Abstract
Helminth pathogens prepare a Th2 type immunological environment in their hosts to ensure their longevity. They achieve this by secreting molecules that not only actively drive type 2 responses but also suppress type 1 responses. Here, we show that the major cysteine proteases secreted from the helminth pathogens Fasciola hepatica (FheCL1) and Schistosoma mansoni (SmCB1) protect mice from the lethal effects of lipopolysaccharide by preventing the release of inflammatory mediators, nitric oxide, interleukin-6, tumor necrosis factor α, and interleukin-12, from macrophages. The proteases specifically block the MyD88-independent TRIF-dependent signaling pathway of Toll-like receptor (TLR)4 and TLR3. Microscopical and flow cytometric studies, however, show that alteration of macrophage function by cysteine protease is not mediated by cleavage of components of the TLR4 complex on the cell surface but occurs by degradation of TLR3 within the endosome. This is the first study to describe a parasite molecule that degrades this receptor and pinpoints a novel mechanism by which helminth parasites modulate the innate immune responses of their hosts to suppress the development of Th1 responses.
Keywords: Enzymes/Peptidases, Immunology/Innate Immunity, Immunology/Toll Receptors, Organisms/Parasite, Parasitology, Proteases/Cysteine Protease
Introduction
To establish successful chronic infections, helminth parasites induce potent Th2 immune responses, which can only occur when Th1 cytokines are suppressed (1). To create this anti-inflammatory environment helminths secrete molecules that prevent dendritic cells and macrophages responding to Toll-like receptor (TLR)2 Th1-stimulating ligands such as LPS and CpG (2–5). It has been proposed that helminth-secreted molecules themselves signal through TLRs, activating a different pattern of kinases compared with LPS, resulting in the extracellular signal-regulated kinase dependent inhibition of interleukin (IL)-12 production (reviewed in Ref. 6). The molecular nature of these Th1-suppressing components, however, remains largely unknown.
Enzyme activity ascribable to the papain family of cysteine proteases (clan CA, family C1), such as cathepsin L and B, has been identified as a major component of the secretions of almost all helminth parasites of humans, livestock, and companion animals (7, 8). These proteases are pivotal to the parasitic lifestyle by mediating fundamental processes such as host invasion, acquisition of nutrients, and the migration through host tissues (reviewed in Ref. 9). They can also have a systemic effect on the immune responses of their hosts by cleaving immunoglobulins and preventing antibody-mediated immune effector function, which protects the parasites from immune elimination (10, 11). We have shown that the predominant secreted product of the human and animal helminth pathogen Fasciola hepatica, a cathepsin L1 cysteine protease (FheCL1), suppressed the onset of protective Th1 immune responses in mice to infections with the respiratory microbe Bordetella pertussis, making them more susceptible to disease. In addition, injection of the cysteine protease immediately prior to immunization of mice with a B. pertussis whole-cell pertussis vaccine prevented the development of a Th1 response to the vaccine (12–14).
The engagement of TLRs is a critical step in the detection of pathogenic infection and promotion of appropriate adaptive immune responses. Here, we show that the FheCL1 protease secreted by the helminth pathogen F. hepatica (liver fluke) and the major cathepsin B1 protease secreted by the related helminth Schistosoma mansoni (blood fluke) inhibit macrophage TLR recognition of bacterial ligands. Delivery of cysteine protease protects mice from LPS-induced septic shock by preventing the release of inflammatory mediators, nitric oxide, TNFα, IL-6, IL-12, by macrophages. Using various TLR agonists we found that the cysteine proteases inactivated MyD88-independent TRIF-dependent signaling pathways of TLR4 and TLR3. This inactivation is not mediated by cleavage of cell surface TLR4 or CD14 but results from specific degradation of TLR3 within the endosome. This study, therefore, describes a novel means by which parasites alter innate immune cell function and prevent the establishment of potent Th1-driven inflammatory responses that would lead to their elimination.
EXPERIMENTAL PROCEDURES
Enzyme Preparation and Animals
Functionally active recombinant F. hepatica cathepsin L1 (FheCL1) and S. mansoni cathepsin B1 (SmB1) were expressed in Pichia pastoris and purified by affinity chromatography on nickel-nitrilotriacetic acid-agarose as described (15, 16). A conformationally intact but proteolytically inactive variant of FheCL1, termed FheCL1Gly26, was prepared by substituting the active site cysteine with a glycine as described (17). 6-to-8-week old female BALB/c mice were purchased from ARC (Perth, Australia) and maintained according to the guidelines of the University of Technology Sydney/Royal North Shore Hospital Animal Care and Ethics committee.
Macrophage Cell Culture
The peritoneal cavity of BALB/c mice was lavaged with 5 ml of supplemented RPMI (Invitrogen). Following determination of viability by trypan blue exclusion, peritoneal exudate cells were adjusted to 5 × 106cells/ml and cultured in six-well plates (Costar, Cambridge, MA). After 2–3 h incubation at 37 °C, nonadherent cells were removed by washing with ice cold 1× phosphate-buffered saline (PBS). The remaining adherent cells were removed with a cell scraper into RPMI. For each experiment, macrophages from 10–15 mice were pooled, and preparations of 1 × 106 cells/ml were stimulated overnight for 12 h (or 6 h for mRNA expression analysis) with varying concentrations of LPS (Escherichia coli 111:B4; Sigma), CpG ODN 1826 (synthesized by Sigma Genosys), poly(IC) (Sigma), flagellin (Calbiochem) and Pam3Cys-Ser-(Lys)4 (EMC Microcollections, Tubingen, Germany).
Quantitation of Cytokine and Nitrite in Macrophage Supernatants
Interferon (IFN)-γ was measured by immunoassay according to the manufacturer's instructions using pairs of commercially available monoclonal antibodies (BD Pharmingen). Concentrations of IL-6, IL-12, and TNFα were determined by commercially available sandwich enzyme-linked immunosorbent assay (ELISA) kits (BD Pharmingen). Production of nitric oxide by macrophages was assessed by measuring the increase in nitrite concentration using the Greiss reagent (Promega).
Mouse Model of Endotoxic Shock
Six-week-old BALB/c mice were intra-peritoneally injected with 10 mg/kg of E. coli LPS (serotype 111:B4; Sigma) and observed hourly over a 70-h period. For the analysis of inflammatory mediators, mice were sacrificed at 2 and 4 h after LPS injection by cervical dislocation. Heart punctures were performed, and blood was collected in heparinized tubes. Plasma was isolated by centrifugation at 13,000 × g for 10 min. Peritoneal lavage was also performed, and macrophages were isolated as described above. Both the sera and lavage supernatants were analyzed for the presence of inflammatory cytokines by ELISA.
Flow Cytometry
Macrophages (1 × 106cells/ml) were stained for 30 min at 4 °C with antibodies specific for CD14 (BD Pharmingen) or the TLR4·MD-2 complex (Santa Cruz Biotechnology, Santa Cruz, CA) along with the relevant isotype controls. Antibodies were diluted in PBS, 0.5% bovine serum albumin and used at the recommended concentrations.
RNA Preparation and RT-PCR
Following incubation of macrophages with TLR agonists, cells were washed with PBS, and total RNA were isolated with Tri-reagent (Sigma) according to the manufacturer's instructions. Reverse transcription reactions were performed with 50 ng of RNA, oligo(dT) primer (synthesized by Sigma Genosys), and avian myeloblastosis virus reverse transcriptase (Promega). First strand cDNA (5 μl) was then subjected to amplification with Taq polymerase (Sigma) using primers specific for β-actin, TNFα, IL-6, IL-12, iNOS, TLR4, IFNβ, interferon-stimulated gene 54, and TLR3 using cycle conditions optimized for each primer set. All PCR products were electrophoretically analyzed on 1% agarose gel and visualized by ethidium bromide staining. In some cases, quantification of PCR products was carried out by densitometric analysis of photographic negatives of agarose gels using Un-Scan-It software (Silk Scientific). The quantification method used in these assays is relative because absolute values of mRNA levels could not be determined, as internal controls for specific mRNAs were not used. Values for mRNA are expressed in relative absorbance units and are standardized per unit of β-actin per sample.
Western Blots
A total of 1 × 106 macrophages/ml were solubilized in 100 μl of ice cold lysis buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and protease and phosphatase inhibitor cocktails (Sigma). Equivalent amounts of protein were resolved by 12.5% SDS-polyacrylamide gel electrophoresis then transferred to Immobilon membrane (Millipore). Membranes were blocked with 5% nonfat dry milk, washed, and incubated overnight at 4 °C with primary antibody specific for TLR3 (1:200 dilution in 5% bovine serum albumin and 5% PBS-Tween 20; Abcam), TRAF3 or TRIF (1:1,000 dilution; Abcam). Following a 3-h incubation at room temperature with a 1:1,000 dilution of anti-rabbit horseradish peroxidase conjugated secondary antibody (Sigma), the blots were visualized with ECL Western blotting detection reagent (Sigma). Loading controls were determined using a primary antibody specific for actin (Sigma).
Microscopy
Six-week-old BALB/c mice were intraperitoneally injected with 50 μg CL1 or PBS. After 2 h, peritoneal macrophages were harvested from lavages by adherence to plastic, as described above. Macrophages from 10 mice were pooled and preparations of 1 × 106 cells/ml were allowed to adhere to glass coverslips. The cells were fixed by incubation with 4% paraformaldehyde and subsequently permeabilized by incubation in the presence of 0.1% Triton-X/PBS. After this, cells were incubated for 1 h at room temperature with primary antibody specific for TLR3, His6 tag, EEA-1 or mannose-6-phosphate receptor (Abcam) diluted 1:50 in 1% fetal calf serum/PBS. Following three washes with 1× PBS, cells were incubated with TRITC-conjugated (Sigma), Alexa Fluor 488-conjugated, or Alexa Fluor 568-conjugated (Molecular Probes) secondary antibody (1:1,000 dilution) and then stained with 4′,6-diamidino-2-phenylindole (1 μg/ml in PBS) to detect cell nuclei. Cells were examined either with a Nikon A1 confocal scanning laser microscope, and images were analyzed using NIS Elements software (Nikon) or a DeltaVision personal DV deconvolution microscope (Applied Precision, Inc.), and the images were analyzed using IMARIS software (Bitplane).
Statistical Analysis
The statistical significance of difference was determined by the two tailed Student's t test. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RESULTS
Helminth Cysteine Protease Rescues Mice from Septic Shock
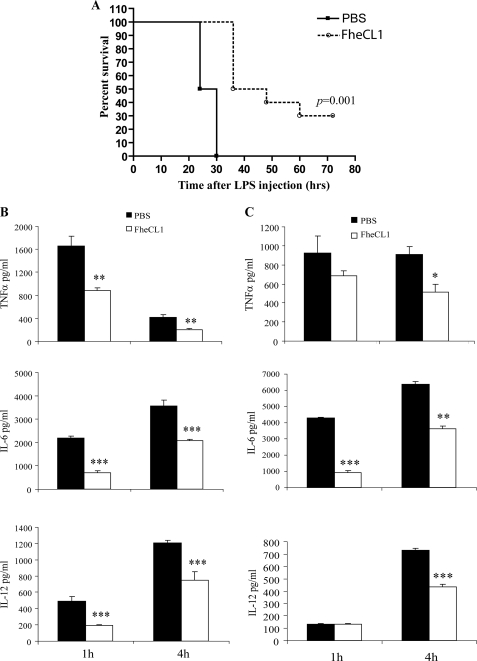
We have previously shown that the rapid suppression of interferon-γ-associated inflammatory responses during infection with F. hepatica is related to the secretion of FheCL1 cysteine proteases (14). Here, we employed a murine sepsis model to examine whether FheCL1 suppressed macrophage-driven Th1 inflammation. When BALB/c mice were injected intraperitoneally with 200 μg (10 mg/kg) of E. coli LPS, fatal endotoxic shock occurred within 30 h (Fig. 1A). However, administration of a single dose (50 μg) of FheCL1, 2 h prior to LPS administration, reduced mortality by 30% (p = 0.001). A significant reduction in the levels of proinflammatory mediators (IL-6, IL-12, TNFα, and IFNγ) of endotoxic shock in both the sera (Fig. 1B) and peritoneal lavage fluid (Fig. 1C) of FheCL1-treated mice was also observed.
FIGURE 1.
Helminth parasite cysteine protease protects mice from LPS-induced lethality. A, mice (n = 15) were pretreated with a single intraperitoneal injection of 50 μl of active cysteine protease enzyme (FheCL1, 1 mg/ml) or PBS 2 h prior to an intraperitoneal injection of LPS (10 mg/kg) and observed over 70 h. Data presented are representative of two independent experiments. Levels of proinflammatory mediators TNFα, IL-6, and IL-12 were measured in the sera (B) and peritoneal lavage fluid of mice (n = 6) (C) at 2 and 4 h after an intraperitoneal injection of LPS (10 mg/kg). Data are from individual mice from two independent experiments.
Helminth Cysteine Proteases Suppress the Activation of Macrophages in Response to TLR4 Stimulation by Lipopolysaccharide
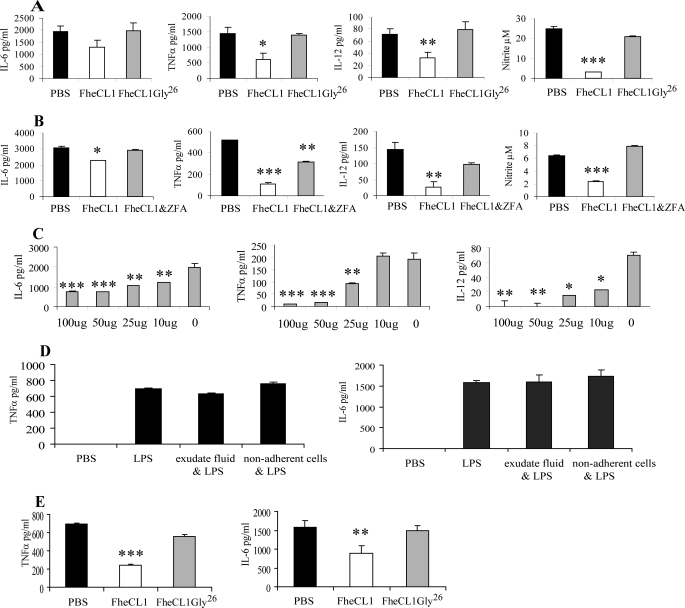
Because macrophages are the main source of pro-inflammatory mediators in the septic shock model, we examined their activation status in FheCL1-treated mice. Macrophages were removed from the peritoneal cavity of mice 2 h after treatment with a single dose of FheCL1 and stimulated ex vivo with LPS for 12 h. We found that the levels of all proinflammatory mediators, nitric oxide, IL-6, TNFα, and IL-12 secreted from these macrophages were significantly reduced compared with macrophages derived from animals pretreated with PBS (Fig. 2A). Suppression of macrophage activation by FheCL1 was negated if the enzyme was complexed with the cysteine protease specific inhibitors Z-Phe-Ala-diazomethylketone (20 μm) or E-64 (1 μm) (Fig. 2B and data not shown) prior to administration to mice. In addition, administration of a proteolytically inactive variant of FheCL1, which has the active site cysteine replaced with a glycine, FheCL1Gly26, did not suppress the response to LPS stimulation (Fig. 2A). Inhibition of macrophage responsiveness to LPS was dependent on the dose of FheCL1 delivered, with decreasing doses showing correspondingly lower suppression of proinflammatory cytokine secretion (Fig. 2C). Using similar experiments, we showed that treatment of mice with the major secreted cathepsin B1 (SmB1) protease of the helminth S. mansoni was also capable of suppressing macrophage proinflammatory cytokine secretion when given in an active form but not when complexed with the inhibitor E-64 (data not shown).
FIGURE 2.
FheCL1-treated macrophages are hyporesponsive to LPS stimulation. A, peritoneal macrophages were isolated from BALB/c mice 2 h after an intraperitoneal injection of PBS, FheCL1 or an inactive mutant of cathepsin L1 (FheCL1Gly26) and stimulated ex vivo with LPS (1 μg/ml) for 12 h. The levels of secretion of IL-6, IL-12, TNFα, and nitrite into the culture supernatants were assessed by ELISA and Greiss assay, respectively. Indicated values are the means + S.E. and representative of three independent experiments. B, F. hepatica cathepsin L1 was inhibited by a 30-min incubation in the presence of cysteine protease-specific inhibitor Z-Phe-Ala-diazomethylketone (20 μm) and then injected intraperitoneally into BALB/c mice. After 2 h, peritoneal macrophages were removed and stimulated in vitro with LPS (1 μg/ml) for 12 h and secretion of proinflammatory mediators measured by ELISA and Greiss assay. Data shown are the mean ± S.E. from triplicate samples. C, BALB/c mice were given a range of concentrations of active FheCL1 (0–100 μg), and 2 h later, peritoneal macrophages were harvested and stimulated ex vivo with LPS (1 μg/ml) for 12 h. ELISA was used to measure the levels of IL-6, IL-12, and TNFα in culture supernatants. Data are presented as the mean ± S.E. of triplicate samples and are representative of three experiments. D, peritoneal macrophages were isolated from naïve BALB/c mice and incubated for 2 h in the presence of peritoneal exudate fluid or nonadherent cells extracted from the peritoneal cavity of mice given a single dose of FheCL1. The quantities of IL-6 and TNFα secreted in response to overnight stimulation with LPS (1 μg/ml) were assessed by ELISA. Data are presented as the mean ± S.E. of triplicate samples and are representative of two experiments. E, peritoneal macrophages were isolated from naïve BALB/c mice and incubated for 2 h in the presence of PBS, FheCL1, or FheCL1Gly26. The cells were subsequently stimulated with LPS (1 μg/ml), and the levels of IL-6 and TNFα in the culture media were measured by ELISA. Data are presented as the mean ± S.E. of triplicate samples and are representative of two experiments.
Peritoneal administration of FheCL1 altered the cellular composition of the peritoneal cavity (data not shown) by increasing the number of lymphocytes to 8% of the cell population as compared with 1% following delivery of PBS. To determine whether the increase in lymphocytes contributed to the suppressive effect on macrophages, peritoneal macrophages isolated from naïve BALB/c mice were cultured in the presence of the nonadherent cellular population of peritoneal exudate isolated from FheCL1-treated mice and also in the presence of the exudate fluid. When these macrophages were subsequently (2 h later) stimulated with LPS, no suppression of IL-6 and TNFα secretion was observed (Fig. 2D). These data exclude the possibility that the increase in lymphocytes, or secretions from these cells, is responsible for the suppression of macrophage inflammatory responses in FheCL1-treated mice.
To further confirm this, macrophages isolated from the peritoneal cavity of naïve BALB/c mice were exposed in vitro to PBS, FheCL1, or FheCL1Gly26 for a period of 2 h. Following removal of the cysteine protease, the macrophages were stimulated with LPS. Similar to that observed with in vivo exposure, macrophages treated with FheCL1 were less responsive to LPS stimulation, secreting significantly reduced levels of both TNFα and IL-6 compared with macrophages treated with PBS (Fig. 2E). The inactive variant FheCL1Gly26 exhibited no effect on the macrophages in vitro, further substantiating the requirement for proteolytic activity. Collectively, these studies demonstrate that helminth cysteine proteases can prevent the activation of macrophages by LPS and that this property is dependent on the hydrolytic activity of the enzymes.
Suppression of Macrophage Function Does Not Occur via the Inactivation of MyD88-signaling Pathways
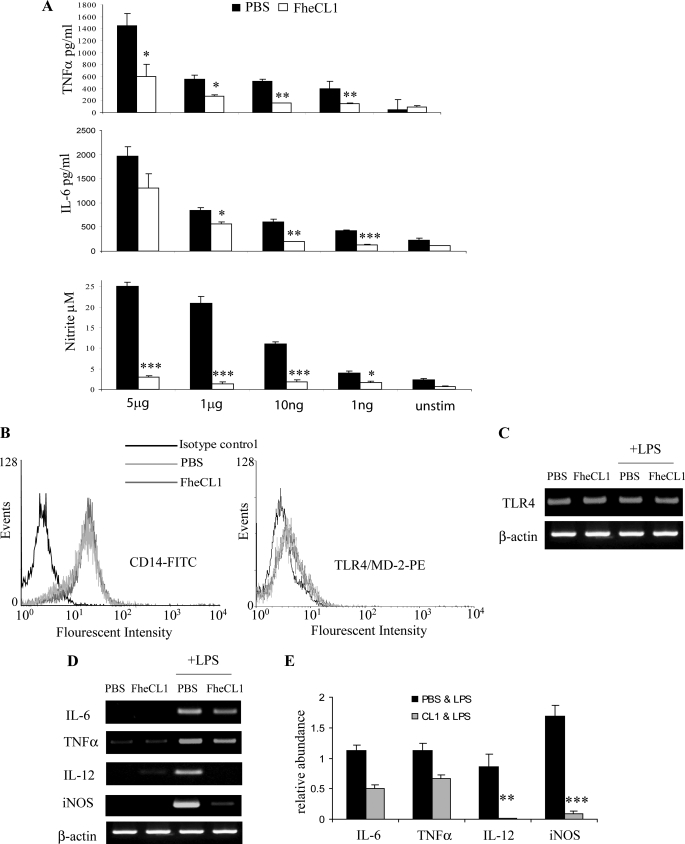
The core macrophage receptors recognizing LPS are CD14, TLR4, and MD-2. Bromelain, a cysteine protease of pineapple and the Arg- and Lys-specific gingipains secreted, respectively, secreted from Porphyromonas gingivalis remove CD14 but not TLR4 from the surface of macrophages (18, 19), making the cells hyporesponsive to LPS stimulation. While the induction of inflammatory mediators in response to low concentrations of LPS (<10 ng/ml) requires the participation of both TLR4·MD-2 complex and CD14, responses to high concentrations of LPS (>1 μg/ml) are independent of CD14 (20). In this study, a single treatment of mice with FheCL1 suppressed the production of inflammatory mediators in response to stimulation with a range of LPS concentrations from 1 ng to 5 μg/ml (Fig. 3A), suggesting that the action of FheCL1 on macrophages does not involve proteolytic removal of CD14. This conclusion was supported by flow cytometry that revealed no difference in the levels of surface CD14 on macrophages isolated from FheCL1-treated and PBS-treated mice (Fig. 3B). Furthermore, we found no reduction in the surface expression of TLR4-MD2 on macrophages from FheCL1-treated mice using flow cytometry (Fig. 3B), and no alteration in TLR4 transcript expression by RT-PCR analysis (Fig. 3C).
FIGURE 3.
FheCL1 does not affect MyD88-dependent signaling through TLR4. A, BALB/c mice were given a single intraperitoneal injection of 50 μl of FheCL1 (1 mg/ml) or PBS and 2 h later peritoneal macrophages were removed. A, these macrophages were then stimulated with varying concentrations of LPS (0–5 μg/ml) for 12 h, and secreted cytokines were measured by ELISA. Data shown are the means + S.E. of triplicates and are representative of two individual experiments. B, macrophages removed from either PBS- or CL1-treated mice were analyzed for the surface expression of CD14 or TLR4/MD-2 by flow cytometry. C, TLR4 mRNA levels in peritoneal macrophages isolated from either PBS- or FheCL1-treated BALB/c mice incubated in the presence or absence of LPS was assessed by RT-PCR. D, peritoneal macrophages isolated from both PBS- and FheCL1-treated mice were stimulated with LPS (1 μg/ml) for 6 h, lysed, and examined for levels of mRNA expression of IL-6, TNFα, IL-12, and iNOS by RT-PCR. Data shown are representative of three independent experiments. E, densitometric analysis of amplification products for IL-6, TNFα, IL-12, and iNOS. Values for mRNA are expressed in relative absorbance units and are standardized per unit of β-actin per sample. The data are obtained from the mean of three independent experiments; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
We investigated whether FheCL1 exhibited its suppressive effect on macrophages by modulating TLR4 signaling pathways. Following interaction between LPS and the CD14· TLR4·MD-2 receptor complex, recruitment of MyD88 activates a series of kinases, ultimately resulting in the translocation of NFκB to the nucleus, where it mediates an increase in inflammatory cytokine gene expression leading to proinflammatory responses (reviewed in Ref. 21). TLR4 recognition of LPS, however, also leads to the activation of a MyD88-independent (TRIF-dependent) pathway that, while inducing NFκB activation, primarily activates the transcription factor IRF3. IRF3 then induces the transcription of IFNβ, which, in turn, leads to STAT1 activation and the subsequent induction of STAT1-dependent genes such as iNOS and interferon-inducible protein 10 (22).
We found a reduction in the expression of IL-6 and TNFα transcripts in macrophages isolated from FheCL1-treated mice compared with PBS-treated mice. Although these changes failed to reach statistical significance (p = 0.08 for both) when analyzed by densitometry, the decrease was reproducible between experiments. In stark contrast, FheCL1 treatment significantly suppressed the induction of both IL-12 (p = 0.0067) and iNOS (p = 0.0005) mRNA expression (Fig. 3, D and E). This differential suppression of gene expression correlates with the reduction of the respective cytokines as seen in Fig. 3A, which clearly shows that the suppression of LPS induced IL-6 and TNFα was far less significant than the reduction in nitric oxide production. This result is relevant because, while the expression of proinflammatory cytokines such as IL-6 and TNFα in response to TLR4 binding is mediated through both TRIF and MyD88 (21), induction of iNOS expression is mostly dependent on TRIF-mediated activation of IFNβ (22). Furthermore, it was recently shown that IL-12 mRNA expression in macrophages is also regulated by IFNβ (23). Therefore, the pronounced suppression of iNOS and IL-12 expression in macrophages caused by FheCL1 treatment was most likely mediated by selective inhibition of TRIF-dependent, IFNβ-mediated TLR signaling.
Suppression of Macrophage Activation by Helminth Cysteine Proteases Occurs via Specific Inhibition of TRIF-dependent Signaling Pathways
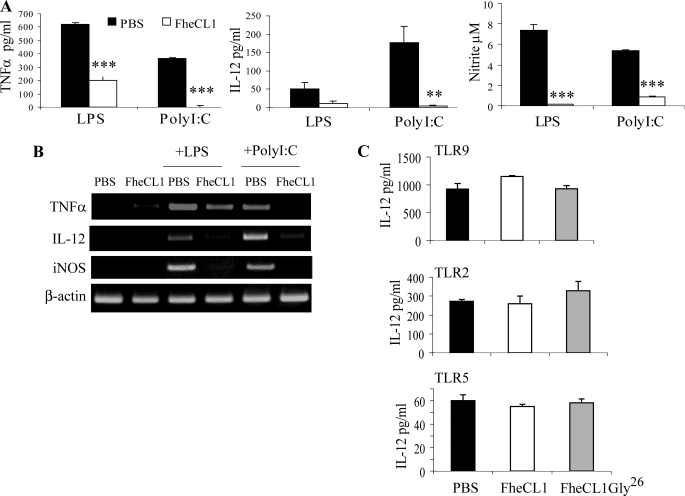
To investigate the possibility that FheCL1 was specifically targeting the TRIF-dependent pathway of macrophage activation, we stimulated macrophages with poly(I·C), a synthetic agonist of TLR3, which signals exclusively via the TRIF-dependent pathway (24). We found that FheCL1-treated macrophages secreted significantly lower levels of IL-12, TNFα, and nitrite when exposed to poly(I·C) for 12 h compared with macrophages taken from PBS-treated or FheCL1Gly26-treated controls (Fig. 4A and data not shown). Furthermore, mRNA transcripts of these inflammatory mediators were not detected in FheCL1-treated macrophages (Fig. 4B). In contrast, FheCL1- or FheCL1Gly26-treatment of mice had no effect on the ability of macrophages to respond to ligands of TLR2 (Pam3Cys-Ser-(Lys)4), TLR5 (flagellin), or TLR 9 (CpG DNA, Fig. 4C and supplemental Fig. 1), all of which mediate their effects solely through MyD88-dependent pathways.
FIGURE 4.
FheCL1 inhibits TRIF-dependent production of inflammatory mediators. A, peritoneal macrophages isolated 2 h after a single 50 μl intraperitoneal injection of either FheCL1 (1 mg/ml) or PBS were stimulated with either LPS (1 μg/ml) or poly(I·C) (25 μg/ml). B, cell supernatants were then analyzed by ELISA, and cell lysates were examined by RT-PCR for levels of mRNA expression of cytokines. C, ELISA was used to measure the quantity of IL-12 in culture media or macrophages stimulated in vitro with the TLR9 ligand CpG (1 μm), the synthetic TLR2 ligand Pam3Cys-Ser-(Lys)4(10 ng/ml) and the TLR5 ligand flagellin (10 ng/ml). Indicated values for ELISA are the means + S.E. of triplicates and are representative of three individual experiments.
Similarly, macrophages isolated from mice after treatment with SmB1 were less responsive to stimulation with both LPS and poly(I·C) but not CpG (supplemental Fig. 2). In addition, mRNA expression of IL-12 and iNOS but not TNFα was absent from LPS-stimulated macrophages. Like FheCL1, SmB1 inhibited the expression of all three mRNAs in response to poly(I·C) (supplemental Fig. 2).
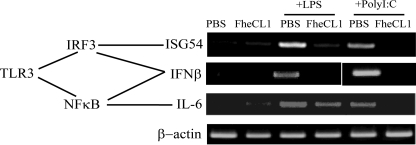
Further RT-PCR analyses on macrophages isolated from FheCL1-treated mice revealed no detectable levels of IFNβ mRNA in response to LPS or poly(I·C) stimulation, in contrast to macrophages from PBS-treated mice that did express IFNβ (Fig. 5). The promoter region of IFNβ has been shown to require the cooperation of both DNA-binding transcription factors IRF3 and NFκB (25). However, activation of IRF3 alone is sufficient for the induction of ISG54 gene expression, and IL-6 is an example of a gene that is induced by activation of NFκB, independently of IRF-3 (26). Macrophages isolated from FheCL1-treated mice show no expression of either ISG54 or IL-6 in response to the ligation of TLR3 with poly(I·C), indicating the absence of activation of both IRF3 and NFκB (Fig. 5). In contrast, exposure of FheCL1-treated macrophages to LPS results in the induction of IL-6 gene expression but not ISG54, suggesting that TLR4/MyD88-dependent induction of NFκB remains intact but the TRIF-dependent activation of IRF3 in response to LPS is inhibited by FheCL1.
FIGURE 5.
FheCL1 treatment of macrophages inhibits TRIF-dependent signaling. Peritoneal macrophages isolated from PBS- or FheCL1-treated mice were stimulated with either LPS (1 μg/ml) or poly(I·C) (25 μg/ml) for 6 h, and expression of IFNβ, ISG54, and IL-6 mRNA were assessed by RT-PCR. These data are representative of three independent experiments.
Suppression of TRIF-dependent Activation Occurs via Degradation of TLR3 by Helminth Cysteine Protease
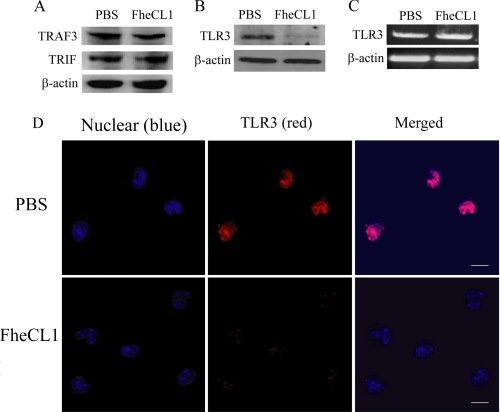
It was shown recently that a serine protease of hepatitis C virus inhibited poly(I·C)-activated signaling via the proteolytic cleavage of the TRIF adaptor protein of TLR3 (27). In addition, the tumor necrosis factor receptor-associated factor 3 (TRAF3), an important link between the recruitment of TRIF and the activation of IRF3, is susceptible to cleavage by protease enzymes (28). However, neither TRIF nor TRAF3 were susceptible to degradation by FheCL1, likely because of their cytosolic location (Fig. 6A). Unlike viral proteases, parasite proteins are taken from the external environment by dendritic cells and macrophages (29, 30) and are trafficked via early endosomes to the lysosomal compartments (30). Considering TLR3 exists in the endolysosomal compartments of cells, we therefore investigated whether FheCL1 may be exerting its suppressive effect on macrophages via the degradation of TLR3. Immunoblot analyses of proteins extracted from macrophages isolated from mice given a single injection of FheCL1 failed to detect TLR3 protein (Fig. 6B) despite there being no alteration in the level of expression of the TLR3 gene, as detected by RT-PCR (Fig. 6C). Confocal laser microscopy analysis confirmed the absence of TLR3 in macrophages treated with FheCL1 (Fig. 6D), with cells stained positively for TLR3 reduced to 20 and 40% of total cells counted in two independent experiments compared with 99.1 and 99.7% of macrophages from PBS-treated mice.
FIGURE 6.
FheCL1 treatment degrades TLR3 in macrophages. Cell lysates were prepared from macrophages isolated from mice, and the presence of TRIF and TRAF3 protein was determined by Western blot (A). B, detection of TLR3 protein in peritoneal macrophages was examined by Western blot, and expression of mRNA for TLR3 was assessed by RT-PCR. C, peritoneal macrophages isolated from PBS-treated (upper panels) or FheCL1-treated (lower panels) mice were immunostained for the presence of TLR3 (red). Cells were also stained with 4′,6-diamidino-2-phenylindole (blue) to detect nuclei and imaged using a Nikon A1 confocal scanning laser microscope. Scale bar is 10 μm. These data are representative of two independent experiments.
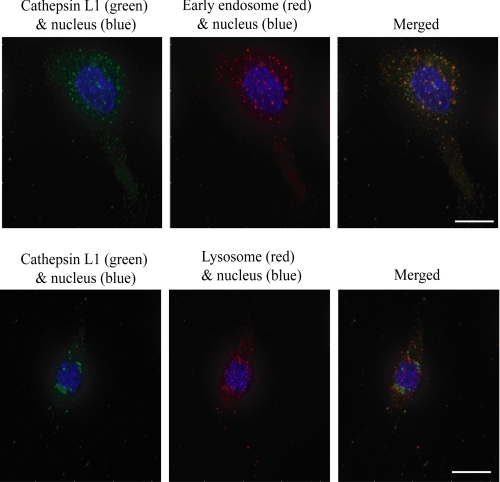
To confirm that the FheCL1 degradation of TLR3 occurs as a result of direct action within the endosome, we examined the subcellular localization of FheCL1 after injection by staining the peritoneal macrophages with an anti-His6 tag antibody. No fluorescence is observed in cells isolated from PBS-treated mice and stained with the anti-His6 tag antibody (data not shown), which confirms its specificity for recombinant FheCL1. While there is some colocalization between FheCL1 and mannose-6-phosphate receptor positive compartments (Fig. 7, lower panel), the majority of FheCL1 appears to be located within the early endosome compartments (Fig. 7, upper panel). These data support our proposal that 2 h after injection into the peritoneal cavity, FheCL1 has been captured by the macrophage and is directly cleaving TLR3 from within the endosome of the macrophage and thus preventing TRIF-dependent signaling.
FIGURE 7.
FheCL1 is targeted to the endolysosomal compartment of macrophages. Peritoneal macrophages were isolated from BALB/c mice 2 h after an intraperitoneal injection of FheCL1. After fixation and permeabilization, cells were stained for His6 tag (FheCL1, green), EEA-1 (early endosomes, red, upper panel) and mannose-6-phosphate receptor (lysosomes, red, lower panel). Cells were also stained with 4′,6-diamidino-2-phenylindole (blue) to detect nuclei and imaged using a DeltaVision personal DV microscope. Scale bar is 10 μm.
DISCUSSION
F. hepatica secretes abundant amounts of FheCL1 protease into the tissues of its host where the enzyme functions in tissue degradation and immunosuppression (9–14). This study broadens our understanding of FheCL1 immune modulation by revealing a novel regulatory role for cysteine proteases in TRIF-dependent signaling. Our data show that parasite proteases specifically degrade TLR3 within the endosome, which diminishes macrophage activation in response to both TLR3 and TLR4 stimulation. However, why degradation of TLR3 results in impaired signaling of the LPS-induced, TLR4-TRIF-dependent pathway remains unanswered. Various levels of cross-talk between the TLR pathways have been described. Significant to the present study are the observations that pretreatment of macrophages with poly(I·C) resulted in the inhibition of LPS induced TRIF-dependent signaling but not the activation of MyD88-dependent pathways (31). These findings highlight an interaction between TLR3 and TLR4 TRIF-dependent pathways, which may be affected by parasite cysteine proteases. In addition, other as yet unidentified mechanisms may be involved which are independently inhibiting the complete activation of TLR4 by LPS.
A number of papain-like cysteine proteases exist within the endolysosomes of mammalian cells exhibiting distinct functions that are dictated by their substrate specificities. In the macrophage endolysosomal compartment, proteolytic cleavage is a prerequisite for TLR9 signaling and requires the activities of both mammalian cathepsins L and S but not cathepsin K (32). By contrast, TLR7 and TLR3, also present within the endolysosome, are not susceptible to cleavage by these proteases. We have previously demonstrated significant differences in the substrate specificities between human and helminth cathepsin L. Most importantly, the parasite enzymes display a broader pH range for activity and greater stability under both acidic and neutral pH (16). These physicobiochemical properties could explain why the parasite but not the resident endolysosomal cathepsin L cleaves TLR3.
In general, macromolecules internalized by macrophages are degraded into antigenic peptides by the range of cathepsins resident in the endosomal and lysosomal compartments. However, our data indicate that FheCL1 is resistant to this proteolytic degradation, which is consistent with our earlier findings showing that the mature enzyme is highly resistant to proteolytic degradation by endopeptidases (33). We have also shown that FheCL1 can degrade cystatins/serpins such as SCCA1 and SCCA2 (34) and therefore this may protect the enzyme from inhibition by cystatins within the lysosome that are known to regulate the activity of resident endolysosomal cathepsins (35).
The cysteine protease used in this study, FheCL1, is secreted by the infective stage of F. hepatica that penetrates the host by traversing the intestinal wall. In correlation with this invasion, we have observed that macrophages become nonresponsive to LPS within 2 days of infection, and Th1 suppression is observed within 7 days, concurrent with the development of a highly Th2-polarized immune response (12, 13 and data not shown). Although the link between protease activity and Th2 inflammation is well documented, the mechanistic basis for this association has not been fully elucidated. The results here indicate that cysteine proteases secreted by helminth parasites prevents cells of the innate immune response promoting a Th1-adaptive response, thus making them more permissive to drive the development of Th2 immune responses. Infection with the helminth S. mansoni illustrates this possibility. The acute stages of schistosomiasis infection leads to the development of a weak Th1 response, but this switches to a potent Th2 response when female worms become fecund and release eggs that get trapped in host liver and intestinal tissues (36). The schistosome cysteine protease, SmB1 (and cathepsin L proteases) is expressed and secreted by the egg stage of the parasite (37) and, therefore, may facilitate this immune switching.
TLRs play a prominent role in the response to endogenous signals provided by the host during injury events. Factors released from damaged cells provide the innate immune system with activation ligands for TLRs. In particular, TLR3, by responding to endogenous RNA from necrotic cells, amplifies the inflammatory response during septic peritonitis and ischemic bowel injury (38). A recent study using TLR3 gene-deficient mice found that Th1-associated factors, such as IL-12, were decreased during infection with S. mansoni, suggesting that the absence of signaling through TLR3 prevented the generation of Th1 cytokines and chemokines. Moreover, the absence of TLR3 correlated with significantly increased Th2 cytokines and chemokines in response to infection (39). Much of the pathology encountered in helminth infection is immune-mediated, and studies demonstrate that increased rates of mortality during acute fasciolosis (40) and schistosomiasis are associated with elevated levels of Th1 cytokines and chemokines (41). Immune-mediated mortality associated with helminth infection can be attributed to the dissemination of bacterial gut flora into the liver with ensuing induction of septic shock (36). As both F. hepatica and S. mansoni compromise the integrity of the intestinal epithelial layer, this may allow for the translocation of gut bacteria into the circulation and liver. Although some studies suggest that naïve liver reactive T-cells are tolerized to liver antigen, it is known that engagement of TLR3 and TLR4, but not TLR2, within the liver promotes the secretion of inflammatory cytokines and chemokines that enhance the infiltration of destructive autoreactive CD8+ T-cells (42). Cysteine proteases secreted by helminth parasites could conceivably restrict TLR3/4-mediated activation to control potentially critical inflammatory-mediated pathology.
Infection with helminth parasites is associated with a suppression of Th1-immune responses, which can result in susceptibility to infection with intracellular pathogens such as B. pertussis, mycobacteria, and human immunodeficiency virus, where a Th1 response is required for protection (12, 43). On the other hand, beneficial aspects of helminth infections in reducing the incidence of Th1-driven inflammatory disease such as colitis and gastric atrophy have been recently highlighted (44). Identification of helminth immunodulatory molecules and the means by which they alter immune effector cell function is, therefore, of both fundamental and practical importance. Only one previous study has demonstrated the capacity for a parasite-secreted molecule to selectively inhibit TRIF-dependent signaling in macrophages. The Sm16 protein is secreted from the S. mansoni cercariae and eggs and has been shown to inhibit signaling from both TLR3 and TLR4 but not TLR2 (45). Because inhibited degradation of the IRAK1 protein in LPS-stimulated macrophages was also observed, the authors concluded that Sm16 exerts its inhibition at a point proximal to the TLR complex, although no mechanism was determined. The identity and activity of Sm16 remains uncharacterized.
This present investigation presents a molecular mechanism for the anti-inflammatory properties of helminth parasites via the degradation of TLR3 and inhibition of TRIF-dependent activation of macrophages. Because proteases are known to be secreted by most helminth parasites (7–9), our study offers an explanation by which these pathogens share a common means of modulating innate immune responses. Moreover, our data may facilitate a novel therapeutic approach for the treatment of Th1 inflammatory disorders particularly in light of the current strategy of developing TLR antagonists as immunotherapy for sepsis and autoimmune disease (reviewed in Ref. 46).
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- TLR
- Toll-like receptor
- LPS
- lipopolysaccharide
- IL
- interleukin
- PBS
- phosphate-buffered saline
- IFN
- interferon
- ELISA
- enzyme-linked immunosorbent assay
- RT
- reverse transcription
- TRAF3
- tumor necrosis factor receptor-associated factor 3
- iNOS
- inducible nitric oxide synthesis
- IRF
- interferon regulatory factor
- TRIF
- TLR domain-containing adapter-inducing interferon-β.
REFERENCES
- 1.Balic A., Harcus Y. M., Taylor M. D., Brombacher F., Maizels R. M. (2006) Int. Immunol. 18, 1421–1431 [DOI] [PubMed] [Google Scholar]
- 2.Carvalho L., Sun J., Kane C., Marshall F., Krawczyk C., Pearce E. J. (2009) Immunol. 126, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siracusa M. C., Reece J. J., Urban J. F., Jr., Scott A. L. (2008) J. Leukoc. Biol. 84, 1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuroda E., Yoshida Y., En Shan B., Yamashita U. (2001) Parasite Immunol. 23, 305–311 [DOI] [PubMed] [Google Scholar]
- 5.Goodridge H. S., Marshall F. A., Else K. J., Houston K. M., Egan C., Al-Riyami L., Liew F. Y., Harnett W., Harnett M. M. (2005) J. Immunol. 174, 284–293 [DOI] [PubMed] [Google Scholar]
- 6.van Riet E., Hartgers F. C., Yazdanbakhsh M. (2007) Immunobiology 212, 475–490 [DOI] [PubMed] [Google Scholar]
- 7.Tort J., Brindley P. J., Knox D., Wolfe K. H., Dalton J. P. (1999) Adv. Parasitol. 43, 161–266 [DOI] [PubMed] [Google Scholar]
- 8.Sajid M., McKerrow J. H. (2002) Mol Biochem. Parasitol. 120, 1–21 [DOI] [PubMed] [Google Scholar]
- 9.Robinson M. W., Dalton J. P., Donnelly S. (2008) Trends Biochem. Sci. 33, 601–608 [DOI] [PubMed] [Google Scholar]
- 10.Berasain P., Carmona C., Frangione B., Dalton J. P., Goñi F. (2000) Exp. Parasitol. 94, 99–110 [DOI] [PubMed] [Google Scholar]
- 11.Smith A. M., Dowd A. J., Heffernan M., Robertson C. D., Dalton J. P. (1993) Int. J. Parasitol. 23, 977–983 [DOI] [PubMed] [Google Scholar]
- 12.Brady M. T., O'Neill S. M., Dalton J. P., Mills K. H. (1999) Infect. Immun. 67, 5372–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Neill S. M., Brady M. T., Callanan J. J., Mulcahy G., Joyce P., Mills K. H., Dalton J. P. (2000) Parasite Immunol. 22, 147–155 [DOI] [PubMed] [Google Scholar]
- 14.O'Neill S. M., Mills K. H., Dalton J. P. (2001) Parasite Immunol. 23, 541–547 [DOI] [PubMed] [Google Scholar]
- 15.Collins P. R., Stack C. M., O'Neill S. M., Doyle S., Ryan T., Brennan G. P., Mousley A., Stewart M., Maule A. G., Dalton J. P., Donnelly S. (2004) J. Biol. Chem. 279, 17038–17046 [DOI] [PubMed] [Google Scholar]
- 16.Stack C. M., Dalton J. P., Cunneen M., Donnelly S. (2005) J. Immunol. Methods 304, 151–157 [DOI] [PubMed] [Google Scholar]
- 17.Stack C. M., Caffrey C. R., Donnelly S. M., Seshaadri A., Lowther J., Tort J. F., Collins P. R., Robinson M. W., Xu W., McKerrow J. H., Craik C. S., Geiger S. R., Marion R., Brinen L. S., Dalton J. P. (2008) J. Biol. Chem. 283, 9896–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J. R., Wu C. C., Hou R. C., Jeng K. C. (2008) Immunol. Invest. 37, 263–277 [DOI] [PubMed] [Google Scholar]
- 19.Sugawara S., Nemoto E., Tada H., Miyake K., Imamura T., Takada H. (2000) J. Immunol. 165, 411–418 [DOI] [PubMed] [Google Scholar]
- 20.Perera P. Y., Mayadas T. N., Takeuchi O., Akira S., Zaks-Zilberman M., Goyert S. M., Vogel S. N. (2001) J. Immunol. 166, 574–581 [DOI] [PubMed] [Google Scholar]
- 21.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 22.Kawai T., Takeuchi O., Fujita T., Inoue J., Mühlradt P. F., Sato S., Hoshino K., Akira S. (2001) J. Immunol. 167, 5887–5894 [DOI] [PubMed] [Google Scholar]
- 23.Thomas K. E., Galligan C. L., Newman R. D., Fish E. N., Vogel S. N. (2006) J. Biol. Chem. 281, 31119–31130 [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 25.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. (2000) Cell 103, 667–678 [DOI] [PubMed] [Google Scholar]
- 26.Andersen J., VanScoy S., Cheng T. F., Gomez D., Reich N. C. (2008) Genes Immun. 9, 168–175 [DOI] [PubMed] [Google Scholar]
- 27.Li K., Foy E., Ferreon J. C., Nakamura M., Ferreon A. C., Ikeda M., Ray S. C., Gale M., Jr., Lemon S. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Z. H., Lee S. E., Kwack K., Yeo W., Lee T. H., Bae S. S., Suh P. G., Kim H. H. (2001) J. Leukoc. Biol. 69, 490–496 [PubMed] [Google Scholar]
- 29.Semnani R. T., Law M., Kubofcik J., Nutman T. B. (2004) J. Immunol. 172, 6229–6238 [DOI] [PubMed] [Google Scholar]
- 30.van Liempt E., van Vliet S. J., Engering A., García Vallejo J. J., Bank C. M., Sanchez-Hernandez M., van Kooyk Y., van Die I. (2007) Mol. Immunol. 44, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 31.Jiang W., Sun R., Wei H., Tian Z. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17077–17082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y. M., Ploegh H. L. (2008) Nat. Immunol. 9, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stack C. M., Donnelly S., Lowther J., Xu W., Collins P. R., Brinen L. S., Dalton J. P. (2007) J. Biol. Chem. 282, 16532–16543 [DOI] [PubMed] [Google Scholar]
- 34.Kanaji S., Tanaka Y., Sakata Y., Takeshita K., Arima K., Ohta S., Hansell E. J., Caffrey C., Mottram J. C., Lowther J., Donnelly S., Stack C., Kadowaki T., Yamamoto K., McKerrow J. H., Dalton J. P., Coombs G. H., Izuhara K. (2007) FEBS Lett. 581, 4260–4264 [DOI] [PubMed] [Google Scholar]
- 35.Kopitar-Jerala N. (2006) FEBS Lett. 57, 6295–6301 [DOI] [PubMed] [Google Scholar]
- 36.Pearce E. J., MacDonald A. S. (2002) Nat. Rev. Immunol. 2, 499–511 [DOI] [PubMed] [Google Scholar]
- 37.Sung C. K., Dresden M. H. (1986) J. Parasitol. 72, 891–900 [PubMed] [Google Scholar]
- 38.Cavassani K. A., Ishii M., Wen H., Schaller M. A., Lincoln P. M., Lukacs N. W., Hogaboam C. M., Kunkel S. L. (2008) J. Exp. Med. 205, 2609–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshi A. D., Schaller M. A., Lukacs N. W., Kunkel S. L., Hogaboam C. M. (2008) Eur. J. Immunol. 38, 3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meeusen E., Lee C. S., Rickard M. D., Brandon M. R. (1995) Parasite Immunol. 17, 37–45 [DOI] [PubMed] [Google Scholar]
- 41.Stadecker M. J., Asahi H., Finger E., Hernandez H. J., Rutitzky L. I., Sun J. (2004) Immunol. Rev. 201, 168–179 [DOI] [PubMed] [Google Scholar]
- 42.Lang K. S., Georgiev P., Recher M., Navarini A. A., Bergthaler A., Heikenwalder M., Harris N. L., Junt T., Odermatt B., Clavien P. A., Pircher H., Akira S., Hengartner H., Zinkernagel R. M. (2006) J. Clin. Invest. 116, 2456–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elias D., Britton S., Kassu A., Akuffo H. (2007) Expert Rev. Anti. Infect. Ther. 5, 475–484 [DOI] [PubMed] [Google Scholar]
- 44.Elliott D. E., Summers R. W., Weinstock J. V. (2007) Int. J. Parasitol. 37, 457–464 [DOI] [PubMed] [Google Scholar]
- 45.Brännström K., Sellin M. E., Holmfeldt P., Brattsand M., Gullberg M. (2009) Infect. Immun. 77, 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tse K., Horner A. A. (2007) Ann. Rheum. Dis. 66, iii77–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.