Abstract
With the recent recognition of non-coding RNAs (ncRNAs) flanking many genes1-5, a central issue is to fully understand their potential roles in regulated gene transcription programs, possibly through different mechanisms6-12. Here, we report that an RNA-binding protein, TLS, serves as a key transcriptional regulatory sensor of DNA damage signals that, based on its allosteric modulation by RNA, specifically binds to and inhibits CBP/p300 HAT activities on a repressed gene target, cyclin D1 (CCND1). Recruitment of TLS to the CCND1 promoter to cause gene-specific repression is directed by single stranded, low copy number ncRNA transcripts tethered to the 5′ regulatory regions of CCND1 that are induced in response to DNA damage signals. Our data suggest that signal-induced ncRNAs localized to regulatory regions of transcription units can act cooperatively as selective ligands, recruiting and modulating the activities of distinct classes of RNA binding co-regulators in response to specific signals, providing an unexpected ncRNA/RNA-binding protein-based strategy to integrate transcriptional programs.
Transcriptional coregulators, including coactivators and corepressors, are required for regulating programs of gene expression in a transcription factor- and gene-specific manner13,14. Among them, the histone acetyltransferases (HATs) CBP and p300, play essential roles as coactivators of multiple classes of signal-dependent transcription factors13,14. To search for cellular factors that might regulate CBP HAT activity, we incubated HeLa whole cell extracts with full-length, flag-tagged CBP immobilized on anti-flag IgG affinity beads (Fig. S1a) and observed marked inhibition of CBP HAT activity on histones (Fig. 1a). Subcellular fractionation studies indicated the presence of two classes of inhibitory activities: one that bound to CBP and was primarily present in nuclear extracts (Fig. 1a, lane 3), and the other, the INHAT complex15, present in both nuclear and cytoplasmic extracts (Fig. S2a).
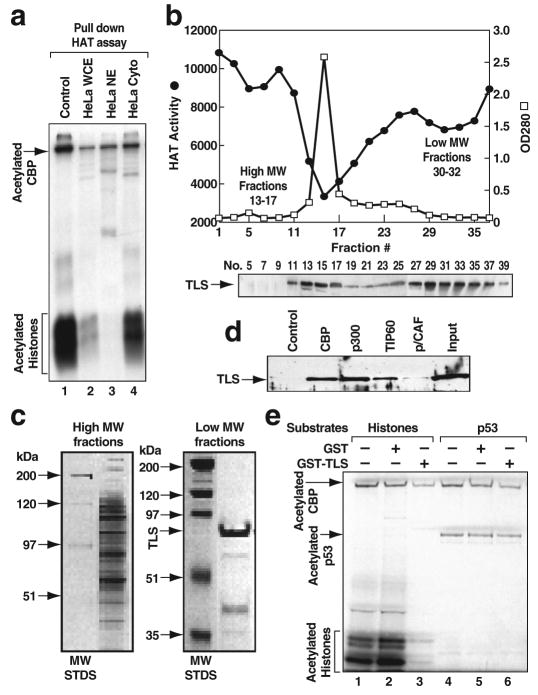
Figure 1. TLS is a specific CBP/p300 HAT inhibitor.
a, CBP HAT activity measured by “pull down” HAT assay. WCE, whole cell extract; NE, nuclear extract; Cyto, cytoplasmic extract. b, Top, CBP HAT inhibitory activity revealed by gel filtration chromatography. MW, molecular weight. Bottom, Profile of TLS detected by Western blotting (WB). c, Representative silver-stained gels of pooled high and low MW fractions. d, TLS interacts with CBP, p300 and TIP60, but not p/CAF. e, The effect of CBP HAT activity by GST-TLS on histones or p53.
The nuclear activity that inhibited CBP in pull-down HAT assays fractionated as two main peaks using gel filtration chromatography (Fig. S1b; Fig. 1b, top). Pooled fractions were further purified using full length, flag-tagged CBP linked to anti-flag IgG beads, based on the observation that inhibitory activity was observed using full-length CBP, but not the isolated HAT domain (Fig. S2b). A large number of proteins were recovered from the high molecular weight (MW) fractions and a major band of approximately 75 kDa in the low MW fractions (Fig. 1c). Using MALDI-re-TOF MS analysis16, this 75 kDa protein was identified in three independent purifications to be TLS (translocated in liposarcoma), an RNA binding protein that has been suggested to play roles in transcription17, RNA processing18 and DNA repair19-22.
These findings were extended by demonstrating that recombinant TLS bound to CBP (Fig. 1d) and strongly inhibited CBP HAT activity on core histones (Fig. 1e, lane 3). GST-TLS partially inhibited acetylation of CBP itself, but not that of p53 (Fig. 1e, lane 6), suggesting that TLS selectively inhibits the ability of the acetylated CBP to transfer acetate to specific substrates. TLS also bound to p300 and TIP60 with similar affinities, but not to p/CAF (Fig. 1d; Fig. S2c). GST-TLS inhibited the HAT activity of p300 (Fig. 2b), but not that of TIP60 (Fig. S2d, e). TLS was also able to inhibit CBP acetylation of histones in nucleosomes prepared from HeLa cell nuclei (Fig. S2f). TLS and its two related proteins EWS and TAFII6823 all proved to be present in high MW fractions that correlate with CBP HAT inhibitory activity (Fig. 1b, bottom; Fig. S3a). Similarly, EWS and TAFII68 were found to bind to CBP and TIP60, but not p/CAF (Fig. S3b, d), and exerted inhibitory effects on CBP/p300 HAT activities (Fig. S3c; data not shown). TLS interacted with several regions of CBP, with the region including the p160-interaction domain24 (1892-2441) serving as the most effective interaction domain (Fig. S4). Pull-down HAT assays showed that recombinant TLS had no effect on the HAT activity of the isolated CBP_HAT region (Fig. S2g), suggesting that the weak interaction of TLS to CBP HAT domain (1099-1877) is not sufficient for HAT inhibitory effects.
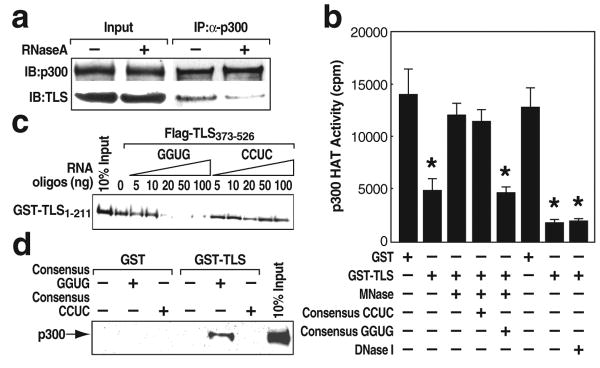
Figure 2. Consensus GGUG-containing RNA oligonucleotide promotes the inhibitory effect of TLS on CBP/p300 HAT activities.
a, Co-immunoprecipitation (IP) of p300 and TLS from RNase A-treated HeLa cells. b, P300 HAT activity was measured using micrococcal nuclease (MNase) or DNase I pre-treated GST and GST-TLS in the presence of GGUG- or CCUC-oligonucleotide. * p<0.02, compared with GST, n=3. c, d, The interaction of TLS N (1-211):C (373-526) termini (c) or GST-TLS:p300 (d) in the presence of GGUG- or CCUC-oligonucleotide. GST and GST-TLS were pre-treated with RNase A. Error bars indicate ± SEM.
We next tested whether the CBP HAT inhibition by TLS was RNA-dependent. A synthetic RNA containing the consensus sequence GGUG (referred to as GGUG-oligonucleotide below) bound to TLS, with mutations of GGUG to CCUC causing impaired binding25 (Fig. S5a). RNase A treatment of TLS, EWS or TAFII68 resulted in dissociation from p300 and CBP, but not TIP60 (Fig. 2a; Fig. S5e, lanes 3-4; Fig. S5f; Fig. S6). Consistently, the inhibitory activity of GST-TLS on p300 HAT was abolished when GST-TLS was pre-treated with the calcium-dependent micrococcal nuclease (MNase), but not with DNase I (Fig. 2b). Following blocking MNase activity with EGTA, addition of the GGUG-, but not CCUC- oligonucleotide, restored the TLS inhibitory effect on p300 HAT activity (Fig. 2b).
Interaction studies demonstrated that the C-terminus of TLS (211-526; 373-526) interacted with the GGUG-oligonucleotide (Fig. S5b), while the N-terminus (1-211) interacted with CBP (Fig. S5c), respectively. Intriguingly, the TLS N-terminus was found to possess a detectably stronger CBP HAT inhibitory activity than the full-length TLS (Fig. S5d) and its interaction with CBP was not disrupted by RNase A treatment (Fig. S5e, lanes 1-2). Furthermore, the TLS N-terminus was capable of interacting with the TLS C-terminus (373-526) in a manner that was inhibited by GGUG-oligonucleotide in a dose-dependent manner (Fig. 2c), while the GGUG-oligonucleotide enhanced the binding of TLS to p300/CBP (Fig. 2d; data not shown). Partial proteolysis assays revealed that the GGUG-oligonucleotide enhanced cleavage of TLS (Fig. S5g). In concert, our findings suggest that an RNA-dependent allosteric modification of TLS relieves the inhibitory function of the TLS C-terminus, allowing the TLS N-terminus to bind to CBP/p300 and allosterically regulate the HAT activity.
CCND1, a cell cycle regulator repressed by DNA damage signals26, is an endogenous CREB target gene27 and is induced in RAW264.7 cells by forskolin (Fig. 3a). Specific murine TLS siRNA (siTLS) caused a marked increase in both basal and forskolin-stimulated CCND1 mRNA levels in these cells (Fig. S7a; Fig. 3a). Overexpression of human TLS could overcome the effect of siTLS (Fig. S7c). Knockdown of p300/CBP using specific siRNAs significantly reduced histone acetylation (AceH3-K9K14) of the CCND1 promoter detected by chromatin immunoprecipitation (ChIP) and CCND1 mRNA levels (Fig. S7a; Fig. 3b-c), indicating required functions of these coactivators on this gene. Wild-type, but not a HAT mutant CBP28, upregulated CCND1 promoter activity (Fig. S7b), suggesting that CCND1 expression is dependent on CBP HAT function.
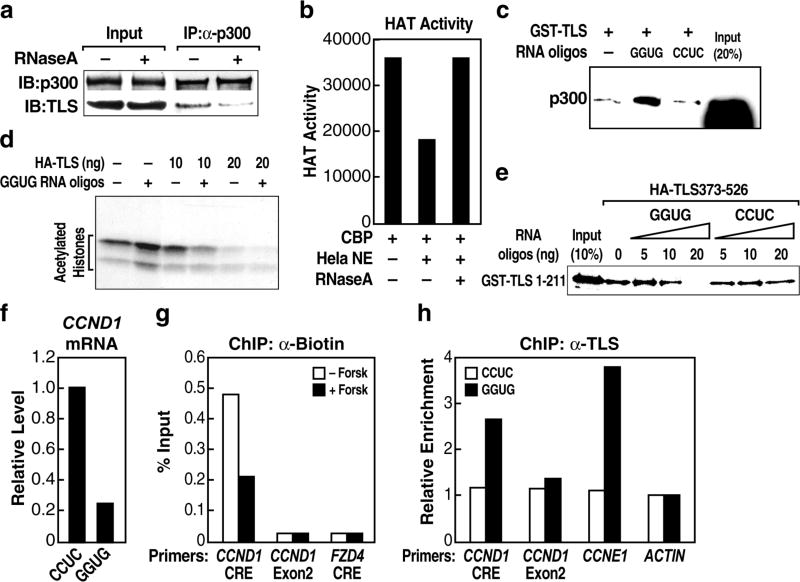
Figure 3. TLS negatively regulates the CBP/p300 HAT-regulated CCND1 gene.
a, CCND1 gene expression from RAW264.7 cells treated with forskolin (Forsk) and TLS siRNA. CTL, control. b, c, Chromatin IP (ChIP) of histone acetylation (AceH3-K9K14) on the CCND1 promoter (b) and CCND1 gene expression (c) in the presence of control or CBP and p300 siRNAs (siCBP/p300). * p<0.01, n=3. d, ChIP with indicated immunoglobulin G (IgG) on the CCND1 promoter upon forskolin treatment. MDM2, control. * p<0.01, n=3. e, ChIP of AceH3 on the CCND1 promoter in the presence of control or TLS siRNA. * p<0.01, n=3. Error bars indicate ± SEM.
When RAW264.7 cells were cultured with carrier (– forskolin) in serum-starved media, both p300 and TLS were bound to CCND1 promoter at the CRE site (Fig. 3d). Forskolin treatment caused TLS to be dismissed from CCND1 promoter (Fig. 3d), despite a slight increase in total cellular TLS levels (Fig. S8a). In contrast, p300 remained bound (Fig. 3d). ChIP analysis revealed hyper-acetylation of histone (AceH3-K9K14) on the CCND1 promoter upon forskolin treatment (Fig. 3d) or knockdown of TLS (Fig. 3e). In concert, our data suggest that TLS acts as a repressor of CCND1. However, we did not observe binding of TLS on all CREB targets (Fig. S8b), suggesting that the negative regulation of CREB target genes by TLS is gene-specific.
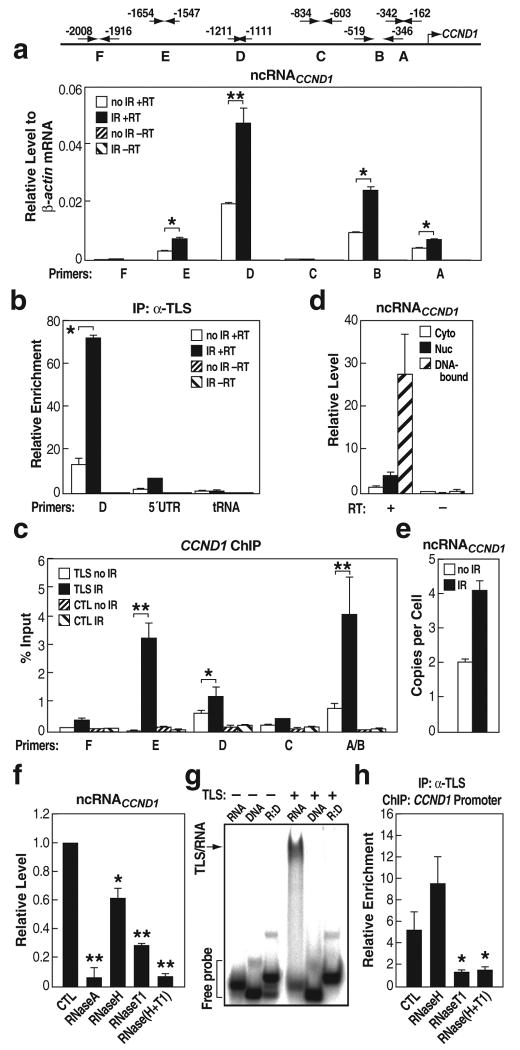
In searching for endogenous regulatory RNAs, we took advantage that expression of CCND1 is down-regulated in response to DNA damage signals, such as ionizing irradiation (IR)29, correlated with decreased histone acetylation (Fig. S9a, b). We considered the possible candidates as previously-unrecognized, local transcripts generated upstream of the CCND1 promoter. As diagrammed in Fig. 4a, first strand synthesis was performed using random primers, followed by real-time PCR using a series of validated specific primer pairs that exhibited similar amplification efficiencies on genomic DNA templates, spanning from -2008 to -162 bp upstream of the established CCND1 transcription start site. These experiments revealed the presence of multiple previously unrecognized, IR-enhanced ncRNAs (A, B, D, E) transcribed from multiple 5′ regulatory regions of CCND1 (ncRNACCND1; Fig. 4a). TLS interacted with these ncRNACCND1s as detected by RNA-immunoprecipitation assays (Fig. 4b; data not shown) and ChIP assay revealed that TLS was recruited to these ncRNACCND1-“expressing” regions in an IR-induced manner (Fig. 4c). In contrast, TLS showed very weak interaction with ncRNACCND1-“non-expressing” regions C and F (Fig. 4c). The level of TLS protein was never upregulated by IR, being either unchanged or, in some experiments, actually downregulated (Fig. S9c).
Figure 4. NcRNACCND1s are predominantly single-stranded, DNA-bound species that bind to TLS.
a, Top, Diagram of ncRNACCND1 detection primers. Bottom, The expression levels of ncRNACCND1s. IR, ionizing irradiation; RT, reverse transcriptase. * p<0.01, ** p<0.002, n=6. b, IP of TLS and detection of associated RNA by RT-real time PCR. D, ncRNACCND1_D; 5′UTR, 5′UTR of CCND1; tRNA, tRNA14TyrATA. * p<0.01, n=3. c, ChIP of TLS on the ncRNACCND1-“expressing” (E, D, and AB) and -“non-expressing” regions (F and C). * p<0.05, ** p<0.01, n=3. d, Subcellular analysis of ncRNACCND1_D. e, The copy number of ncRNACCND1_D. f, The expression levels of ncRNACCND1_D upon indicated ribonucleases treatments. * p<0.05, ** p<0.001, compared with control, n=3. g, Gel shift analysis of TLS interactions with RNA, complementary DNA or RNA:DNA hybrid (R:D). RNA, -454s derived from ncRNACCND1_B. h, ChIP of TLS on the CCND1 promoter upon indicated ribonucleases treatments. * p<0.01, compared with CTL, n=3. Error bars indicate ± SEM.
Subcellular and chromatin fractionation studies revealed that ncRNACCND1 was mainly chromatin-bound (Fig. 4d). Real-time PCR analyses using several RNA species for which copy numbers have been well established as standards, revealed that ncRNACCND1 was, remarkably, present at low copy number (e.g., region D at ∼2 copies/cell under basal conditions and ∼4 copies/cell following IR treatment; Fig. 4e). To test whether ncRNACCND1 might be present, in part, as an RNA:DNA hybrid, we evaluated the effects of RNase H treatment, finding that ncRNACCND1 was partially diminished by RNase H treatment (Fig. 4f). A portion of ncRNAs was also diminished by RNase T1, which digests single stranded RNA (ssRNA). The combination of RNase H and RNase T1 caused a complete loss of ncRNAs (Fig. 4f). This suggests that a portion of the ncRNA exists, at least transiently, as ssRNA, in addition to a portion present as an RNA:DNA hybrid. Intriguingly, TLS did not bind to the corresponding DNA sequence, nor to an RNA:DNA hybrid of the tested sequences (Fig. 4g; data not shown). ChIP for TLS on CCND1 promoter was performed following digestion with RNase H, RNase T1, or both. As shown in Fig. 4h, RNase T1 blocked TLS recruitment, while RNase H treatment had no inhibitory effect. These data argue against RNA:DNA hybrids serving as the landing pads for TLS. Our data also revealed the presence of bidirectional ncRNA transcripts, further induced by IR (Fig. S10); in contrast, the adjacent 5′ UTR of CCND1 mRNA exhibited a decreased level in response to IR (Fig. S10).
Northern blotting analysis, using non-overlapped probes (∼200 nt each) targeting the 5′ regulatory regions of CCND1, showed species of ∼330, ∼200 nt and larger transcripts (Fig. S11). The observations of clear variability in the lengths of these RNAs, and the bands being always multiple or diffuse, suggest diverse Pol II entry sites, or/and imprecise processing. NcRNACCND1 proved to be pol II-regulated and polyadenylated, but not capped (Fig. S12).
To investigate the potential function of ncRNACCND1, we identified specific siRNAs to the ncRNACCND1-“expressing” regions A (siA), D (siD) and E (siE); the ncRNACCND1-“non-expressing” regions C (siC) and F (siF); and the antisense 5′UTR of CCND1 (si5′UTR). SiA specifically knocked-down ncRNACCND1 in region A without affecting that in region D, and conversely, siD knocked-down ncRNACCND1 in region D but not A (Fig. S13), suggesting that multiple ncRNA transcripts were present, either as separate transcripts or as a result of rapid processing. Both strands of ncRNACCND1 were targeted by siA or siD (Fig. S13). SiA, siD and siE (Fig. 5a, left), but not siC, siF and si5′UTR (Fig. S15a), significantly enhanced the levels of endogenous CCND1 mRNA. In contrast, the level of CCNE1 mRNA expression was not affected by either siA or siD (Fig. 5a, right). Cotransfection of siA, siD and siE (siADE) showed similar effects on CCND1 mRNA level compared with single siRNA transfection (Fig. 5a, left). As a control, the siRNA targeting CCND1 coding region (siCCND1) specifically blocked CCND1 expression (Fig. 5a, left). These results argue against a trans-acting role for ncRNACCND1. SiD also enhanced the activity of CCND1 promoter-driven reporter containing the ncRNACCND1-“expressing” region (Fig. S14). Consistent with their putative local biological roles, the siA or siD (Fig. 5b), but not siC or siF (Fig. S15b), caused a decrease in TLS recruitment to CCND1 promoter at region A under IR treatment. In contrast, the recruitment of p300 was unaffected by either siA or siD (Fig. 5b). Similar data was observed in the absence of IR (data not shown). Neither siA nor siD reduced the level of TLS protein (Fig. S15c). These data suggest that ncRNACCND1s combinatorially/cooperatively cause repression of CCND1 transcription unit.
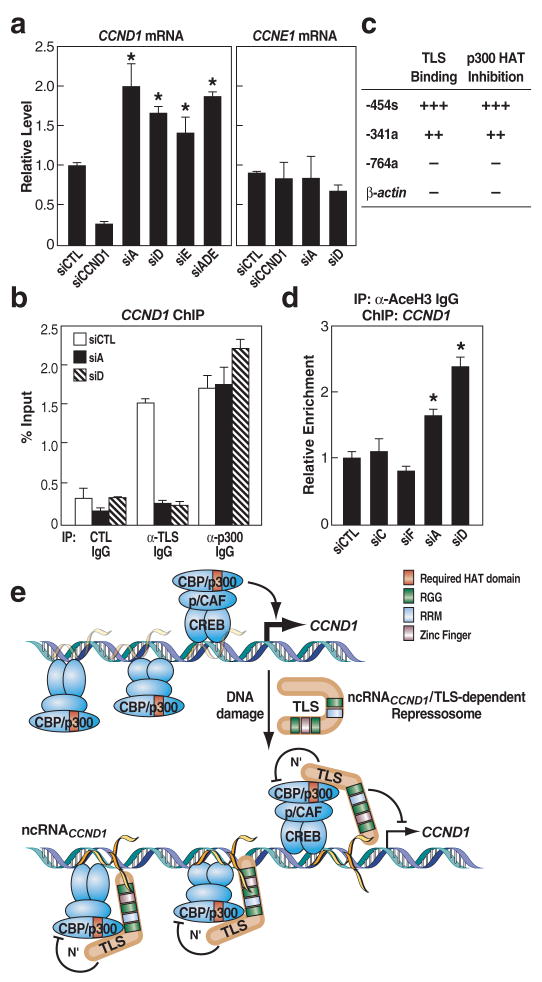
Figure 5. NcRNACCND1 negatively regulates CCND1 transcription by recruiting TLS to the CCND1 promoter.
a, The expression levels of CCND1 and CCNE1 in the presence of siRNA targeting ncRNACCND1_A (siA), _D (siD), _E (siE) or cotransfection of these siRNAs (siADE), or targeting CCND1 coding region (siCCND1). * p<0.01, compared with control siRNA, n=6. b, ChIP of TLS and p300 on the CCND1 promoter in the presence of siA or siD upon IR. c, RNA oligonucleotides tested for TLS binding and p300 HAT inhibition. d, ChIP of AceH3 on the CCND1 promoter in the presence of indicated siRNAs. * p<0.05, n=3. e, Model. Error bars indicate ±SEM.
Real-time PCR studies revealed existence of ncRNACCND1s (D and A, but not C) in both high and low MW fractions (Fig. S16a; data not shown). RNA oligonucleotides corresponding to the ncRNACCND1-“expressing” regions (e.g., -454s; -341a) were capable of binding to TLS and inhibiting p300 HAT function (Fig. 5c; Fig. S16b, c). In contrast, a series of other RNA oligonucleotides evaluated, including oligonucleotides based on β-actin mRNA sequence and the ncRNACCND1-“non-expressing” region C (-764a), were unable to bind to TLS (Fig. S16d) or inhibit p300 HAT functions (Fig. 5c). Moreover, siA and siD, but not siC or siF, resulted in an increase of histone acetylation (AceH3-K9K14) on CCND1 promoter (Fig. 5d).
We suggest a model in which ncRNAs serve as molecular “ligands” for a specific RNA binding protein, TLS, causing an allosteric effect to release it from an inactive conformation. This in turn permits gene-specific TLS:CBP/p300 interactions resulting in inhibition of CBP/p300 HAT functions and repression of transcription (Fig. 5e). It is tempting to speculate that other RNA binding coregulators exert functional roles on gene transcription by being analogously recruited to the transcription units through gene-specific ncRNAs.
Methods Summary
RAW264.7 and HeLa cells were maintained in DMEM (GIBCO) supplemented with 10% fetal calf serum (FCS, Gemini). Plasmids and siRNAs were transfected using Lipofectamine 2000 (Invitrogen) as directed. Specific antibodies were obtained from BD Biosciences (anti-TLS), Santa Cruz Biotechnology (anti-CBP and anti-p300) and Millipore (anti-acetylated histone H3). The sequences of siRNA, RNA and DNA oligonucleotides and details of other assays are described in the Full Methods.
Full Methods
Materials and reagents
Antibodies were obtained from Santa Cruz Biotechnology (anti-p/CAF, anti-TAFII68 and anti-EWS), Upstate Biotechnology (anti-TIP60), and Synaptic Systems (anti-cap). Small interfering RNAs (siRNAs) were obtained from Qiagen: siA, 5′-GGCGCCUCAGGGAUGGCUU-3′; siD, 5′-AAUUCAGUCCCAGGGCAAA-3′; siE, 5′-GACCCGGAAUAUUAGUAAU-3′; siC, 5′-GGCUAGAAGGACAAGAUGA-3′; siF, 5′-GAGUGGGCGAGCCUCUUUA-3′; si5′UTR, 5′-GGACUUUGCAACUUCAACA-3′; siCCND1, SI02654547; siCTL, 5′-AAUUCUCCGAACGUGUCAC-3′; siTLS, 5′-CAGAGUUACAGUGGUUAUG-3′ and 5′-UUCUCUGGGAAUCCUAUUA-3′.
HAT assays
HeLa extracts, histones (Sigma) or mononucleosomes (from HeLa cell), and [14C] acetyl-CoA were incubated with baculovirus-expressed CBP in solution HAT assays as described30. Pull down HAT assays were performed by capturing baculovirus-expressed, flag-tagged CBP on anti-flag agarose beads (Sigma). Beads were incubated with HeLa extracts for 1 h, washed three times with HAT assay buffer, and then incubated with histones and [14C] acetyl-CoA. CBP and histones were subsequently resolved by SDS-PAGE and acetylation was detected by autoradiography.
Biochemical purification and protein identification
Hela nuclear extract were dialyzed against 0.1 M NaCl containing dialysis buffer (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 0.5 mM DTT), applied onto a 500 ml column of Sephacryl S-300 equilibrated and fractionated into 43 fractions, which were analyzed with HAT assay. Fractions with the inhibitory activity were further incubated with baculovirus-expressed flag-tagged CBP bound anti-flag agarose beads and extracted with the 0.3 M NaCl extraction buffer and separated on SDS-PAGE gel. The protein bands were analyzed by matrix-assisted laser desorption/ionization reflectron time-of-flight (MALDI-re-TOF) mass spectrometry (MS) (UltraFlex TOF/TOF; BRUKER; Bremen, Germany) as described16. Selected peptide ions (m/z) were taken to search a “non-redundant” human protein database (National Center for Biotechnology Information; Bethesda, MD) to identify the proteins.
Gel shift assays
[32P]-RNA or DNA oligonucleotides (200,000 cpm) were heated at 95 °C for 2 min and immediately placed on ice. RNA and its complementary DNA oligonucleotides were heated at 95 °C for 2 min, and annealed down to room temperature. The probes were then incubated in the reaction buffer containing baculovirus expressed TLS, 10 mM Tris-HCl, pH 7.5, 5% glycerol, 10 mM EDTA, 1 mM DTT, and 5 μg of yeast tRNA at 25 °C for 15 min. The samples were then analyzed on 6% PAGE gel. The gel was dried and analyzed by autoradiography. The RNA and DNA oligonucleotides sequences were:
GGUG-oligonucleotide, 5′-UUGUAUUUUGAGCUAGUUUGGUGAC-3′;
CCUC-oligonucleotide, 5′-UUGUAUUUUGAGCUAGUUUCCUCAC-3′;
-454s (or RNA in Fig. 4g), 5′-UCUGCCGGCUUGGAUAUGGGGUGUC-3′;
-341a, 5′- CCCGGGAUUUAGGGGGUGAGGUGGA-3′;
-764a, 5′- UCCAGCAGCAGCCCAAGAUGGUGGC-3′;
β-actin, 5′- UGGCAUCGUGAUGGACUCCGGUGAC-3′;
DNA, 5′- GACACCCCATATCCAAGCCGGCAGA-3′.
RNA extraction and real-time PCR
HeLa cells were lysed in RSB-100 buffer (100 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 40 μg/ml digitonin) followed by centrifugation at 2,000 g for 8 min. The supernatant fraction was collected as cytosolic fraction. The cell pellet was then resuspended in RSB-100 containing 0.5% Triton X-100 (RSB-100T). After centrifugation at 2,000 g for 8 min, the supernatant was collected as nuclear fraction. The resulting cell pellet was resuspended in RSB-100T and sonicated (Fisher Sonic Dismembrator, Model 300). The soluble DNA-bound RNA fraction was collected after centrifugation at 4,000 g for 15 min. RNA was extracted using Trizol (Invitrogen) and treated with RNase-free DNase I (DNA-free; Ambion). Reverse transcription (RT) was performed using random hexamer or gene specific primer. Reaction without transcriptase was performed as no RT control. Real-time PCR was performed using the Mx3000P (Stratagene).
RNase A, micrococcal nuclease (MNase), DNase I, RNase H and RNase T1 treatment
Whole cell extracts of GST proteins were treated with RNase A (25 μg/50 μl, Sigma), and incubated on ice for 20 min. GST-TLS in whole cell extracts was sequentially treated with 10 μg of micrococcal nuclease (Roche) in 100 mM sodium glycine (pH 8.6) and 10 mM CaCl2 at 37 °C for 4 min, 0 °C for 1 min, and room temperature for 20 min, and terminated by addition of 10 mM EGTA, followed with or without incubation of 100 pmol/20 μl of RNA oligonucleotides. GST-TLS was treated with DNase I (1 μg/50 μl) in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2 and 50 μg/ml BSA, at 37 °C for 30 min. For co-immunoprecipitation and RT-real time PCR, the cell fractionation extracts containing the DNA-bound RNA were obtained as described before and treated with 50 ng/μl of RNase A (Sigma), 1 U/10 μl of RNase H (Invitrogen), or 1 U/10 μl of RNase T1 (Ambion) at room temperature for 30 min.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde and stopped by glycine solution (125 mM). The cells were then sequentially washed in ice-cold buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5) and buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cell pellets were re-suspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1 X protease inhibitor cocktail) and sonicated. The soluble chromatin was then diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1, 1 X protease inhibitor cocktail). Protein A/G-sepharose beads were added and incubated for 1 h at 4 °C for pre-clearing. Specific antibody was added to the supernatant and incubated at 4 °C. The next day, protein A/G-sepharose beads were added for 2 h incubation at 4 °C. Beads were harvested by centrifugation and washed sequentially in TSE I buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), TSE II buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), buffer III (0.25M LiCl, 1% NP-40, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and TE buffer. DNA fragments were eluted in 1% SDS, 0.1 M NaHCO3 at 65 °C overnight and purified with a QIAquick Spin Kit (Qiagen, CA).
RNA-immunoprecipitation (RNA-IP) assay
Whole cell extracts were obtained in NETN buffer (125 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.1, 0.5% NP40, 10% glycerol, 1 X protease inhibitor cocktail) without cross-linking, followed by sonication and pre-clearing as described in ChIP assay. Conjugated antibody/protein A/G-sepharose beads were pre-treated with RNase inhibitor and then added for a further incubation at 4 °C overnight. Beads were then washed for 10 min each at 4 °C in NETN buffer for at least six times. Bound RNA was then eluted from the beads by directly adding Trizol (Invitrogen) to the beads, followed by RNA extraction and RT-real time PCR as described previously.
Supplementary Material
Acknowledgments
We thank A. Gettings for help with mass spectrometric analysis. We thank M. Hiramatsu, W. Sato and C. Nelson for providing excellent technical assistance. We thank A. Matsushita, M. Matsubara and T. Oyoshi for useful discussion. We thank J. Hightower and M. Fisher for figure and manuscript preparation. This work was supported by the Fujisawa Foundation, Takeda Science Foundation, the Naito Foundation, Sankyo Foundation Life Science, and grants-in-aid (no. 17054036 and no. 18055029) from the Ministry of Education, Culture, Sports, Science, and Technology in Japan to RK, by NIH grants CA52599 and HL59694 to CKG, NCI Cancer Center Support Grant P30 CA08748 to PT, and by DK39949 and NS34934 to MGR. MGR is an HHMI investigator.
References
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–6. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 15 Spec No. 2006;1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petruk S, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–23. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 11.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum Mol Genet. 2005;14(Spec No 1):R113–20. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 14.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 15.Seo SB, et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 16.Sebastiaan Winkler G, et al. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–9. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 17.Uranishi H, et al. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivator. J Biol Chem. 2001;276:13395–401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–4. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 19.Hicks GG, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–9. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda M, et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. Embo J. 2000;19:453–62. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baechtold H, et al. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–42. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–21. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- 23.Ron D. TLS-CHOP and the role of RNA-binding proteins in oncogenic transformation. Curr Top Microbiol Immunol. 1997;220:131–42. doi: 10.1007/978-3-642-60479-9_8. [DOI] [PubMed] [Google Scholar]
- 24.Kurokawa R, et al. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–3. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 25.Lerga A, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–16. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 26.Miyakawa Y, Matsushime H. Rapid downregulation of cyclin D1 mRNA and protein levels by ultraviolet irradiation in murine macrophage cells. Biochem Biophys Res Commun. 2001;284:71–6. doi: 10.1006/bbrc.2001.4950. [DOI] [PubMed] [Google Scholar]
- 27.Impey S, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Murata T, et al. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum Mol Genet. 2001;10:1071–6. doi: 10.1093/hmg/10.10.1071. [DOI] [PubMed] [Google Scholar]
- 29.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 30.Korzus E, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–7. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.