hydrolysis assays was prepared by large scale RecA-mediated DNA strand exchange reactions as described (22). The concentrations of DNA substrates were determined by absorbance at 260 nm using the conversion factors 108 μm nucleotides A260−1 and 151 μm nucleotides A260−1 for single-stranded and double-stranded DNA, respectively. DNA concentrations are expressed as the total concentration of nucleotides.
DNA Strand Exchange Reactions
The DNA strand exchange reactions were carried out at 37 °C in solutions containing 25 mm buffer (varied as indicated to alter the pH), 1 mm DTT, 3 mm potassium glutamate, 5% (w/v) glycerol, and an ATP regeneration system consisting of 3 mm P-enolpyruvate and 10 units/ml pyruvate kinase. The magnesium acetate concentration was 5 mm for reactions containing RecA ΔC17 or E38K/ΔC17. Reactions containing wild type RecA or RecA E38K had 10 mm magnesium acetate. The buffers used were MES (33% anion, pH 6.0), MES (55% anion, pH 6.4), HEPES (17% anion, pH 7.0), Tris-OAc (100% cation, pH 7.3), Tris-OAc (80% cation, pH 7.8), Tris-OAc (56% cation, pH 8.0), CHES (12% anion, pH 8.5), CHES (33% anion, pH 8.8), CHES (70% anion, pH 9.3), and CHES (100% anion, pH 9.6). The reported pH values reflect the final pH obtained after addition of all reaction components. Typically, 3.5 μm RecA and 10 μm cssDNA (M13mp7 or M13mp18) were preincubated in the reaction buffer and regeneration system for 10 min before addition of 3 mm ATP and one-tenth the cssDNA concentration of SSB. The reactions were then started by adding 20 μm homologous linear double-stranded DNA after 20 min of incubation at 37 °C with ATP. The reactions were incubated at 37 °C for 2 h, unless otherwise specified in the figures. Reaction aliquots (10 μl) were deproteinized by addition of a mixture containing 1.2 μl of 10% SDS, 0.3 μl of 0.5 m EDTA, and 0.6 μl of 20 mg/ml proteinase K and incubated for 30 min at 37 °C. Aliquots were then mixed with 2.5 μl of loading buffer (15% Ficoll, 0.25% bromphenol blue, 0.25% xylene cyanol FF), loaded on a 1% agarose gel, and electrophoresed at 20–30 mA overnight at room temperature. The DNA was visualized by ethidium bromide staining and exposure to UV light. Gel images were captured with a Fotodyne FOTO/Analyst® CCD camera, PC Image acquisition software, and a FOTO/Convertible dual transilluminator. The intensity of DNA bands was measured using the software package TotalLab version TL100 from Nonlinear Dynamics. In the strand exchange reaction featuring a shift in pH from 7 to 8.1, the shift was carried out by adding 5% of the total reaction volume of 0.5 m CHES (100% anion) after the 45-min time point. The pH values of each reaction were measured after the reactions were initiated and after the pH was shifted.
ATP Hydrolysis Assays
A coupled spectrophotometric enzyme assay (62, 63) was used to measure the ATPase activities of the E38K/ΔC17 on circular ssDNA and nicked circular dsDNA. The regeneration of ATP from ADP and P-enolpyruvate was coupled to the oxidation of NADH and monitored by observing the decrease in absorbance at 380 nm over time. An NADH extinction coefficient of ϵ380 = 1.21 mm−1cm−1 was used to calculate the amount of ATP hydrolyzed over time. The assays were carried out in a Varian Cary 300 dual beam spectrophotometer equipped with a temperature controller and a 12-position cell changer. The cell path lengths are 1 or 0.5 cm and bandpass was 2 nm.
The reactions were carried out at 37 °C with 25 mm buffer (varied as indicated to alter pH). The concentration of magnesium acetate was 10 or 5 mm for wild type or for RecA E38K/ΔC17 protein, respectively. The reactions contained 1 mm DTT, 3 mm potassium glutamate, 5% (w/v) glycerol, an ATP regeneration system (3 mm P-enolpyruvate and 10 units/ml pyruvate kinase), and a coupling system (10 units/ml lactate dehydrogenase, 2 mm NADH for 1-cm path length cuvettes, or 3 mm NADH for 0.5 cm path length cuvettes). Wild type or RecA E38K/ΔC17 protein, at a concentration of 2 μm, was incubated in the reaction solution with 4 μm M13mp7 cssDNA or 4.2 μm M13mp7 nicked circular double-stranded DNA, and the regeneration/coupling system for 10 min before 3 mm ATP (and 0.4 μm SSB in the reactions containing cssDNA only) was added to initiate the reaction.
ATP Hydrolysis during DNA Strand Exchange and RecA K72R Challenge Assays
The coupled spectrophotometric enzyme assay described above was used to measure the rate of RecA-catalyzed ATP hydrolysis during DNA strand exchange. The reactions were carried out at 37 °C in solutions containing 25 mm buffer (varied as indicated to change the pH), 5% (w/v) glycerol, an ATP regeneration system of 4 mm P-enolpyruvate, and 10 units/ml pyruvate kinase, a coupling system of 4.5 mm NADH, and 10 units/ml lactate dehydrogenase, 3 mm potassium glutamate, and 10 or 5 mm magnesium acetate for wild type or RecA E38K/ΔC17 protein, respectively. The buffers used were Tris-OAc (80% cation, pH 7.8), HEPES (17% anion, pH 7.0), and CHES (12% anion, pH 8.5). Wild type or RecA E38K/ΔC17 protein at a concentration of 2 μm was incubated with the reaction solution, regeneration system, coupling system, and 4 μm M13mp18 cssDNA for 10 min before 3 mm ATP and 0.4 μm SSB were added to start the ATP hydrolysis reaction. Hydrolysis was measured for 20 min before the addition of 8 μm M13mp18 ldsDNA, which started the DNA strand exchange reaction. An equivalent volume of TE buffer was added in place of ldsDNA as a control in one reaction per experiment. ATP hydrolysis was monitored for another 30 min before the addition of 4 μm RecA K72R protein or an equivalent volume of its storage buffer as a control. ATP hydrolysis was monitored for approximately another 100 min.
Samples of 15 μl were removed from the reaction solution at various times during the DNA strand exchange reaction and added to a solution of 4.5 μl of 15% (w/v) Ficoll dissolved in 20 mm Tris-OAc (80% cation, pH 7.8) and 3 μl of 10% SDS. The deproteinized samples were loaded and run on 0.8% agarose gels as described above. Images of the DNA bands and quantitation of band intensities were obtained as described above.
ATP Hydrolysis during DNA Strand Exchange and RecA K72R Challenge and pH Shift Assays
The reaction solution, ATP regeneration system, coupling systems, and protein and DNA concentrations are the same as in the ATP hydrolysis during DNA strand exchange assays described above. HEPES buffer (17% anion) was used to set the starting pH to 7.0. RecA E38K/ΔC17 protein was incubated with the reaction solution, the regeneration system, the coupling system, and cssDNA for 10 min at 37 °C. ATP and SSB were added to start the ATP hydrolysis reaction. The ATP hydrolysis reaction proceeded for 10 min before the addition of ldsDNA, or a TE control, to start the DNA strand exchange reaction. RecA K72R protein at 4 μm was added to the reaction mixture 15 min after the ldsDNA. An equivalent volume of RecA K72R storage buffer was added as a control to those reactions without RecA K72R. The rate of ATP hydrolysis was measured for another 15 min before an empirically determined amount of 1 m CHES (100% anion, pH ∼10.7) was added to the reaction to abruptly shift the pH from 7.0 to 8.5. In this case, 18.56 μl of CHES were added to each 495-μl reaction. Those reactions without a pH shift received 1 m HEPES (17% anion, pH 7.0) as a control. The rate of ATP hydrolysis was measured for ∼150 more min. Samples for agarose gel electrophoresis, images of agarose gels, and DNA band intensities were obtained as described above.
Electron Microscopy Experiments
A modified Alcian method was used to visualize RecA filaments. Activated grids were prepared as described previously (42). Reaction mixtures contained 25 mm buffer (HEPES, 17% anion at pH 7.0, or Tris-OAc, 56% cation at pH 8.0), 5% (w/v) glycerol, 3 mm potassium glutamate, and an ATP regeneration system of 10 units/ml of creatine phosphokinase and 12 mm phosphocreatine. Reactions with wild type RecA contained 10 mm magnesium acetate, whereas reactions with RecA E38K/ΔC17 protein contained 5 mm magnesium acetate. Wild type or mutant RecA protein at 2.67 μm was incubated with 8 μm M13mp18 cssDNA for 10 min at 37 °C in the presence of all reaction components except ATP, SSB, or ATPγS. The reaction was initiated with the addition of ATP and SSB (3 mm and 0.8 μm, respectively) followed by another 10-min incubation at 37 °C. The RecA filaments were stabilized with 3 mm ATPγS and incubated for 3 min at 37 °C. An 8-μl sample of the reaction mixture described above was diluted 14 times in a solution consisting of 10 mm HEPES, 200 mm ammonium acetate, and 10% (w/v) glycerol (with the pH adjusted to 7.0 or 8.0 depending on the reaction solution conditions) followed by adsorption to the activated carbon film for 3 min. The grid was then touched to another drop of the above HEPES buffer and floated on a drop of the same buffer for 1 min. The sample was then stained by touching to a drop of 5% uranyl acetate solution and floated on another drop of the same solution for 30 s. Finally, the grid was washed by touching to a drop of double distilled water followed by two immersions in 10 ml of double distilled water and one immersion in 10 ml of 100% ethanol. After the sample was dried under a heat lamp for a minute, it was rotary-shadowed in an Edwards evaporator with platinum. The protocol is designed for visualization of complete reaction mixtures. Imaging and photography were carried out with a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 CCD camera. To determine the proportion of RecA nucleoprotein filaments that were either full or discontinuous, at least 280 RecA nucleoprotein filaments from at least five separate regions of two grids were counted at the identical magnification for each sample. A filament was considered “continuous” if it completely encompassed the circular DNA molecule with no visible break in the regular striations of the filament. A filament was considered “discontinuous” if it met one or both of the following criteria: it had at least one gap in which no RecA was visible, or the regular striations of the filament were disrupted creating an angular rather than a smooth circular filament. “Miscellaneous” filaments include any discrete RecA nucleoprotein filament that does not meet the criteria for the continuous or discontinuous filaments. For example, RecA filaments that form on linear ssDNA fall into this category.
The cytochrome c method was used to visualize M13mp18 circular single-stranded DNA to check the percentage of circular molecules. Samples were prepared as described previously (64) except the spreading solution was assembled by combining 32 μl of 1 m sodium carbonate, 40 μl of 0.126 m EDTA, 400 μl of 37% formaldehyde, 292 μl of 100% formamide, and 10 μl of 5 m potassium hydroxide. The final DNA concentration was 0.004 μg/μl.
RESULTS
Experimental Design
The purpose of this study was to characterize the RecA E38K/ΔC17 mutant and exploit its properties to further examine the mechanism of DNA strand exchange. For comparison, the work includes the wild type and also the E38K and ΔC17 single mutant RecA proteins.
We altered the DNA strand exchange reaction conditions slightly for each RecA protein to reflect its reaction optimum. To promote its most efficient DNA strand exchange reaction, the wild type RecA protein requires 10 mm Mg2+, 7 mm beyond that needed to chelate the ATP. The RecA ΔC17 mutant, in contrast, promotes the same reaction most efficiently with a reduced amount of Mg2+ (3 mm) (41). We found that the optimal range of Mg2+ concentrations for RecA E38K/ΔC17-catalyzed activities ranged from 3 to 5 mm (data not shown). The Mg2+ concentration used for the assays involving RecA E38K/ΔC17 in this study was 5 mm.
RecA E38K/ΔC17 Forms Filaments on cssDNA
The ability of wild type RecA protein to catalyze ATP hydrolysis is DNA-dependent; thus, ATP hydrolysis assays are often used to infer the extent of RecA binding to DNA. Visual confirmation that RecA protein forms extended filaments on DNA can be obtained by electron microscopy. The electron microscopy experiments were carried out with stoichiometric amounts of RecA protein and cssDNA so that the background of the micrographs would not be cluttered by excess RecA.
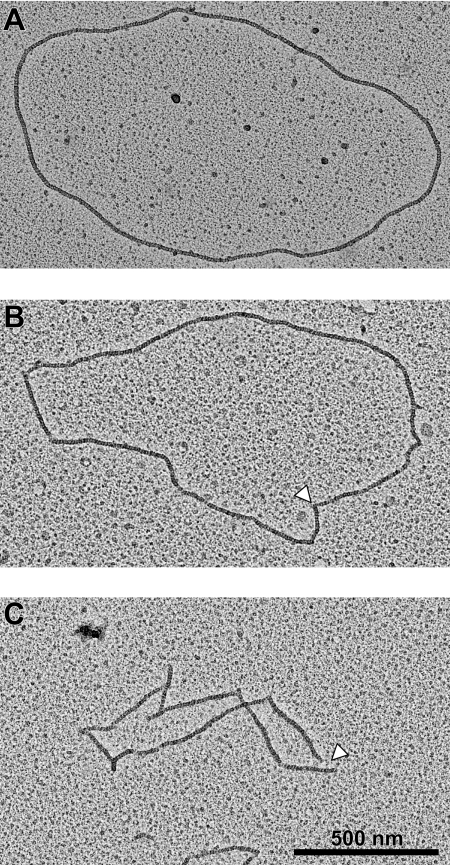
In brief, both wild type and E38K/ΔC17 RecA proteins primarily form filaments with short discontinuities on cssDNA at pH 7 (Table 1; Fig. 1). The double mutant protein exhibits a greater propensity to form discontinuous filaments at pH 8.0. The discontinuities range in size from minor disruptions that make an otherwise circular filament angular (Fig. 1B) to clearly visible gaps in the filament (Fig. 1C). The filaments with visible gaps make up the largest percentage of all the observed nucleoprotein filaments (38.9%) formed by RecA E38K/ΔC17 at this pH. Despite the filament discontinuities at pH 8.0, the EM data confirm that this mutant RecA protein forms filaments on cssDNA at pH 7.0 and 8.0, which is consistent with ATP being hydrolyzed by RecA at these pH values.
TABLE 1.
Morphologies of wild type and E38K/ΔC17 RecA filaments on cssDNA at pH 7.0 and 8.0
The wild type and mutant RecA filaments observed by electron microscopy were counted and grouped as described under “Experimental Procedures.” The reactions contained 8 μm M13mp18 cssDNA, 2.67 μm RecA, 3 mm ATP, 3 mm ATPγS, 0.8 μm SSB, and 25 mm buffer at the pH indicated. Continuous filaments encompassed the entire length of the cssDNA with no disruptions in the striated pattern of the filament. Discontinuous filaments contained disruptions in the filament that created abrupt angles in the filament or gaps in which no RecA striations could be seen. Miscellaneous filaments included RecA filaments that formed on linear ssDNA and any other discrete filament that did not fall into one of the other categories. The percentages of each filament type for wild type and mutant RecA were almost identical at pH 7.0. At pH 8.0, RecA E38K/ΔC17 protein forms predominantly discontinuous filaments, and wild type RecA forms predominantly continuous filaments.
| pH 7.0 |
pH 8.0 |
|||
|---|---|---|---|---|
| No. of filaments | % | No. of filaments | % | |
| Wild type | ||||
| Continuous | 20 | 2.7 | 317 | 41.5 |
| Discontinuous | 571 | 77.5 | 301 | 39.4 |
| Miscellaneous | 146 | 19.8 | 146 | 19.1 |
| Totals | 737 | 100.0 | 764 | 100.0 |
| E38K/ΔC17 | ||||
| Continuous | 2 | 0.3 | 8 | 2.8 |
| Discontinuous | 576 | 80.2 | 198 | 68.8 |
| Miscellaneous | 140 | 19.5 | 82 | 28.5 |
| Totals | 718 | 100.0 | 288 | 100.0 |
FIGURE 1.
RecA E38K/ΔC17 forms filaments on cssDNA. The following electron micrographs of RecA nucleoprotein filaments were obtained at a magnification of ×15,000 as described under “Experimental Procedures.” See Table 1 legend for concentrations of the reactants. A, continuous, wild type RecA filament on cssDNA at pH 8.0. There are no visible gaps or angles in this filament. B, discontinuous RecA E38K/ΔC17 filament on cssDNA. Small disruptions in this filament (see arrowhead) make it angular, rather than circular, but there are no visible gaps in the filament. C, discontinuous RecA E38K/ΔC17 filament at pH 8.0 that contains visible gaps in the filament (see arrowhead). The highest percentages of RecA E38K/ΔC17 filaments at pH 8.0 (38.9%) are of this form.
RecA E38K/ΔC17 Can Promote a Complete DNA Strand Exchange Only at Higher pH Values
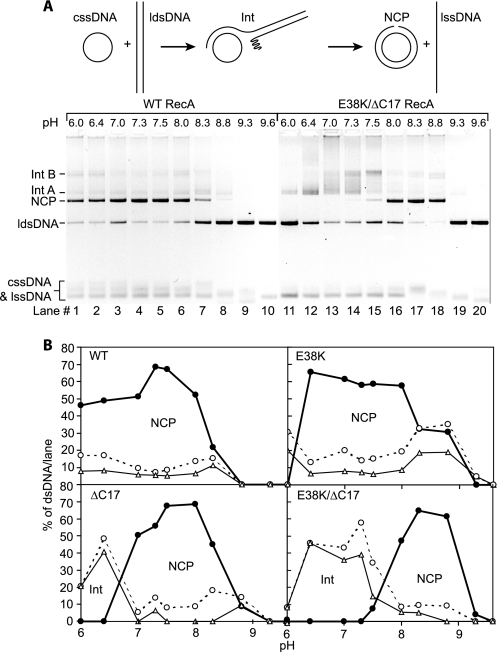
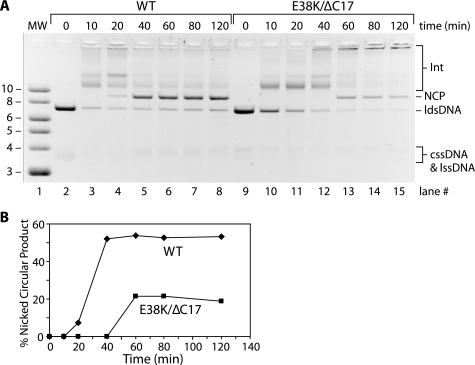
We examined whether the RecA E38K/ΔC17 double mutant can promote the fundamental RecA protein-mediated DNA strand exchange reaction (Fig. 2). As seen in earlier studies, the wild type RecA protein promotes an efficient DNA strand exchange reaction over most of the pH range examined, from pH 6 to above pH 8.3 (Fig. 2A, lanes 1–8). The RecA E38K/ΔC17 mutant does not efficiently carry out the strand exchange reaction within the optimal pH range for the wild type protein (pH 7–7.5). Instead, the optimal pH values for the double mutant RecA are shifted to a narrow range between pH 8 and 8.8 (Fig. 2A, lanes 16–18). Smaller shifts in the pH reaction profiles were seen for each of the single mutant proteins (Fig. 2B), as was documented previously for the C-terminal truncation mutant alone (42).
FIGURE 2.
RecA E38K/ΔC17 double mutant can promote a three-strand exchange reaction within a limited pH range. A, shown at the top is a schematic of DNA strand exchange between cssDNA and a homologous linear duplex. RecA forms a filament on the cssDNA. The ldsDNA is then aligned with the bound single strand, and exchange is initiated to form a branched intermediate (Int). Strand exchange is propagated unidirectionally, 5′ to 3′ relative to the ssDNA in the circle, until the nicked circular product (NCP) is generated, with release of the displaced single strand (lssDNA). The DNA strand exchange pH profiles for wild type (WT) and E38K/ΔC17 are illustrated in the photograph of the agarose gel. The DNA bands in the gel are labeled according to the schematic shown above. Reactions were carried out as described under “Experimental Procedures.” The reaction solutions contained 10 μm M13mp7 cssDNA, 3.5 μm RecA, 10 μm M13mp7 ldsDNA, 3 mm ATP, and 2 μm SSB. The Mg2+ concentration is 10 mm for wild type RecA and 5 mm for RecA E38K/ΔC17. B, DNA bands for the intermediates and products of DNA strand exchange catalyzed by RecA wild type, E38K, ΔC17, and E38K/ΔC17 were quantitated as a percent of the total intensity of the ldsDNA substrate, intermediates, and product bands in each lane. The product (NCP, closed circle), earlier intermediate or “Int A” (Int, open triangle), and total joint molecules (open circle) are graphed as a function of pH. The Mg2+ concentration was 5 mm for RecA ΔC17 and RecA E38K/ΔC17. The Mg2+ concentration was 10 mm for wild type and RecA E38K. The reactions were carried out as described above for A.
The RecA E38K/ΔC17 double mutant also accumulates DNA strand exchange reaction intermediates at lower pH values that are not converted to final products. The initial pairing of duplex DNA with ssDNA bound in a RecA nucleoprotein filament results in a joint molecule intermediate with a short segment of hybrid DNA. This intermediate has a higher electrophoretic mobility than more advanced intermediate species (Fig. 2A, Int A) (65). Extension of the initial paired regions incorporates more DNA into the hybrid region, and the late intermediates migrate more slowly in the gel prior to the formation of final products (Fig. 2A, Int B) (65). The extension generally requires ATP hydrolysis (17), although limited and slow extension occurs without ATP hydrolysis under some conditions (particularly under the low Mg2+ conditions used for the C-terminal deletion proteins (22)). The RecA E38K/ΔC17 double mutant protein promotes only the initial pairing reaction at lower pH values. Final products appear only at pH values at or above 7.3. Thus, at the lower pH values, the double mutant protein is proficient at DNA pairing but cannot promote the extension of the hybrid DNA region.
An Abrupt Shift in pH Allows Resolution of DNA Strand Exchange Intermediates Formed by the RecA E38K/ΔC17 Mutant at pH 7
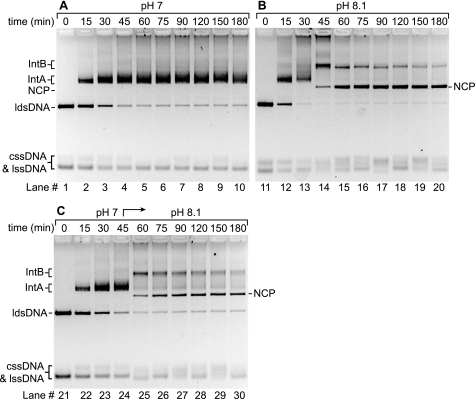
To determine whether the DNA strand exchange intermediates observed at low pH values could be converted to final products, we designed a pH shift experiment. The reaction was begun at pH 7.0 and then abruptly shifted by the addition of buffer to pH 8.1. As shown in Fig. 3A, a DNA strand exchange reaction carried out at pH 7 stalls at the earliest joint molecule intermediate. No products were seen even after 3 h. At pH 8.1, the same reaction generated final reaction products after 45 min of incubation (Fig. 3B, lane 14). Another reaction was started at pH 7 and then shifted from pH 7 to pH 8.1 after 45 min of incubation. The pH shift resulted in the appearance of the complete hybrid product within 15 min (Fig. 3C, lane 25). This result indicates that the intermediate generated at pH 7 is the early intermediate appearing in the reaction promoted by wild type RecA protein. The ability of the mutant RecA protein to catalyze homologous pairing at lower pH values, but only to complete strand exchange at higher pH values, demonstrates a mutational separation of the DNA pairing and the extended strand exchange phases of the reaction.
FIGURE 3.
Abrupt shift in pH allows resolution of DNA strand exchange intermediates formed by the E38K/ΔC17 mutant at pH 7. Reactions were carried out as described under “Experimental Procedures.” The reaction solutions contained 25 mm HEPES (17% anion) for the pH 7 reaction and 25 mm CHES (17% anion) for the pH 8.1 reaction, and 5 mm Mg2+. The DNA and protein concentrations are described in the legend to Fig. 2. The reaction time points were taken at 0, 15, 30, 45, 60, 75, 90, 120, 150, and 180 min after the addition of ldsDNA. The pH shift from pH 7 to 8.1 was carried out by adding 5% of the total reaction volume of 0.5 m CHES (100% anion) after the 45-min time point was removed. The pH values of each reaction were measured after the reactions were initiated and after the pH was shifted. Int, intermediate; NCP, nicked circular product.
dsDNA-dependent ATPase Activity of RecA E38K/ΔC17 Is Highly pH-sensitive
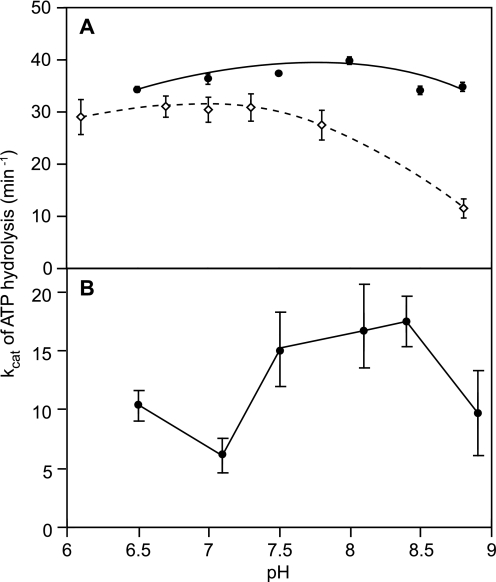
The extension phase of DNA strand exchange generally requires ATP hydrolysis although the initial pairing step does not, so it seems likely that some element of the coupling between ATP hydrolysis and extension of paired intermediates may be affected in the double mutant protein. To further investigate this idea, we measured the rates of RecA E38K/ΔC17-catalyzed ATP hydrolysis over a pH range of 6.3 to 8.8 on cssDNA and nicked circular dsDNA substrates (Fig. 4). Because the measurements were carried out with excess RecA and excess ATP, the data were plotted as apparent kcat values calculated by dividing the rate observed (μm min−1) by the concentration of RecA-binding sites on the DNA.
FIGURE 4.
pH affects dsDNA-dependent ATPase activity but not ssDNA-dependent ATPase activity of the RecA E38K/ΔC17 protein. The reactions were carried out as described under “Experimental Procedures.” A, comparison of the pH profiles of ATP hydrolysis catalyzed by the wild type (open diamonds) and E38K/ΔC17 RecA proteins (closed circles). The reactions contained 4 μm M13mp18 cssDNA, 2 μm RecA protein, 3 mm ATP, and 0.4 μm SSB. The wild type and RecA E38K/ΔC17 reactions contained 10 and 5 mm Mg2+, respectively. The pH values of the reaction solutions were measured after ATP hydrolysis was complete. B, kcat of RecA E38K/ΔC17-catalyzed ATP hydrolysis on dsDNA. The reactions contained 4.2 μm M13mp7 nicked circular dsDNA, 2 μm E38K/ΔC17 RecA, and 3 mm ATP. The rates of ATP hydrolysis were measured once the RecA proteins had achieved steady state hydrolysis. The kcat values were calculated as the rate of ATP hydrolysis when RecA saturates the DNA divided by the concentration of RecA-binding sites on the DNA.
The rates of ATP hydrolysis promoted by RecA E38K/ΔC17 bound to ssDNA did not vary much between pH 6 and 9, although there was a slight increase in hydrolysis rate at pH 8 (Fig. 4A). Also, the double mutant RecA protein hydrolyzed ATP with a somewhat higher kcat than the wild type protein at all tested pH values (Fig. 4A). Above pH 8.5, the wild type RecA protein hydrolyzed ATP more slowly, but the double mutant was unaffected.
RecA E38K/ΔC17-mediated ATP hydrolysis became much more pH-dependent when the protein was bound to dsDNA (Fig. 4B). Rates of ATP hydrolysis were slower at pH values below 7.5, with an apparent kcat of ∼6 min−1 at pH 7. At higher pH values, the rates of hydrolysis increased, with a maximum of ∼17 min−1 at pH 8.4. For wild type RecA-catalyzed ATP hydrolysis on dsDNA, the maximal kcat is ∼20 min−1 and occurs at pH 6, whereas ATP hydrolysis above pH 7 appears negligible due to slow nucleation at high pH values (42, 66, 67). This means that the pH profile for E38K/ΔC17 RecA-catalyzed ATP hydrolysis on dsDNA is shifted to higher pH values, just like the pH profile for DNA strand exchange. However, the altered pH profile for ATP hydrolysis on dsDNA does not explain the pH profile for DNA strand exchange. Circular ssDNA is also present in DNA strand exchange reactions, and the pH profile for RecA E38K/ΔC17-catalyzed ATP hydrolysis on cssDNA is not shifted to higher pH values compared with wild type RecA. There must be another reason for the altered pH profile of RecA E38K/ΔC17-catalyzed DNA strand exchange.
RecA E38K/ΔC17 Undergoes a Shift Indicating Entry into the P- state at Both pH 7.0 and pH 8.0
The commencement of DNA strand exchange is accompanied by a change in filament state, signaled by changes in multiple filament properties (17, 18, 54, 59, 68, 69), that is designated the P state (59). A failure to enter the P filament state during DNA strand exchange could explain why RecA E38K/ΔC17 cannot catalyze extended branch migration at lower pH values. One change that is diagnostic of an entry to the P state is a sudden drop in the rate of ATP hydrolysis observed upon addition of homologous duplex DNA to RecA filaments formed on ssDNA (18). In our experiments, there was a clear drop in the rate of ATP hydrolysis when homologous ldsDNA was added to reactions with either the wild type RecA or the RecA E38K/ΔC17 protein. This was seen both at pH 7 and at pH 8 (data not shown). By this criterion, RecA E38K/ΔC17 enters the P-state, or at least alters its functional state in some way, even under conditions that do not support full DNA strand exchange.
The ATP hydrolysis assays carried out during DNA strand exchange indicated that the double mutant RecA protein continues to hydrolyze ATP after homologous ldsDNA is added to the nucleoprotein filament. Although ATP hydrolysis takes place at pH 7.0 at rates nearly comparable with the wild type protein, the extended branch migration reaction that requires ATP hydrolysis does not take place.
These results indicate that ATP hydrolysis has been uncoupled from extended branch migration in a pH-dependent fashion. In effect, the RecA E38K/ΔC17 double mutant has a mutational separation between the DNA pairing step, which still proceeds at low pH values, and the extended exchange phase of strand exchange, which does not. We decided to exploit these properties to more closely examine the coupling between ATP hydrolysis and DNA strand exchange with the double mutant protein.
Kinetics of RecA E38K/ΔC17-catalyzed DNA Strand Exchange Are Slow Compared with Wild Type RecA
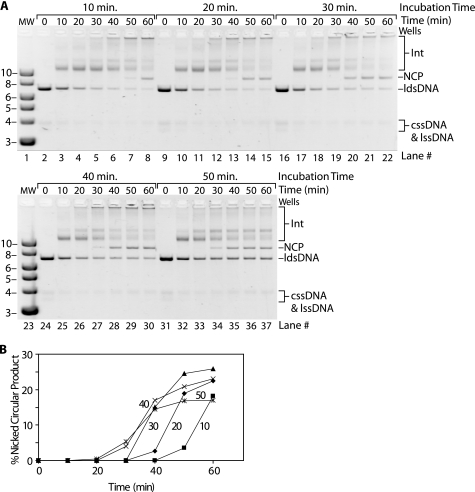
Even at pH values that are permissive for complete DNA strand exchange, the RecA E38K/ΔC17 protein promotes the reaction with kinetics of product formation that are considerably slower than those observed for wild type RecA protein (Fig. 5). At pH 7.8, wild type RecA protein produces nicked circular product in less than 20 min with these long M13-based DNA substrates (Fig. 5, A, lane 4, and B). At its optimal pH of 8.5, RecA E38K/ΔC17 requires 60 min to produce nicked circular product (Fig. 5, A, lane 13, and B). In this experiment, we knew that the ATP regeneration system would be used up prior to the observed generation of strand exchange products, and we decided to determine whether the delay reflected a requirement for the build-up of ADP.
FIGURE 5.
Kinetics of RecA E38K/ΔC17-catalyzed DNA strand exchange are slow compared with wild type RecA even at permissive pH values. Reactions were carried out as described under “Experimental Procedures” and contained 10 μm M13mp18 cssDNA, 20 μm M13mp18 ldsDNA cut with BsmBI, 3.5 μm RecA protein, 1 μm SSB protein, and 3 mm ATP. The pH values of the reaction buffers were 7.8 and 8.5 for wild type (WT) and RecA E38K/ΔC17 proteins, respectively. A, progress of DNA strand exchange catalyzed by wild type and RecA E38K/ΔC17 proteins by agarose gel electrophoresis. The RecA proteins were incubated in their respective reaction solutions with cssDNA, ATP regeneration system, ATP, and SSB for 10 min before ldsDNA was added to begin DNA strand exchange. Aliquots were removed and stopped at the times indicated above each lane. The cssDNA and ldsDNA substrates, as well as the joint molecule intermediates (Int) and nicked circular product (NCP) bands are all visible in the gel. B, intensities of the nicked circular product bands are presented as percentages of the total intensity of all dsDNA-containing bands in each lane, including the ldsDNA substrate, joint molecule intermediate, and nicked circular product bands. The joint molecule intermediates that remained in the wells were also included in this calculation. The percent nicked circular product is plotted versus reaction time where time 0 corresponds to addition of the ldsDNA substrate. Wild type RecA is represented with diamonds; RecA E38K/ΔC17 is represented with squares.
Increasing the Ratio of ADP to ATP Can Speed the Kinetics of RecA E38K/ΔC17 Protein-catalyzed DNA Strand Exchange at High pH Values
Lengthening the time RecA E38K/ΔC17 protein is preincubated with the DNA strand exchange reaction solution and cssDNA before homologous ldsDNA is added leads to more advanced depletion of the ATP regeneration system before strand exchange begins. We found that this also reduces the time it takes to form nicked circular product (Fig. 6). With a 10-min incubation period, nicked circular product is first seen after 50 min (Fig. 6, A, lane 7, and B). With 20- and 40-min incubation periods, the first detection of nicked circular products occurs at the 40- and 30-min time points, respectively (Fig. 6, A, lanes 13 and 27, respectively, and B). In contrast, if ATP regeneration is extended by using a higher concentration of P-enolpyruvate, the time required for appearance of DNA strand exchange products is correspondingly increased (data not shown).
FIGURE 6.
Increasing the incubation time before ldsDNA is added to the reaction speeds the kinetics of RecA E38K/ΔC17 protein-catalyzed DNA strand exchange. Reactions were carried out as described under “Experimental Procedures” and contained 10 μm M13mp18 cssDNA, 20 μm M13mp18 ldsDNA cut with BsmBI, 3.5 μm RecA protein, 1 μm SSB protein, and 3 mm ATP. The pH of the reaction buffer was 8.5. RecA E38K/ΔC17 protein was incubated in the reaction solution with cssDNA, ATP regeneration system, ATP, and SSB at 37 °C for the amount of time indicated above each time course before ldsDNA was added to begin DNA strand exchange. The protocol was otherwise identical to that described in the legend to Fig. 5. A, progress of DNA strand exchange catalyzed by RecA E38K/ΔC17 protein by agarose gel electrophoresis. B, percent nicked circular product (NCP) is plotted versus reaction time where time 0 corresponds to addition of the ldsDNA substrate. Each plot is labeled according to the incubation time before ldsDNA addition. The plot symbols represent the following incubation times: squares, 10 min; diamonds, 20 min; triangles, 30 min; ×, 40 min; asterisks, 50 min. Int, intermediate.
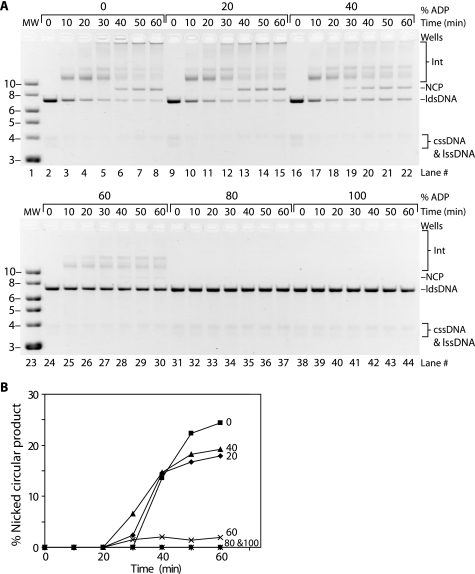
These results could indicate that P-enolpyruvate is directly inhibiting DNA strand exchange or that maintaining a low ADP to ATP ratio is actually inhibiting the reaction. In the absence of a regeneration system, we found that increasing the ratio of ADP to ATP alone speeds the kinetics of DNA strand exchange (Fig. 7). Of the ADP percentages screened, the reaction with 40% ADP produced the most intense product band within 30 min of ldsDNA addition (Fig. 7A, lane 19). Omission of the regeneration system alone sped the appearance of nicked circular product from the 50-min time point (Fig. 6A, lane 7) to the 40-min time point (Fig. 7, A, lane 6 and B). These results suggested that maintaining a low ratio of ADP to ATP was responsible for slowing nicked circular product formation rather than some inhibitory effect of P-enolpyruvate.
FIGURE 7.
Increasing the ratio of ADP to ATP can speed the kinetics of RecA E38K/ΔC17 protein-catalyzed DNA strand exchange. Reactions were carried out as described under “Experimental Procedures” and contained 10 μm M13mp18 cssDNA, 20 μm M13mp18 ldsDNA cut with BsmBI, 3.5 μm RecA protein, and 1 μm SSB protein. The pH of the reaction buffer was 8.5. ADP, ATP, and SSB were added as a mixture to each reaction. The percent ADP is indicated above each time course; the remaining percentage of nucleotide cofactor consisted of ATP. The total nucleotide concentration was 3 mm. The ATP regeneration system was omitted. The protocol was otherwise identical to that described in the legend to Fig. 5. A, progress of DNA strand exchange catalyzed by RecA E38K/ΔC17 protein by agarose gel electrophoresis. The percent ADP present at the beginning of each reaction is labeled above each time course. B, percent NCP is plotted versus reaction time where time 0 corresponds to addition of the ldsDNA substrate. Each plot is labeled according to the percent ADP added to each reaction. The plot symbols represent the following percentages of ADP: squares, 0%; diamonds, 20%; triangles, 40%; ×, 60%; asterisks, 80%; circles, 100%. Int, intermediate; NCP, nicked circular product.
Shifting the ratio of ADP to ATP also has an effect on RecA filament disassembly. Raising the concentration of ADP favors filament disassembly even when RecA filaments are bound to cssDNA (68, 70, 71). Disassembly of RecA subunits from a protein filament is an ATP hydrolysis-dependent activity (69, 72). Because RecA E38K/ΔC17 protein-catalyzed DNA strand exchange is uncoupled from ATP hydrolysis at nonpermissive pH values, it may follow that filament disassembly is also uncoupled from ATP hydrolysis.
RecA Protein Filament Disassembly Occurs during DNA Strand Exchange Catalyzed by Wild Type RecA Protein
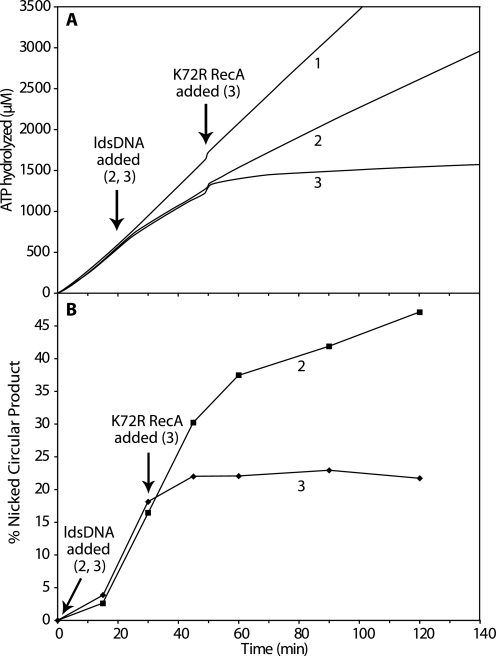
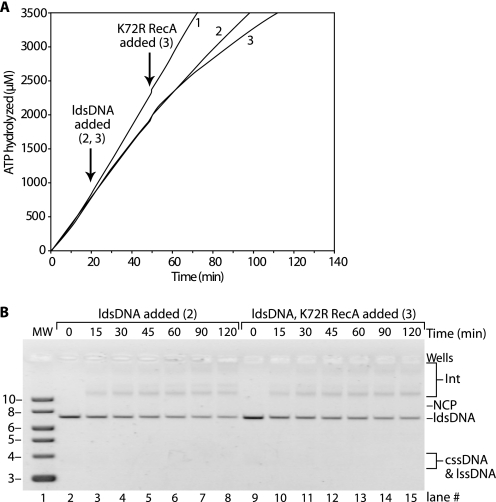
Disassembly of RecA subunits from a filament can be indirectly observed as an exchange of hydrolysis-proficient RecA protein subunits in a filament for the hydrolysis-deficient RecA K72R protein, which produces a concurrent drop in the rate of ATP hydrolysis (54). If no disassembly occurs, the bound RecA continues to hydrolyze ATP uninterrupted after a challenge with RecA K72R. The use of RecA K72R can be illustrated with a DNA strand exchange reaction using wild type RecA protein, with RecA K72R protein added to the reaction after DNA strand exchange has been initiated (Fig. 8). Reaction 1 is a control that shows RecA-catalyzed ATP hydrolysis with cssDNA being the sole DNA substrate. In Reaction 2, homologous ldsDNA is added after 20 min to begin the DNA strand exchange reaction. There is an expected drop in the rate of ATP hydrolysis to a new steady state as the RecA filaments enter the “pairing-enhanced” or “P” functional state during exchange (18, 59). The rate of ATP hydrolysis drops much more, and the decline continues for some time after the addition of RecA K72R to Reaction 3, indicating that wild type RecA filaments disassemble during the course of DNA strand exchange.
FIGURE 8.
RecA nucleoprotein filament disassembly occurs during DNA strand exchange catalyzed by wild type RecA protein. Reactions were carried out as described under “Experimental Procedures” and contained 4 μm M13mp18 cssDNA, 8 μm M13mp18 ldsDNA cut with BsmBI, 2 μm wild type RecA protein, 0.4 μm SSB protein, 3 mm ATP, and 4 μm RecA K72R protein. The pH of the reaction buffer was 7.8. Wild type RecA protein was incubated in the reaction solution with cssDNA, ATP regeneration system, a system to couple ATP hydrolysis to NADH oxidation, ATP, and SSB at 37 °C for 20 min, although the rate of ATP hydrolysis was measured. The ldsDNA substrate was added to start DNA strand exchange, and the rate of ATP hydrolysis was measured for another 30 min. RecA K72R protein was added to the reaction, and the rate of ATP hydrolysis was measured for another 100 min. The RecA K72R protein does not hydrolyze ATP and does not disassemble from DNA. As wild type RecA filaments disassemble during DNA strand exchange, RecA K72R protein binds in their place, and the rate of ATP hydrolysis decreases. A, amount of ATP hydrolyzed in micromolars is plotted versus time in minutes. Reaction 1 contains equivalent volumes of TE or storage buffer in place of ldsDNA or RecA K72R, respectively. Reaction 2 consisted of Reaction 1 components and ldsDNA. Reaction 3 contained Reaction 2 components plus RecA K72R protein. B, amount of nicked circular product produced in Reactions 2 (squares) and 3 (diamonds) is plotted versus time as in Figs. 5–7. Reaction 1 did not produce any nicked circular product because there was no ldsDNA to initiate DNA strand exchange.
Disassembly of RecA E38K/ΔC17 Filaments during DNA Strand Exchange Is Inhibited at Nonpermissive pH Values
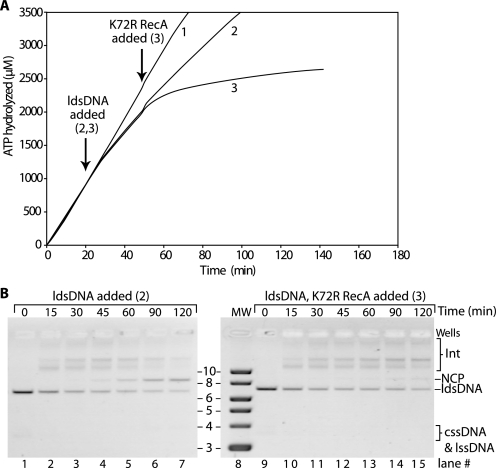
At the nonpermissive pH of 7.0, RecA E38K/ΔC17 protein hydrolyzes ATP during DNA strand exchange (Fig. 9A, Reaction 2) and will make joint molecule intermediates, but no nicked circular product (Fig. 9B) as observed in Fig. 2. When RecA K72R protein is added to the DNA strand exchange reaction mixture, the drop in the rate of ATP hydrolysis (Fig. 9A, Reaction 3) is very small, much smaller than that seen for wild type RecA protein during DNA strand exchange (Fig. 8A, Reaction 3). This indicates that disassembly of RecA E38K/ΔC17 filaments is inhibited during homologous DNA pairing reactions promoted by the double mutant protein at nonpermissive pH values.
FIGURE 9.
Disassembly of RecA E38K/ΔC17 filaments during DNA strand exchange is inhibited at nonpermissive pH values. Reactions were carried out as described under “Experimental Procedures” and contained 4 μm M13mp18 cssDNA, 8 μm M13mp18 ldsDNA cut with BsmBI, 2 μm RecA E38K/ΔC17 protein, 0.4 μm SSB protein, 3 mm ATP, and 4 μm RecA K72R protein. The pH of the reaction buffer was 7.0. The protocol was otherwise identical to that described in the legend to Fig. 8. Fig. 9A is identical to Fig. 8A except that RecA E38K/ΔC17 is used in place of wild type RecA. Reaction 1 contains equivalent volumes of TE or storage buffer in place of ldsDNA or RecA K72R, respectively. Reaction 2 consisted of Reaction 1 components and ldsDNA. Reaction 3 contained Reaction 2 components plus RecA K72R protein. B, progress of DNA strand exchange catalyzed by RecA E38K/ΔC17 protein by agarose gel electrophoresis. The gel picture is included to show that joint molecule intermediates are made even though nicked circular product is not made. Aliquots of 15 μl were stopped at the times indicated above each lane. Reaction 1 is not included as no joint molecules were made because no ldsDNA was added to begin DNA strand exchange. Int, intermediate; NCP, nicked circular product.
Disassembly of RecA E38K/ΔC17 Filaments during DNA Strand Exchange Is Restored at Permissive pH Values
At the permissive pH of 8.5, RecA E38K/ΔC17 protein hydrolyzes ATP (Fig. 10A) and produces nicked circular DNA (Fig. 10B, lanes 4–7) as the product of DNA strand exchange. When RecA K72R protein is added to the reaction mixture, there is a rapid decrease in the rate of ATP hydrolysis (Fig. 10A, Reaction 3), much like that seen when RecA K72R protein is added to wild type RecA filaments promoting DNA strand exchange (Fig. 8A, Reaction 3). This result indicates that disassembly of RecA E38K/ΔC17 filaments, as well as nicked circular product formation, has been re-coupled to ATP hydrolysis at the permissive pH of 8.5. From Figs. 8 to 10, we see that nicked circular product formation during DNA strand exchange only occurs when the RecA protein filament undergoes disassembly. The lack of nicked circular product formation when filament disassembly is inhibited suggests that disassembly is necessary for product formation and that disassembly might be the ATP hydrolysis-dependent step that couples hydrolysis to DNA strand exchange. We note that the RecA E38K/ΔC17 protein will promote a four-strand exchange reaction at permissive pH values, as well as the three-strand exchange reactions described here (data not shown).
FIGURE 10.
Disassembly of RecA E38K/ΔC17 filaments is restored during DNA strand exchange at pH 8.5. Reactions were carried out as described under “Experimental Procedures” and contained 4 μm M13mp18 cssDNA, 8 μm M13mp18 ldsDNA cut with BsmBI, 2 μm RecA E38K/ΔC17 protein, 0.4 μm SSB protein, 3 mm ATP, and 4 μm RecA K72R protein. The pH of the reaction buffer was 8.5. The protocol was otherwise identical to that described in the legend to Fig. 8. Fig. 10A is identical to Fig. 8A except that RecA E38K/ΔC17 is used in place of wild type RecA. Reaction 1 contains equivalent volumes of TE or storage buffer in place of ldsDNA or RecA K72R, respectively. Reaction 2 consisted of Reaction 1 components and ldsDNA. Reaction 3 contained Reaction 2 components plus RecA K72R protein. B shows the progress of DNA strand exchange catalyzed by RecA E38K/ΔC17 protein by agarose gel electrophoresis. The gel picture shows that nicked circular product formation is restored at pH 8.5. Aliquots of 15 μl were stopped at the times indicated above each lane. Reaction 1 is not included as no nicked circular product was made because no ldsDNA was added to begin DNA strand exchange. Int, intermediate; NCP, nicked circular product.
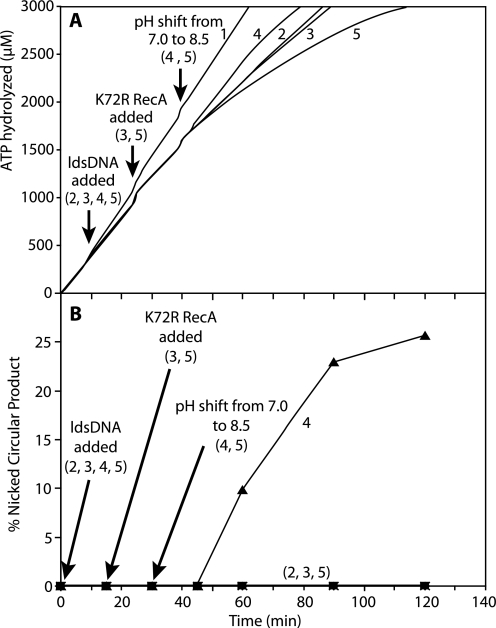
Abrupt Shift in pH from 7.0 to 8.5 Restores RecA E38K/ΔC17 Filament Disassembly and Nicked Circular Product Formation during DNA Strand Exchange
In Fig. 3, we found that joint molecule intermediates formed by RecA E38K/ΔC17 protein during DNA strand exchange at pH 7.0 could be converted to nicked circular product if the pH of the reaction solution was abruptly shifted to a permissive pH. If RecA filament disassembly is necessary for nicked circular product formation, we expect that enhanced disassembly would also be restored when the pH is shifted to a permissive value. To test this idea, we set up an experiment in which DNA strand exchange is initiated at the nonpermissive pH of 7.0, and disassembly is observed before and after an abrupt shift in pH from 7.0 to 8.5 (Fig. 11). We again observe a very small decrease in the rate of ATP hydrolysis when RecA K72R protein is added to the reaction at pH 7.0 (compare Fig. 11A, Reactions 2 and 3), which is consistent with the findings presented in Fig. 9. However, the rate of ATP hydrolysis decreases rapidly after the pH is shifted to 8.5 (Fig. 11A, Reaction 5), indicating that filament disassembly is restored. In the reaction without the RecA K72R protein challenge that undergoes the pH shift (Fig. 11A, Reaction 4), nicked circular product formation is restored within 30 min of the shift (Fig. 11B, Reaction 4). These data demonstrate even more clearly the tight correlation between RecA filament disassembly and nicked circular product formation during DNA strand exchange with this mutant RecA protein. This strengthens the conclusion that disassembly of RecA filaments is required for nicked circular product formation during DNA strand exchange.
FIGURE 11.
Disassembly of RecA E38K/ΔC17 filaments during DNA strand exchange at pH 7. 0 is restored by an abrupt shift of pH to 8.5. Reactions (550 μl) were carried out as described under “Experimental Procedures” and contained 4 μm M13mp18 cssDNA, 8 μm M13mp18 ldsDNA cut with BsmBI, 2 μm RecA E38K/ΔC17 protein, 0.4 μm SSB protein, 3 mm ATP, and 4 μm RecA K72R protein. Reactions were begun at pH 7.0. RecA E38K/ΔC17 protein was incubated in the reaction solution with cssDNA, ATP regeneration system, coupling system, ATP, and SSB at 37 °C for 10 min, although the rate of ATP hydrolysis was measured. The ldsDNA substrate was added to start DNA strand exchange, and the rate of ATP hydrolysis was measured for another 15 min. RecA K72R protein was added to the reaction. After another 15 min, 18.56 μl of 1 m CHES (100% anion, pH ∼10.7) were added to shift the pH from 7.0 to 8.5. The rate of ATP hydrolysis was measured for ∼150 more min. A, amount of ATP hydrolyzed in micromolar is plotted versus time in minutes. Reaction 1 contains equivalent volumes of TE or storage buffer in place of ldsDNA or RecA K72R, respectively, and 1 m HEPES (17% anion, pH 7.0) was added as a control for the pH shift buffer. Reaction 2 was the same as Reaction 1 except that it contained ldsDNA. Reaction 3 was the same as Reaction 2 except that it contained RecA K72R protein. Reaction 4 was the same as Reaction 2 except that its pH was shifted to 8.5. Reaction 5 contained ldsDNA, RecA K72R, and pH shift buffer. B, amount of nicked circular product produced in Reactions 2–5 is plotted versus time as in Figs. 5–7. Reaction 1 did not produce any nicked circular product because there was no ldsDNA to initiate DNA strand exchange. Reactions 2 (squares), 3 (diamonds), and 5 (×) produced only joint molecule intermediates (data not shown). Nicked circular product (NCP) produced in Reaction 4 is represented by triangles.
All of the apparent rates of ATP hydrolysis catalyzed by wild type and RecA E38K/ΔC17 during DNA strand exchange (Figs. 8–11) are summarized in Table 2. For many of the intervals listed in Table 2, ATP is not being hydrolyzed at a constant rate. For example, in (Fig. 10, Reaction 2), the rate of ATP hydrolysis continues to decrease over time after ldsDNA is added to the reaction even without the addition of RecA K72R. Thus, the rates in the table are apparent or average rates during the time interval indicated and should be used only for comparisons among each other. The rates in the table recapitulate numerically, and the drop in the rate of ATP hydrolysis represented graphically in Figs. 8–11 when a RecA filament actively undergoing disassembly is challenged with RecA K72R.
TABLE 2.
Rates of RecA-catalyzed ATP hydrolysis during DNA strand exchange.
The apparent rates of ATP hydrolysis were catalyzed by wild type and RecA E38K/ΔC17 proteins during DNA strand exchange from Figs. 8 to 11. All reagent concentrations and reaction conditions are described under “Experimental Procedures” and in each figure legend. The additions made to each reaction after ATP and SSB are listed in parentheses next to each reaction. The length of the last interval for each reaction spans the first time point listed in parentheses to the time point ∼5 min before the ATP regeneration system is exhausted or until the 120-min time point.
| Rate of ATP hydrolysis |
|||
|---|---|---|---|
| After ATP/SSB added (0–20 min) | After ldsDNA added (21–50 min) | After K72R RecA added (50–120 min) | |
| μm/min | |||
| Fig. 8, WT RecA, pH 7.8 | |||
| Reaction 1 | 29.5 | 36.0 | 35.3 |
| Reaction 2 (ldsDNA) | 28.6 | 22.5 | 18.1 |
| Reaction 3 (ldsDNA + K72R RecA) | 27.6 | 22.5 | 2.6 |
| Fig. 9, E38K/C17 RecA, pH 7.0 | |||
| Reaction 1 | 43.0 | 50.4 | 51.2 |
| Reaction 2 (ldsDNA) | 41.9 | 37.7 | 31.2 |
| Reaction 3 (ldsDNA + K72R RecA) | 42.0 | 38.7 | 23.4 |
| Fig. 10, E38K/C17 RecA, pH 8.5 | |||
| Reaction 1 | 47.2 | 51.1 | 47.0 |
| Reaction 2 (ldsDNA) | 48.0 | 37.5 | 27.7 |
| Reaction 3 (ldsDNA + K72R RecA) | 48.0 | 36.1 | 6.2 |
| Rate of ATP hydrolysis |
||||
|---|---|---|---|---|
| After ATP/SSB added (0–9 min) | After ldsDNA added (10–24 min) | After K72R RecA added (25–39 min) | After pH shift (40–120 min)a | |
| μm/min | ||||
| Fig. 11, E38K/C17 RecA, pH shift | ||||
| Reaction 1 | 38.1 | 46.1 | 47.6 | 47.8 |
| Reaction 2 (ldsDNA) | 36.6 | 37.4 | 33.7 | 29.9 |
| Reaction 3 (ldsDNA + K72R RecA) | 39.0 | 39.2 | 33.1 | 27.7 |
| Reaction 4 (ldsDNA + pH shift) | 37.9 | 38.6 | 34.2 | 38.3 |
| Reaction 5 (ldsDNA + K72R RecA + pH shift) | 39.4 | 39.5 | 32.9 | 17.9 |
a These intervals span the beginning time indicated in parentheses to ∼5 min before the ATP regeneration system runs out or to 120 min.
DISCUSSION
On the Relationship of ATP Hydrolysis, Filament Disassembly, and Extended DNA Strand Exchange
This study has two conclusions. First, the RecA E38K/ ΔC17 protein demonstrates a mutational separation between the DNA pairing and extended exchange phases of DNA strand exchange. At pH values below 7.3, DNA pairing proceeds but extension of the paired region does not occur. The entire reaction proceeds at pH values above 7.3. The clear separation reinforces the concept that these phases of the strand exchange reaction represent distinct processes. The separation of phases also allowed a new dissection of the relationship between the extension phase of DNA strand exchange, ATP hydrolysis, and filament disassembly. This led to the second conclusion as follows: for the RecA E38K/ΔC17 mutant protein, a strong correlation exists between the disassembly of RecA filaments and the completion of DNA strand exchange. This correlation provides evidence that filament disassembly plays a role in the mechanism by which ATP hydrolysis is coupled to DNA strand exchange.
This is not the first time that a linkage between DNA strand exchange and RecA filament disassembly has been proposed (21, 49, 51–53). In fact, dissociation of RecA filament subunits during DNA strand exchange has been previously documented (54). Whereas considerable evidence has been compiled linking ATP hydrolysis and DNA strand exchange (17, 49, 55, 73, 74), no evidence has appeared to indicate whether the role of the observed filament dissociation is determinant or incidental. This study begins to fill this void, presenting for the first time a set of conditions (the low pH, nonpermissive conditions with RecA E38K/ΔC17) in which ATP hydrolysis continues but dissociation of RecA filaments is suppressed. In effect, ATP hydrolysis has been uncoupled from RecA subunit dissociation in this mutant protein under the nonpermissive conditions. Under these same conditions, DNA strand exchange is also suppressed. The result is a correlation that uniquely strengthens the view that filament dissociation and DNA strand exchange are mechanistically linked.
A role for filament disassembly is readily interpreted within the context of either the RecA redistribution model (51) or the earlier Howard-Flanders model (53) for coupling ATP hydrolysis to DNA strand exchange. In both models, the ATP hydrolysis-dependent process of RecA filament disassembly is the key event that explains the requirement for ATP hydrolysis in DNA strand exchange. In the redistribution model, disassembly of RecA monomers allows a filament to be remodeled, filling in discontinuities in the filament that present a barrier to DNA strand exchange (21, 51, 52). The required subunit dissociation need not occur at the DNA branch point and subunit dissociation per se is not coupled to movement of the branch. In the Howard-Flanders model (modified so that it does not include an unsubstantiated four-strand DNA pairing intermediate (49, 56)), disassembly of subunits from a RecA filament creates a conformational change in the RecA or the associated DNA that allows the DNA strand switch to occur (49, 53).
A necessity for filament disassembly in DNA strand exchange is not considered in the facilitated DNA rotation model. It is assumed that the disassembly process recycles RecA monomers but plays no substantive role in the DNA strand exchange process itself (49). It remains possible that the progress of strand exchange is augmented by coupling ATP hydrolysis to rotation of DNA strands around a RecA filament. However, based on the findings of this study, the facilitated rotation model must, at a minimum, be modified to include disassembly of RecA subunits as a potentially integral part of DNA strand exchange.
There is always some uncertainty associated with the extrapolation from the properties of a mutant protein to the mechanism of the wild type protein. A few caveats derived from the data are thus in order. First, although we document a correlation between RecA subunit exchange in the filament and the completion of DNA strand exchange, subunit dissociation alone is apparently not sufficient to promote this latter stage of the reaction. At the permissive pH values, the strand exchange reaction is completed, but even under optimal conditions, RecA E38K/ΔC17 promotes the reaction at a rate that is much slower than the reaction promoted by the wild type protein (17). However, the rates of ATP hydrolysis are similar between the mutant and wild type proteins. This indicates that the mechanism coupling ATP hydrolysis to strand exchange is somehow altered. Meanwhile, the pH-facilitated dissociation of RecA E38K/ΔC17 subunits from filaments is observed at all times after the linear dsDNA is added, long before products appear (data not shown).
Second, if dissociation of RecA subunits at the DNA branch point represents the key event in driving branch movement, then that dissociation, accompanied by subunit replacement behind the branch with RecA K72R, should allow strand exchange to go to completion. However, when the RecA K72R challenge occurs 15 min after strand exchange has been initiated, it virtually eliminates the generation of strand exchange products. Incorporation of RecA K72R into the filament thus blocks strand exchange. This is readily explained in the context of a RecA dissociation model for strand exchange (53) if RecA is dissociating and re-associating at several points along the cssDNA where the filament is not contiguous. The branch would presumably be moved only at one dissociation point. Dissociation at a point downstream of the branch and replacement with RecA K72R would generate a segment of filament incapable of dissociation in the path of the branch. The reaction would then halt when it reached the RecA K72R segment. A contiguous filament, if it contained segments of RecA K72R, would be insufficient to sustain the strand exchange reaction. Such a segment need not be large to halt the reaction. A filament with a random mixture of wild type and RecA K72R subunits will not generate full strand exchange products if as little as one-third of the subunits are K72R (54). These results argue for an active role of RecA subunit dissociation at the branch point (53), rather than a redistribution role of RecA dissociation with the objective of filling in filament gaps to create a contiguous filament (21, 51, 52). Given the significant DNA strand exchange (1–1.5 kbp) observed in the absence of ATP hydrolysis (19–22), the active role of RecA filament disassembly may be the final release of the displaced DNA strand or some other process that renders the exchange effectively irreversible.
Finally, the new results do not explain why the entire RecA filament hydrolyzes ATP (16). During DNA strand exchange, every RecA monomer in the filament is hydrolyzing an ATP every 3 s, but only a small fraction of those events, predominantly those occurring in the last RecA subunit at the 5′-proximal end of a filament segment, result in dissociation. A role for the ATP hydrolysis that occurs along the length of a filament segment without resulting in dissociation remains a mechanistic issue in this system.
RecA Filament Structure and Coordination of ATP Hydrolytic Cycles between Adjacent Subunits
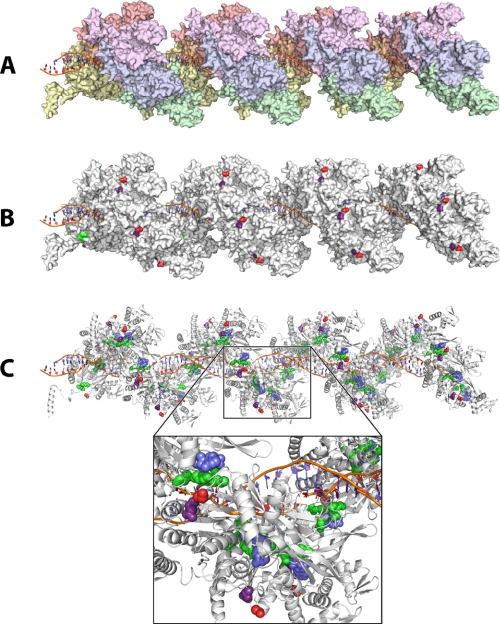
In Fig. 12, we further explore the role of Glu-38 and the C terminus in coupling ATP hydrolysis to DNA strand exchange by examining the structure of RecA protein bound to DNA (46). In Fig. 12A, the surface of a RecA filament is depicted, with individual subunits colored differentially. The filament is a right-handed helix with the DNA (Fig. 12A, whose phosphate backbone is colored orange) bound inside the helix, along the helical axis. In Fig. 12B, all the subunits are white, with Glu-38 and Ser-333 (the most C-terminal residue of each monomer in the construct) colored purple and red, respectively. This shows that both Glu-38 and the base of the C-terminal flap are close to each other, on the exterior surface of the filament, and oriented away from the DNA. Both residues are also close to the subunit-subunit interface. In Fig. 12C, the ATP residue at each subunit-subunit interface is colored blue and revealed within a ribbon rendering. Several amino acid residues previously implicated either directly in ATP hydrolysis or in coordinating hydrolysis between adjacent subunits are highlighted in green in Fig. 12C, and all are clustered near the bound ATP. These include Ser-69 (74), Lys-72 (21, 22, 75), Glu-96 (76, 77), Lys-248 (77), and Lys-250 (73). Because Glu-38 and the C terminus are on the surface of the RecA filament, away from the ATP cofactor and the DNA, they must affect the coupling between ATP hydrolysis and DNA strand exchange indirectly. This may be achieved via an effect on overall subunit conformation. In every structure of RecA to date, the C terminus is either disordered or omitted from the constructs used in structure determination. Presumably, the flexible C terminus has the capacity to interact with several surfaces on a RecA subunit, thus affecting its conformation. The proximity of residue Glu-38 and the C terminus to the subunit-subunit interface also make it possible that they affect the interactions between two adjacent RecA subunits. This could have a profound effect on ATP hydrolysis because of the extensive coordination between residues on both subunits needed to carry out hydrolysis.
FIGURE 12.
Structure-function in RecA related to ATP hydrolysis and DNA strand exchange. A, RecA filament on linear duplex DNA with 24 subunits, colored so that adjacent subunits are distinguishable. B, same RecA filament, with the Glu-38 (purple) and Ser-333 (red) residues colored in each subunit. Note that Ser-333 is the final residue in the published structure (46), whereas the RecA E38K/ΔC17 mutant described in this report also includes Thr-334 and Pro-335. C, ribbon rendering with bound ATP (blue) and some key active site residues highlighted (Ser-69, Lys-72, Glu-96, Lys-248, and Lys-250; green). The Glu-38 and Ser-333 residues are colored as in B. The inset provides a closer look at the spatial arrangement of these residues.
The pH dependence of the coupling and uncoupling of ATP hydrolysis and DNA strand exchange leads directly to questions about which amino acid residue or residues are being affected by the pH changes in this mutant protein. We propose that an ionizable side chain plays a key role in the productive coupling of ATP hydrolysis and DNA strand exchange. Productive coupling requires that this residue be in the unprotonated state. In the double mutant RecA, neither the Lys residue placed at position 38 nor the base of the C terminus (proline 335 present in our protein but not in the crystal structure) is likely to be the ionizable residue that brings about the pH-dependent transition evident in Fig. 2. A Lys residue at the protein surface is unlikely to have its pKa perturbed sufficiently to ionize near pH 7, and the α-carboxyl group of proline should already be ionized.
The most attractive candidate for the ionizable residue is histidine 97. This is based on several considerations. First, although we are cognizant of the potential for a perturbed pKa, His residues have pKa values nearest to the observed transition. Second, there are only two His residues in the EcRecA protein, and the other His at position 163 can be substituted with a nonionizable Trp residue and retain nearly normal function (78–80). In contrast, H97A mutations are nonfunctional, although H97R mutations have a reported SOS constitutive phenotype, suggesting formation of a very stable filament (81). His-97 is at the subunit-subunit interface, although it is not close enough to Glu-38 or Ser-333 for direct interaction. At least one of the nitrogens of the His-97 side chain is solvent-accessible in the protein-DNA structure (46). Interaction of His-97 with a nearby carboxyl group brought closer by a conformational change in the double mutant could bring about an increase in the observed pKa of this residue.
One way to determine whether key parts of the pH-rate profile observed with RecA E38K/ΔC17 is due to His-97 is to replace the His with other amino acid residues and determine whether the pH behavior is altered. Based on in vivo work indicating that the proteins were likely to be at least partially active (81, 82), we expressed and purified the RecA H97R and H97N mutant proteins.3 In contrast to expectations, neither of these proteins binds to ssDNA, forms filaments, or hydrolyzes ATP. Both mutant proteins are entirely inactive. Although these observations reinforce the idea that His-97 is a critical residue, the mutants do not deliver proteins with activities that can further shed light on His-97 function.
Further study of the ATP hydrolysis-dependent activities of the RecA E38K/ΔC17 double mutant may yield more information about how ATP hydrolysis is coupled to DNA strand exchange. The double mutant is particularly useful in this capacity because its ATP hydrolysis can be uncoupled from DNA strand exchange at will by altering the pH of the reaction solution, without affecting the fundamental DNA pairing process. This allows for more control over the activity being studied.
Acknowledgment
We thank Elizabeth Wood for cloning the recA E38K/ΔC17 mutant gene and providing for its overexpression.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32335 (to M. M. C.).
This paper is dedicated to the memory of our longtime friend and colleague, Ross B. Inman.
R. Britt, unpublished observations.
- ssDNA
- single-stranded DNA
- DTT
- dithiothreitol
- dsDNA
- double-stranded DNA
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- MES
- 4-morpholineethanesulfonic acid
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- SSB
- single strand DNA-binding protein
- ldsDNA
- linear double-stranded DNA
- cssDNA
- circular ssDNA.
REFERENCES
- 1.Cox M. M. (2000) Prog. Nucleic Acids Res. Mol. Biol. 63, 311–366 [DOI] [PubMed] [Google Scholar]
- 2.Cox M. M. (2003) Annu. Rev. Microbiol. 57, 551–577 [DOI] [PubMed] [Google Scholar]
- 3.Lusetti S. L., Cox M. M. (2002) Annu. Rev. Biochem. 71, 71–100 [DOI] [PubMed] [Google Scholar]
- 4.Roca A. I., Cox M. M. (1997) Prog. Nucleic Acid Res. Mol. Biol. 56, 129–223 [DOI] [PubMed] [Google Scholar]
- 5.Sandler S. J., Satin L. H., Samra H. S., Clark A. J. (1996) Nucleic Acids Res. 24, 2125–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S., Yu X., Seitz E. M., Kowalczykowski S. C., Egelman E. H. (2001) J. Mol. Biol. 314, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 7.Sung P. (1994) Science 265, 1241–1243 [DOI] [PubMed] [Google Scholar]
- 8.Sigurdsson S., Trujillo K., Song B., Stratton S., Sung P. (2001) J. Biol. Chem. 276, 8798–8806 [DOI] [PubMed] [Google Scholar]
- 9.Ogawa T., Yu X., Shinohara A., Egelman E. H. (1993) Science 259, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 10.Yu X., Jacobs S. A., West S. C., Ogawa T., Egelman E. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Register J. C., 3rd, Griffith J. (1985) J. Biol. Chem. 260, 12308–12312 [PubMed] [Google Scholar]
- 12.Shan Q., Bork J. M., Webb B. L., Inman R. B., Cox M. M. (1997) J. Mol. Biol. 265, 519–540 [DOI] [PubMed] [Google Scholar]
- 13.Pugh B. F., Cox M. M. (1988) J. Mol. Biol. 203, 479–493 [DOI] [PubMed] [Google Scholar]
- 14.Cox M. M. (2004) in The Bacterial Chromosome ( Higgins N. P. ed) pp. 369–388, American Society of Microbiology, Washington, D. C. [Google Scholar]
- 15.Jain S. K., Cox M. M., Inman R. B. (1994) J. Biol. Chem. 269, 20653–20661 [PubMed] [Google Scholar]
- 16.Brenner S. L., Mitchell R. S., Morrical S. W., Neuendorf S. K., Schutte B. C., Cox M. M. (1987) J. Biol. Chem. 262, 4011–4016 [PubMed] [Google Scholar]
- 17.Bedale W. A., Cox M. (1996) J. Biol. Chem. 271, 5725–5732 [DOI] [PubMed] [Google Scholar]
- 18.Schutte B. C., Cox M. M. (1987) Biochemistry 26, 5616–5625 [DOI] [PubMed] [Google Scholar]
- 19.Cox M. M., Lehman I. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 6018–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox M. M., Lehman I. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehrauer W. M., Kowalczykowski S. C. (1993) J. Biol. Chem. 268, 1292–1297 [PubMed] [Google Scholar]
- 22.Shan Q., Cox M. M., Inman R. B. (1996) J. Biol. Chem. 271, 5712–5724 [DOI] [PubMed] [Google Scholar]
- 23.Kim J. I., Cox M. M., Inman R. B. (1992) J. Biol. Chem. 267, 16438–16443 [PubMed] [Google Scholar]
- 24.Kim J. I., Cox M. M., Inman R. B. (1992) J. Biol. Chem. 267, 16444–16449 [PubMed] [Google Scholar]
- 25.Kowalczykowski S. C., Krupp R. A. (1987) J. Mol. Biol. 193, 97–113 [DOI] [PubMed] [Google Scholar]
- 26.Morrical S. W., Cox M. M. (1990) Biochemistry 29, 837–843 [DOI] [PubMed] [Google Scholar]
- 27.Lavery P. E., Kowalczykowski S. C. (1992) J. Biol. Chem. 267, 9315–9320 [PubMed] [Google Scholar]
- 28.Cox M. M., Lehman I. R. (1982) J. Biol. Chem. 257, 8523–8532 [PubMed] [Google Scholar]
- 29.Eggler A. L., Lusetti S. L., Cox M. M. (2003) J. Biol. Chem. 278, 16389–16396 [DOI] [PubMed] [Google Scholar]
- 30.Umezu K., Kolodner R. D. (1994) J. Biol. Chem. 269, 30005–30013 [PubMed] [Google Scholar]
- 31.Lohman T. M., Overman L. B. (1985) J. Biol. Chem. 260, 3594–3603 [PubMed] [Google Scholar]
- 32.Beernink H. T., Morrical S. W. (1999) Trends Biochem. Sci. 24, 385–389 [DOI] [PubMed] [Google Scholar]
- 33.Hobbs M. D., Sakai A., Cox M. M. (2007) J. Biol. Chem. 282, 11058–11067 [DOI] [PubMed] [Google Scholar]
- 34.Morimatsu K., Kowalczykowski S. C. (2003) Mol. Cell 11, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 35.Sakai A., Cox M. M. (2009) J. Biol. Chem. 284, 3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churchill J. J., Anderson D. G., Kowalczykowski S. C. (1999) Genes Dev. 13, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churchill J. J., Kowalczykowski S. C. (2000) J. Mol. Biol. 297, 537–542 [DOI] [PubMed] [Google Scholar]
- 38.Spies M., Kowalczykowski S. C. (2006) Mol. Cell 21, 573–580 [DOI] [PubMed] [Google Scholar]
- 39.Wang T. C., Chang H. Y., Hung J. L. (1993) Mutat. Res. 294, 157–166 [DOI] [PubMed] [Google Scholar]
- 40.Ennis D. G., Levine A. S., Koch W. H., Woodgate R. (1995) Mutat. Res. 336, 39–48 [DOI] [PubMed] [Google Scholar]
- 41.Lusetti S. L., Shaw J. J., Cox M. M. (2003) J. Biol. Chem. 278, 16381–16388 [DOI] [PubMed] [Google Scholar]
- 42.Lusetti S. L., Wood E. A., Fleming C. D., Modica M. J., Korth J., Abbott L., Dwyer D. W., Roca A. I., Inman R. B., Cox M. M. (2003) J. Biol. Chem. 278, 16372–16380 [DOI] [PubMed] [Google Scholar]
- 43.Datta S., Ganesh N., Chandra N. R., Muniyappa K., Vijayan M. (2003) Proteins 50, 474–485 [DOI] [PubMed] [Google Scholar]
- 44.Story R. M., Steitz T. A. (1992) Nature 355, 374–376 [DOI] [PubMed] [Google Scholar]
- 45.Story R. M., Weber I. T., Steitz T. A. (1992) Nature 355, 318–325 [DOI] [PubMed] [Google Scholar]
- 46.Chen Z., Yang H., Pavletich N. P. (2008) Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 47.Tateishi S., Horii T., Ogawa T., Ogawa H. (1992) J. Mol. Biol. 223, 115–129 [DOI] [PubMed] [Google Scholar]
- 48.Benedict R. C., Kowalczykowski S. C. (1988) J. Biol. Chem. 263, 15513–15520 [PubMed] [Google Scholar]
- 49.Cox M. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 50.Cox M. M. (1995) J. Biol. Chem. 270, 26021–26024 [DOI] [PubMed] [Google Scholar]
- 51.Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 52.Kowalczykowski S. C., Krupp R. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3478–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howard-Flanders P., West S. C., Stasiak A. (1984) Nature 309, 215–219 [DOI] [PubMed] [Google Scholar]
- 54.Shan Q., Cox M. M. (1997) J. Biol. Chem. 272, 11063–11073 [DOI] [PubMed] [Google Scholar]
- 55.MacFarland K. J., Shan Q., Inman R. B., Cox M. M. (1997) J. Biol. Chem. 272, 17675–17685 [DOI] [PubMed] [Google Scholar]
- 56.Shan Q., Cox M. M. (1998) Mol. Cell 1, 309–317 [DOI] [PubMed] [Google Scholar]
- 57.Craig N. L., Roberts J. W. (1981) J. Biol. Chem. 256, 8039–8044 [PubMed] [Google Scholar]
- 58.Lohman T. M., Green J. M., Beyer R. S. (1986) Biochemistry 25, 21–25 [DOI] [PubMed] [Google Scholar]
- 59.Haruta N., Yu X., Yang S., Egelman E. H., Cox M. M. (2003) J. Biol. Chem. 278, 52710–52723 [DOI] [PubMed] [Google Scholar]
- 60.Messing J. (1983) Methods Enzymol. 101, 20–78 [DOI] [PubMed] [Google Scholar]
- 61.Neuendorf S. K., Cox M. M. (1986) J. Biol. Chem. 261, 8276–8282 [PubMed] [Google Scholar]
- 62.Morrical S. W., Lee J., Cox M. M. (1986) Biochemistry 25, 1482–1494 [DOI] [PubMed] [Google Scholar]
- 63.Lindsley J. E., Cox M. M. (1990) J. Biol. Chem. 265, 9043–9054 [PubMed] [Google Scholar]
- 64.Schnos M., Inman R. B. (2000) Mol. Biotechnol. 16, 77–86 [DOI] [PubMed] [Google Scholar]
- 65.Holmes V. F., Scandellari F., Benjamin K. R., Cozzarelli N. R. (2002) J. Biol. Chem. 277, 38945–38953 [DOI] [PubMed] [Google Scholar]
- 66.Pugh B. F., Cox M. M. (1987) J. Biol. Chem. 262, 1326–1336 [PubMed] [Google Scholar]
- 67.Weinstock G. M., McEntee K., Lehman I. R. (1981) J. Biol. Chem. 256, 8829–8834 [PubMed] [Google Scholar]
- 68.Shan Q., Cox M. M. (1996) J. Mol. Biol. 257, 756–774 [DOI] [PubMed] [Google Scholar]
- 69.Arenson T. A., Tsodikov O. V., Cox M. M. (1999) J. Mol. Biol. 288, 391–401 [DOI] [PubMed] [Google Scholar]
- 70.Cox M. M., Soltis D. A., Lehman I. R., DeBrosse C., Benkovic S. J. (1983) J. Biol. Chem. 258, 2586–2592 [PubMed] [Google Scholar]
- 71.Lee J. W., Cox M. M. (1990) Biochemistry 29, 7677–7683 [DOI] [PubMed] [Google Scholar]
- 72.Lindsley J. E., Cox M. M. (1989) J. Mol. Biol. 205, 695–711 [DOI] [PubMed] [Google Scholar]
- 73.Cox J. M., Li H., Wood E. A., Chitteni-Pattu S., Inman R. B., Cox M. M. (2008) J. Biol. Chem. 283, 24909–24921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nayak S., Bryant F. R. (1999) J. Biol. Chem. 274, 25979–25982 [DOI] [PubMed] [Google Scholar]
- 75.Gruenig M. C., Renzette N., Long E., Chitteni-Pattu S., Inman R. B., Cox M. M., Sandler S. J. (2008) Mol. Microbiol. 69, 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campbell M. J., Davis R. W. (1999) J. Mol. Biol. 286, 437–445 [DOI] [PubMed] [Google Scholar]
- 77.Cox J. M., Abbott S. N., Chitteni-Pattu S., Inman R. B., Cox M. M. (2006) J. Biol. Chem. 281, 12968–12975 [DOI] [PubMed] [Google Scholar]
- 78.Stole E., Bryant F. R. (1994) J. Biol. Chem. 269, 7919–7925 [PubMed] [Google Scholar]
- 79.Stole E., Bryant F. R. (1995) J. Biol. Chem. 270, 20322–20328 [DOI] [PubMed] [Google Scholar]
- 80.Stole E., Bryant F. R. (1997) Biochemistry 36, 3483–3490 [DOI] [PubMed] [Google Scholar]
- 81.Campbell M. J., Davis R. W. (1999) J. Mol. Biol. 286, 417–435 [DOI] [PubMed] [Google Scholar]
- 82.McGrew D. A., Knight K. L. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 385–432 [DOI] [PubMed] [Google Scholar]