Abstract
There is strong evidence for a genetic contribution to epilepsy, but it is commonly assumed that this genetic contribution is limited to ‘generalized’ epilepsies, and that most forms of ‘partial’ epilepsy are nongenetic. In a linkage analysis of a single family containing 11 affected individuals, we obtained strong evidence for localization of a gene for partial epilepsy. This susceptibility gene maps to chromosome 10q, with a maximum two-point lod score for D10S192 of 3.99 at θ=0.0. All affected individuals share a single haplotype for seven tightly linked contiguous markers; the maximum lod score for this haplotype is 4.83 at θ=0.0. Key recombinants place the susceptibility locus within a 10 centimorgan interval.
Epilepsy is a condition in which seizures tend to recur in the absence of acute precipitating factors1. It is one of the most common neurologic disorders, affecting approximately 4% of individuals at some time in their lives2. There is strong evidence for a genetic contribution but until recently, little progress has been made in identifying specific loci which affect susceptibility. This slow progress is due in part to inherent complexity in the genetic contributions. In most forms of epilepsy, the distribution of affected individuals within families does not conform to a simple mendelian model, suggesting that environmental factors or modifying genes may be required for disease expression in susceptible individuals. There are likely to be both genetic and nongenetic influences on susceptibility, and the important genetic influences are likely to differ across families or clinically defined subgroups.
In three human epilepsy syndromes, evidence exists for linkage of susceptibility loci to specific chromosomal regions. A locus for benign familial neonatal convulsions (BFNC) was originally found on chromosome 20q (ref. 3), and another locus has been identified on chromosome 8q (ref. 4). The neuronal nicotinic acetylcholine receptor α4 subunit (CHRNA4) has been suggested as a candidate for the chromosome 20 mutation in BFNC (ref. 5). One form of progressive myoclonus epilepsy (Unverricht-Lundborg type) was localized to chromosome 21 q (ref. 6). Unlike the other two disorders with linkage evidence, the third disorder, juvenile myoclonic epilepsy (JME), has an uncertain mode of inheritance, with reduced penetrance and a range of phenotypic expressions within families. In families of probands with JME, Greenberg et al. found evidence for linkage of epilepsy to the HLA region of chromosome 6 (ref. 7). Two subsequent studies confirmed the linkage8,9, but another found evidence against it10.
The syndromes with linkage evidence account for only a small proportion of all epilepsy. Little is known about genetic mechanisms underlying familial aggregation in the remainder. All three of the syndromes with linkage evidence are generalized epilepsies, in which seizures are presumed to involve the entire brain from the outset11,12. Most localization-related (partial or focal) epilepsies12, in which seizures begin in a specific brain region, are assumed to be nongenetic. However, relatives of probands with partial epilepsy have increased risk of epilepsy, compared with the general population13,14, suggesting a genetic influence on at least some partial epilepsies. In two rare forms of partial epilepsy, benign rolandic epilepsy with centrotemporal spikes15 and a form of frontal epilepsy16, an autosomal dominant mode of inheritance has been suggested. However, linkage evidence to support the existence of a susceptibility gene has not been obtained for either syndrome.
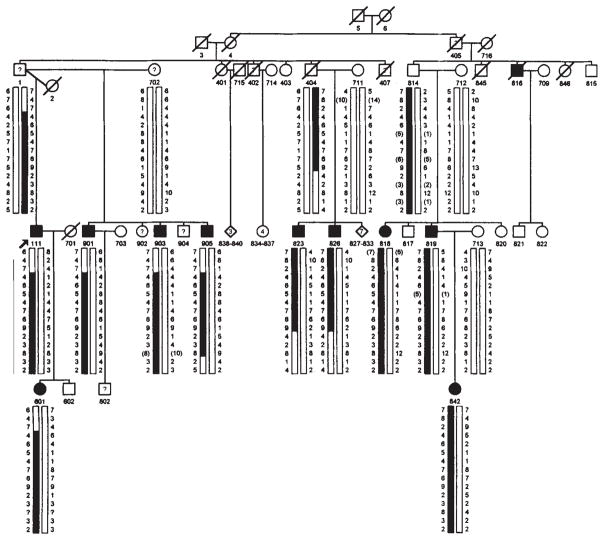
We have identified a family in which individuals in three successive generations are affected with partial epilepsy, in a pattern strongly suggestive of autosomal dominant inheritance with reduced penetrance (Fig. 1). Linkage analysis of this family provides evidence for an epilepsy susceptibility gene located within a 10 centimorgan (cM) interval on chromosome 10.
Fig. 1.
Pedigree of family. Filled symbols denote individuals with idiopathic/cryptogenic epilepsy. Symbols containing ‘?’ denote those with symptomatic epilepsy, acute symptomatic seizures, or possible but unconfirmed epilepsy, all of whom were classified as unknown in the analysis. Individuals currently younger than age 20 who have never had seizures are omitted from the pedigree. Marker genotypes are shown in the same order as in Table 2. Genotypes are shown only for individuals with idiopathic/cryptogenic epilepsy and their parents. Alleles shown in parentheses are inferred from the genotypes of other relatives, including some whose genotypes are omitted from the figure.
Description of the family
In an ongoing study of genetic contributions to epilepsy, we collected information on seizure disorders in the families of 1,957 adult probands with epilepsy ascertained from voluntary organizations17. We collected data systematically on the parents, siblings, half-siblings and offspring of each proband. Only 42 (2%) families contained ≥3 first-degree relatives with epilepsy (including the proband), and very few of these appeared to be segregating a highly penetrant, autosomal dominant susceptibility allele. In one family, however, the proband reported that his daughter had epilepsy, and that his father and all five of his paternal half-siblings had had seizures. We extended the pedigree to include all branches containing individuals reported to have had seizures (Fig. 1). Each family member was classified by seizure disorder (epilepsy, isolated unprovoked seizure, febrile convulsion or other acute symptomatic seizure)1, and those with epilepsy were classified according to seizure type11, epilepsy syndrome12, age at onset and presumed aetiology, according to established criteria1.
Seventeen individuals in the family have had seizures, of whom 14 had epilepsy and the remaining three had acute symptomatic seizures (Table 1). Three of those with epilepsy have histories of central nervous system (CNS) disease or insults presumed to be causal (brain tumour, cerebral palsy, head trauma) and were therefore classified as having remote symptomatic epilepsy. In the remaining 11 cases, epilepsy was classified as idiopathic/cryptogenic1. Epilepsy was clearly localization-related in all but one of these cases; the remaining person has had only nocturnal seizures and thus could not be classified. Six (55%) of those with idiopathic/cryptogenic epilepsy and one with remote symptomatic epilepsy reported nonspecific auditory disturbances as a simple partial component of their seizures (such as a ringing noise that grew louder, or humming like a machine) (Table l). None of the interictal EEGs showed an epileptiform abnormality. The age at onset of idiopathic/cryptogenic epilepsy ranged from eight to 19 years. All family members had normal intelligence, with the exception of one individual (#407) who was reported to have had epilepsy associated with cerebral palsy, and to have been severely retarded. In family members with epilepsy, seizures occurred infrequently. Seven (64%) of the 11 individuals with idiopathic/cryptogenic epilepsy had been seizure-free for ≥3 years prior to interview. Ten of the 11 subjects with idiopathic/cryptogenic epilepsy received phenytoin as a primary anti-seizure medication; the remaining subject received carbamazapine.
Table 1.
Clinical diagnoses and information used for diagnosis in subjects with seizures
| Sources of information | |||||
|---|---|---|---|---|---|
| ID | Interviews | EEG | Neurological examination | Age at onset | Seizure classificationsa (aetiology) |
| Idiopathic/cryptogenic epilepsy | |||||
| 111 | Self, father | Yes | Yes | 8 | SP, SGTC, auditory features |
| 601 | Self, father | Yes | Yes | 12 | SP (possible), CP, SGTC, auditory features |
| 816 | Brother, son, daughter, spouseb | No | No | 16 | CP, SGTC |
| 818 | Self, mother, father | No | No | 12 | SGTC |
| 819 | Self, mother, father | No | Yes | 13 | CP, SGTC, auditory features |
| 823 | Self, mother, father | Yes | Yes | 19 | Nocturnal GTC |
| 826 | Self, mother, father | Yes | No | 12 | CP, SGTC |
| 842 | Self, mother | Yes | Yes | 8 | CP, SGTC, auditory features |
| 901 | Self, mother | Yes | Yes | 10 | P, SGTC |
| 903 | Self, mother | Yes | Yes | 12 | CP, SGTC, auditory features |
| 905 | Self, mother | Yes | Yes | 17 | P, SGTC, auditory features |
| Symptomatic epilepsy | |||||
| 402 | Spouse, brother, sister-in-lawb | No | No | 56 | P, SGTC (neoplasm) |
| 407 | Brother, sister-in-lawb | No | No | 1 | Unknown seizure type (cerebral palsy) |
| 902 | Self, mother | Yes | Yes | 25 | SP, SGTC, auditory features (head injury) |
| Acute symptomatic seizures | |||||
| 001 | Self | Yes | Yes | 55 | (Alcohol-related seizures) |
| 702 | Self | No | No | 2 | (Febrile convulsion) |
| 904 | Self, mother | No | No | 1 | (Febrile convulsions) (2) |
| Epilepsy possible but uncertain | |||||
| 405 | Two sonsb | No | No | ? | unknown |
| 802 | Mother | No | No | 4 | SP |
Seizure types: SP, simple partial; CP, complex partial; SGTC, secondarily generalized tonic-clonic; GTC, generalized tonic-clonic; P, partial, unknown whether simple or complex.
Deceased subject.
Linkage Analysis
We carried out linkage analysis using the ‘fast’ modification of the LINKAGE computer package18–20. All of the assumptions regarding phenotype definition and genetic parameters (mode of inheritance, and penetrance and frequency of the susceptibility allele) were made a priori, without any information about the genetic marker phenotypes. Only those with idiopathic/cryptogenic epilepsy were classified as affected; those with acute symptomatic seizures or remote symptomatic epilepsy were classified as unknown. Two additional individuals were classified as unknown because although epilepsy was suspected, the available information was insufficient for definitive diagnosis. We assumed an autosomal dominant model with a frequency of 0.001 for the susceptibility allele, and a risk of 0.01 in non-gene carriers. These assumptions were based on a preliminary segregation analysis20. Lifetime cumulative incidence of idiopathic/cryptogenic epilepsy in gene carriers in this family was estimated to be 71%. All but one of those affected with idiopathic/cryptogenic epilepsy were ≥20 years old at observation (current age or age at death), and none of those affected had onset after age 20. Based on these observations, we assumed a uniform penetrance of 71% in gene carriers ≥20 years, and classified those younger than age 20 who were currently unaffected as unknown.
In preliminary analyses, we found one branch of the family (individuals 402 and 714 and their offspring) in which the marker alleles did not segregate consistently with known familial relationships. This nuclear family was excluded from further analysis. (It would not have contributed information to the linkage analysis in any case, because the father had epilepsy associated with a neoplasm and was classified as unknown, and none of the offspring was affected.)
After analysing the data for 110 markers, we obtained preliminary evidence for linkage with D10S222 (two-point lod score 2.69 at θ=0.00). In addition, one of two flanking markers, D10S201, also suggested linkage with a maximum two-point lod score of 2.79 (θ=0.06). To explore the region near Dl0S222 further, we selected 12 additional tightly linked markers from the 1993–94 Généthon map22. We determined genotypes and inferred haplotypes for these additional markers in the family (Fig. 1).
Table 2 shows the two-point lod scores for a series of 15 tightly linked microsatellite markers on chromosome 10. The results show a maximum lod score of 3.99 for D10S192. All ten living affected individuals share a single haplotype for the seven contiguous markers spanning 10 cM from D10S200 to D10S205. Among ten genotyped individuals over age 20 who were classified as unaffected and at 50% risk of being gene carriers, only one carries the haplotype. The two-point lod score for the seven-locus haplotype is 4.83. This figure is a close approximation to the maximum multipoint LOD score for these seven loci. The lod score for the haplotype dropped only slightly (from 4.83 to 4.46) when 50% penetrance was assumed, indicating that it was not very sensitive to the penetrance assumption. We also note that three obligate carriers in the older generations were classified as unaffected (individuals 4,404 and 814) and two were classified as unknown (individuals 1 and 405). This illustrates the difficulty in obtaining valid information on the epilepsy histories of older relatives, as we have noted previously23.
Table 2.
Maximum two-point lod scores (Zmax) and maximum likelihood recombination fractions (θs) for chromosome 10q markers
| Marker locus | Ha | Map locationb | Zmax | θ |
|---|---|---|---|---|
| D10S201 | 0.82 | 106 | 2.79 | 0.06 |
| D10S583 | 0.87 | 120 | 1.94 | 0.07 |
| D10S185 | 0.82 | 121 | 3.06 | 0.06 |
| D10S200 | 0.65 | 121 | 0.97 | 0.00 |
| D10S574 | 0.75 | 122 | 3.77 | 0.00 |
| D10S198 | 0.70 | 127 | 2.46 | 0.00 |
| D10S603 | 0.68 | 128 | 2.35 | 0.00 |
| D10S192 | 0.83 | 129 | 3.99 | 0.00 |
| D10S222 | 0.68 | 131 | 2.69 | 0.00 |
| D10S205 | 0.89 | 131 | 3.01 | 0.01 |
| D10S566 | 0.74 | 131 | 1.34 | 0.08 |
| D10S540 | 0.74 | 132 | 1.76 | 0.08 |
| D10S530 | 0.62 | 132 | 1.46 | 0.00 |
| D10S597 | 0.68 | 136 | 0.68 | 0.14 |
| D10S554 | 0.14 | 136 | 0.75 | 0.18 |
| Haplotypec | 4.83 | 0.00 |
Heterozygosity=1-Σρi2, where pi=frequency of each marker allele calculated from the family data using ILINK18–20.
Map location in map units relative to p terminus, adapted from Généthon map22.
Presumed disease allele-bearing haplotype at contiguous loci D10S200 through D10S205 (Fig. 1) coded as allele ‘2’, all others coded as allele ‘1’, For linkage analysis, the frequency of allele ‘2’ was assumed to be 0.001.
Inspection of the haplotypes in affected individuals reveals meiotic recombination events that define the minimal region that must contain the susceptibility locus (Fig. 1). The proximal recombination site is between D10S185 and D10S200; the distal site is between D10S205 and D10S566. These recombination events place the epilepsy susceptibility locus within the 10 cM interval between markers D10S185 and D10S566.
Discussion
Our evidence for linkage provides the first demonstration of a major genetic effect on susceptibility to idiopathic/cryptogenic localization-related epilepsy. In the current International League Against Epilepsy (ILAE) classification of epilepsy syndromes12, the term ‘idiopathic’ is reserved for syndromes of presumed genetic origin, and the term ‘cryptogenic’ for syndromes presumed to be nongenetic but with insufficient evidence to assign a specific aetiology. This distinction is not meaningful in the absence of evidence of a clear genetic basis for those defined as ‘idiopathic’ and a clear nongenetic basis for those defined as ‘cryptogenic’. For most of the syndromes currently classified as ‘idiopathic’, clear evidence of a genetic basis, either from linkage studies or demonstration of a specific mode of inheritance, is lacking. In contrast, affected individuals in the family we studied have a form of epilepsy that would be classified as ‘cryptogenic’ according to the ILAE system12, yet our linkage data provide strong evidence of a genetic susceptibility.
Because of the complexity in the genetic contributions to the epilepsies, clinical syndromes cannot be divided into two broad classes based on a genetic susceptibility. Even in the presence of strong evidence of a genetic contribution to a given syndrome, the susceptibility genotype may be present in only a proportion of those affected, while others have a different genetic mechanism or a nongenetic cause. On the other hand, phenotypic expression may be influenced by other genes or environmental exposures, and thus some of those with a given genotype may manifest a different seizure disorder or epilepsy syndrome. Thus, we believe it more appropriate to use one term (either ‘idiopathic’ or ‘cryptogenic’) to describe cases in whom evidence to establish aetiology is lacking, and to address the question of genetic susceptibility separately.
As affected individuals in this family did not have a recognized ILAE syndrome, we decided a priori to define as affected, for purposes of genetic analysis, anyone in the family who had epilepsy in the absence of a known or suspected exogenous cause. The 11 individuals in the family who met these criteria had similar seizure types and a narrow range of age at onset of epilepsy. This consistency in clinical features is unlikely to result from the effects of modifying genes or shared environmental exposures because the clinical similarity is as great in affected family members who are distantly related and geographically dispersed as in those who are more closely related. Thus the mutation in this family probably influences both susceptibility to epilepsy and its specific clinical features. Although we did not have sufficient data (neuroimaging or depth electrode studies) to localize precisely the epileptogenic abnormality in affected subjects, the auditory features observed in 55% of those affected suggest that the effect of the mutation is localized to a narrowly delimited functional brain region (such as the neocortical temporal lobe).
The human genome database (Welch Library, Johns Hopkins University) lists more than 50 genes whose localization overlaps with the cytological localization (10q22–q24) of the epilepsy susceptibility gene in this family. This list is probably a small fraction of the genes residing in this region, and obviously might not include the gene involved in epilepsy susceptibility in this family. Several previously identified genes are of potential interest, however. There are two neurotransmitter receptors (β-1 and α-2A adrenergic) and several coding sequences with homology to other known receptors. The region contains several genes that are involved in the metabolism of glutamate that could affect the availability of substrates for this important excitatory neurotransmitter. Other genes such as calcium/calmodulin-dependent protein kinase γ may affect epilepsy susceptibility by modulating key metabolic and regulatory pathways. Further evaluation of the relevance of these candidate genes will require more precise localization of both the epilepsy locus on chromosome l0q and the genes themselves.
Because our analysis was restricted to a single family, we cannot estimate the proportion of familial epilepsy that can be attributed to alleles at this locus, which will require the study of additional families. However, even if the mutation found in our family is rare, identification of its product and pathophysiologic effect may help to elucidate basic epileptogenic mechanisms. Study of additional families will also be important for investigating the effects of allelic and locus heterogeneity, and the range of phenotypic manifestations of the susceptibility gene. Linkage to chromosome 10 may be restricted to families with partial epilepsy with auditory features, like that in the family presented here. Even within this type of epilepsy, locus heterogeneity is possible, and some families may not show linkage despite having similar clinical features. On the other hand, alternative alleles at the same locus may raise risk for different types of epilepsy. If this is true, then linkage to chromosome 10 may not be restricted to families with partial epilepsy with auditory features. We are currently investigating these possibilities through linkage analysis in other families.
Methodology
Clinical data collection
Seizure disorders were classified according to the criteria of Hauser et al. Epilepsy was defined as a lifetime history of recurrent (≥2) seizures that were not precipitated by acute structural or metabolic insults to the CNS. Subjects with epilepsy who had a history of an insult to the CNS before, and occurring ≥7 days prior to, the first unprovoked seizure were classified as remote symptomatic, those with no identified cause were classified as idiopathic/cryptogenic. Seizures precipitated by acute alterations in homeostasis (such as high fever, metabolic disturbance) or acute insults to the CNS were excluded from the definition of epilepsy and classified as acute symptomatic1.
The data used for classification included semistructured interviews to screen for a history of seizures, diagnostic interviews and neurological examinations in those who screened positive for afebrile seizures, electroencephalograms (EEGs) in those known or suspected to have epilepsy, and reviews of medical records. We administered semistructured interviews personally to each family member over age 12, to obtain information about vital status and age of all first-degree relatives, and to screen for seizure occurrence in the subject and his or her children. Those who screened positive for afebrile seizures were interviewed in their homes by a field neurologist (K.S.L. or K.J.N.), using a semistructured diagnostic interview. For deceased relatives reported to have had seizures, the diagnostic interview was administered by the neurologist to the family informant who had the most complete information about the relative’s medical history. Living relatives who had had afebrile seizures were also given neurological examinations by the field neurologists, to detect evidence of localized or lateralized neurological dysfunction suggestive of localizaton-related epilepsy. Those known or suspected to have epilepsy were given EEGs at collaborating hospitals according to a standardized protocol (that is, two-hour tracings during wake and sleep, using both referential and bipolar montages, including photic stimulation and hyperventilation as activating procedures unless contraindicated). The EEG tracings were read independently by two expert neurologists (W.A.H. and T.A.P.). All data were collected with the full informed consent of the study subjects. Diagnoses were made at consensus meetings attended by expert neurologists (W.A.H., T.A.P., M.L.S., K.J.N., K.S.L.), where all data collected on each subject were reviewed. The reviewers were blinded to family membership by deleting identifying information, and reviewing subjects from different families in random order. All of the diagnoses were completed without knowledge of the genetic marker information.
Genotype determination
Blood specimens were collected in acid citrate dextrose tubes, delivered to the laboratory and processed within three days of collection. For the initial genomic screen, we selected primers for 225 microsatellite markers spaced at 10–20 cM intervals22–24. Genotypes for these markers were determined from purified blood DNA, based on the electrophoretic mobility of the PCR amplification product made using a 32P-end-labelled primer. Genotypes were determined from autoradiograms with the assistance of a computerized digital film analyser (Bioimage) and recorded into a database automatically. Genotypes were determined blind to diagnosis, but with the pedigree structure available.
Acknowledgments
We are grateful to D. King, R. Glazer, W. Jimenez, M. Vyazmensky, M. Bernstein, E. Pavlou, M. De Vera, and J. Mabutas for assistance with data collection, and to R. Mayeux, Z. Stein and N. Schupf for critical comments on the manuscript. This study was supported by NIH2RO1 -NS20656 (R.O.), and a grant from the Klingenstein Foundation (K.C.W.).
References
- 1.Hauser WA, Annegers JF, Kurland LT. Prevalence of epilepsy in Rochester, Minnesota, 1940–1980. Epilepsia. 1991;31:429–445. doi: 10.1111/j.1528-1157.1991.tb04675.x. [DOI] [PubMed] [Google Scholar]
- 2.Hauser WA, Annegers JF, Kurland LT. Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935–1984. Epilepsia. 1993;34:453–468. doi: 10.1111/j.1528-1157.1993.tb02586.x. [DOI] [PubMed] [Google Scholar]
- 3.Leppert M, et al. Benign familial neonatal convulsions linked to genetic markers on chromosome 20. Nature. 1989;337:647–648. doi: 10.1038/337647a0. [DOI] [PubMed] [Google Scholar]
- 4.Lewis TB, Leach RJ, Ward K, O’Connell P, Ryan SG. Genetic heterogeneity in benign familial neonatal convulsions: identification of a new locus on chromosome 8q. Am J hum Genet. 1993;53:670–675. [PMC free article] [PubMed] [Google Scholar]
- 5.Steinlein O, et al. Refinement of the localization of the gene for neuronal nicotinic acetylcholine receptor a4 subunit (CHRNA4) to human chromosome 20q13.2–q13.3. Genomics. 1994;22:493–495. doi: 10.1006/geno.1994.1420. [DOI] [PubMed] [Google Scholar]
- 6.Lehesjoki A-E, et al. Localization of a gene for progressive myoclonus epilepsy to chromosome 21 q22. Proc natn Acad Sci USA. 1991;88:3696–3699. doi: 10.1073/pnas.88.9.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg KA, et al. Juvenile myoclonic epilepsy may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet. 1988;31:185–192. doi: 10.1002/ajmg.1320310125. [DOI] [PubMed] [Google Scholar]
- 8.Weissbecker KA, Durner M, Janz D, Scaramelli A, Sparkes RS, Spence MA. Confirmation of linkage between juvenile myoclonic epilepsy locus and the HLA region of chromosome 6. Am J Med Genet. 1991;38:32–86. doi: 10.1002/ajmg.1320380109. [DOI] [PubMed] [Google Scholar]
- 9.Durner M, Sander T, Greenberg DA, Johnson K, Beck-Mannagetta G, Janz D. Localization of idiopathic generalized epilepsy on chromosome 6p in families ascertained through juvenile myoclonic epilepsy patients. Neurology. 1991;41:1651–1655. doi: 10.1212/wnl.41.10.1651. [DOI] [PubMed] [Google Scholar]
- 10.Whitehouse WP, et al. Linkage analysis of idiopathic generalized epilepsy (IGE) and marker loci on chromosome 6p in families of patients with juvenile myoclonic epilepsy: no evidence for an epilepsy locus in the HLA region. Am J hum Genet. 1993;53:652–662. [PMC free article] [PubMed] [Google Scholar]
- 11.Commission on Classification and Terminololgy of the International League Against Epilepsy. Proposal for revised clinical and electroencephalographic classification of epileptic seizures. Epilepsia. 1981;22:489–501. doi: 10.1111/j.1528-1157.1981.tb06159.x. [DOI] [PubMed] [Google Scholar]
- 12.Commission on Classification and Terminology of the International League Against Epilepsy. A revised proposal for the classification of epilepsy and epileptic syndromes. Epilepsia. 1989;30:268–278. [PubMed] [Google Scholar]
- 13.Ottman R. Genetics of the partial epilepsies: a review. Epilepsia. 1989;30:107–111. doi: 10.1111/j.1528-1157.1989.tb05290.x. [DOI] [PubMed] [Google Scholar]
- 14.Ottman R, Annegers JF, Hauser WA, Kurland LT. Seizure risk in offspring of parents with generalized vs. partial epilepsy. Epilepsia. 1989;30:157–161. doi: 10.1111/j.1528-1157.1989.tb05448.x. [DOI] [PubMed] [Google Scholar]
- 15.Heijbel J, Blom S, Rasmuson M. Benign epilepsy of childhood with centrotemporal EEG foci: a genetic study. Epilepsia. 1975;16:285–93. doi: 10.1111/j.1528-1157.1975.tb06059.x. [DOI] [PubMed] [Google Scholar]
- 16.Scheffer IE, et al. Autosomal dominant frontal epilepsy misdiagnosed as sleep disorder. Lancet. 1994;343:515–517. doi: 10.1016/s0140-6736(94)91463-x. [DOI] [PubMed] [Google Scholar]
- 17.Ottman R, Susser M. Data collection strategies in genetic epidemiology: the Epilepsy Family Study of Columbia University. J clin Epidemiol. 1992;45:721–727. doi: 10.1016/0895-4356(92)90049-s. [DOI] [PubMed] [Google Scholar]
- 18.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 19.Lathrop GM, Lalouel JM. Efficient computations in multilocus linkage analysis. Am J hum Genet. 1988;42:498–505. [PMC free article] [PubMed] [Google Scholar]
- 20.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 21.Ottman R, Sherman S. Genetic analysis of epilepsy in families ascertained from voluntary organizations [abstract] Epilepsia. 1990;31:611. [Google Scholar]
- 22.Gyapay G, et al. The 1993–94 Généthon human genetic linkage map. Nature Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- 23.Ottman R, Lee JH, Hauser WA, Risch N. Birth cohort and familial risk of epilepsy: the effect of diminished recall in studies of lifetime prevalence. Am J Epidemiol. doi: 10.1093/oxfordjournals.aje.a117425. (in the press) [DOI] [PubMed] [Google Scholar]
- 24.Weber JL, May PE. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 25.Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucl Acids Res. 1989;17:6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]