Abstract
Sensory nerves may dampen inflammatory processes through the release of the neuropeptide calcitonin gene-related peptide (CGRP). CGRP mediates immunosuppressive activities through up-regulation of interleukin-10 or, alternatively, through an interleukin-10-independent pathway that is associated with rapid induction of the transcriptional inducible cAMP early repressor (ICER). In this work, we further investigated the molecular mechanisms of immune modulation by CGRP. Using TLR2-stimulated dendritic cells, we show that inhibition of tumor necrosis factor-α production by CGRP is dependent on up-regulation of endogenous ICER. Dendritic cell expression of ICER was selectively induced by CGRP and elevation of cellular cAMP levels but not by numerous pro- and anti-inflammatory cytokines. Treatment of dendritic cells with CGRP did not interfere with the induction of Tnfa gene expression but caused premature repression of TLR2-induced transcriptional activity. ATF-2 was rapidly phosphorylated and recruited to the Tnfa promoter following ligation of TLR2. Concomitant administration of CGRP completely prevented binding of ATF-2 to the Tnfa promoter, whereas recruitment of ICER was markedly elevated. In contrast, CGRP did not influence TLR2-stimulated binding of NF-κB p65. Together, these results are consistent with a model suggesting that CGRP causes rapid up-regulation of ICER, which in turn competes with ATF-2 for binding to the Tnfa promoter, leading to repression of gene expression.
Keywords: Cytokines/Induction, Cytokines/Tumor Necrosis Factor, DNA/Transcription, Gene/Transcription, Immunology/Innate Immunity, Immunology/Toll Receptors, Signal Transduction/Adenylate Cyclase, Neuropeptides
Introduction
Neural pathways conveying the sensation of pain are known to modulate inflammatory processes (1, 2). Stimulation of C-type pain fibers in inflamed tissues is caused by tryptase that is released from mast cells found in close contact with sensory nerves (3, 4). Activation of these nerves is part of the pain reflex and leads to the release of neuropeptides such as CGRP.2 In addition to its function as a potent vasodilator and hypotensive agent, CGRP mediates various anti-inflammatory and immunosuppressive activities and therefore may play an important role in the neuronal control of inflammation. CGRP influences adhesive and migratory capacities of immune cells, including T cells, eosinophilic granulocytes, and dendritic cells (5–7). Stimulation of dendritic cells or macrophages with CGRP reduces the expression of major histocompatibility complex class II and co-stimulatory proteins, inhibits the antigen-presenting capacity of these cells, and impairs production of inflammatory cytokines (8–12). In addition, treatment of mice with CGRP was reported to inhibit delayed-type and contact hypersensitivity responses (9, 13). Similarly, inflammation and organ injury in models of acute endotoxemia and chronic colitis were found to be attenuated by CGRP administration (12, 14–16).
The receptor for CGRP is a multiprotein complex consisting of the seven-transmembrane domain calcitonin receptor-like receptor and an accessory protein called RAMP1 (receptor activity-modifying protein-1) (17, 18). This membrane-bound receptor complex is linked to the cytosolic CGRP receptor component protein (19). Signal transduction through the CGRP receptor complex is initiated by activation of receptor-associated heterotrimeric G proteins. In most cells, the CGRP receptor is coupled to Gαs proteins, leading to elevation of cellular cAMP levels. Alternatively, exposure of cells to CGRP may activate phospholipase Cβ1 via Gαq/11 proteins, causing calcium mobilization (20).
CREB, CREM, and ATF proteins comprise the bZIP class of transcription factors, binding to CRE sites in target promoters and enhancers (21, 22). Elevation of cAMP levels induces the transactivating capacity of these factors by protein kinase A-mediated phosphorylation of critical serine residues, but activation may also be achieved by various other kinases, including protein kinase C, casein kinase II, and MAPKs. In the bZIP family, the CREM gene is unique because it encodes multiple isoforms that may act as either transcriptional activators or repressors. The CREM isoforms are generated by RNA splicing and transcriptional initiation from alternative promoters. Transcription of the repressor protein ICER is driven by an intronic promoter of the CREM gene that contains a cluster of four CRE sites and is responsive to cAMP (23). The protein structure of ICER encompasses mainly the bZIP DNA-binding domain but lacks a transactivating domain (21). Thus, ICER may repress transcription either by heterodimerization with bZIP-containing transcriptional activators or by competition with these factors for DNA binding.
CGRP may mediate certain immunosuppressive activities through the up-regulation of IL-10 production in target cells (10, 11). Alternatively, we have recently described an IL-10-independent anti-inflammatory pathway of CGRP that causes reduced secretion of cytokines like TNFα and CCL4 and is associated with the rapid and cAMP-dependent induction of ICER expression (12). In this study, we further investigate the molecular mechanisms of immune modulation by this IL-10-independent pathway. Our results provide strong evidence that the anti-inflammatory activity of CGRP in TLR-stimulated dendritic cells is dependent on the up-regulation of endogenous ICER, leading to the premature repression of inflammatory gene expression, most likely by preventing promoter recruitment of ATF-2.
EXPERIMENTAL PROCEDURES
BMDC Preparation and Stimulation
Bone marrow cells of C57BL/6 origin were cultured in medium supplemented with 20 ng/ml granulocyte/macrophage colony-stimulating factor (PeproTech, Rocky Hill, NJ) to generate BMDC. Cultures received fresh medium containing granulocyte/macrophage colony-stimulating factor every 3 days, and cells were used for experiments at day 10. Cells were stimulated with 100 ng/ml ultrapure LPS from Salmonella minnesota R595 (List Biological Laboratories) or 1 μg/ml P3Cys (EMC Microcollections) in RPMI 1640 medium containing 10% fetal calf serum for the indicated time periods to activate TLR4 or TLR2, respectively. Murine cytokines IL-4, IL-6, IL-10, IFN-γ, and TNFα (R&D Systems) were used at 20 ng/ml, CGRP and inhibitory CGRP (Bachem) at 100 nm, and the adenylyl cyclase activator forskolin (Sigma) at 50 μm.
Analysis of Tnfa Transcription and Protein Expression
The rate of transcription of the Tnfa gene was determined by real-time quantitative primary transcript RT-PCR. The principle of the assay is to use primers that are placed in adjacent exons and introns of a given gene, thereby generating amplicons from the unspliced populations of RNAs. RNA was prepared using the RNeasy® mini kit (Qiagen). For cDNA synthesis, the QuantiTec® reverse transcription kit (Qiagen) was used. DNA contamination of RNA preparations was monitored by omitting reverse transcriptase from control reactions and was not detectable in any of the experiments reported. Expression levels of primary Tnfa transcripts were normalized to those of GAPDH and were displayed as -fold change relative to samples of unstimulated cells used as calibrator. The primers were as follows: Tnfa, 5′-CCG GGA CCT CAT AGC CA-3′ (sense) and 5′-GCA AAT CGG CTG ACG GTG TG-3′ (antisense); and GAPDH, 5′-TCC AGT ATG ACT CCA CTC-3′ (sense) and 5′-ATT TCT CGT GGT TCA CAC-3′ (antisense). PCR products were quantified on an ABI 7300 cycler (Applied Biosystems).
Protein concentrations of TNFα in culture supernatants or cellular lysates were determined by enzyme-linked immunosorbent assay (R&D Systems). TNFα protein expression of individual cells was detected by flow cytometry analysis of fixed and permeabilized cells with rat anti-mouse TNFα antibody (BD Biosciences) using standard procedures.
Analysis of ICER mRNA Expression
ICER mRNA was detected by RT-PCR of titrated amounts of template cDNA using specific primers (sense, 5′-ATG GCT GTA ACT GGA GAT GAA ACT-3′; and antisense, 5′-CTA ATC TGT TTT GGG AGA GCA AAT GTC-3′). As a control, mouse GAPDH was amplified (sense primer, 5′-CAA TGC ATC CTG CAC CAC CAA; and antisense primer, 5′-GTC ATT GAG AGC AAT GCC AGC-3′). GAPDH primer sequences were separated by introns to control for contaminations with genomic DNA.
For quantitative RT-PCR analysis, MasterMix Plus for SYBR® Green I (Eurogentec) was used. The expression levels of ICER-I/ICER-Iγ mRNA (sense, 5′-ACC AGG AAG CCT GCA CAG TC-3′; and antisense, 5′-TCT TCT TCC TGC GAC ACT CC-3′) were normalized to those of β-actin (sense, 5′-ACC CAC ACT GTG CCC ATC TAC-3′; and antisense, 5′-AGC CAA GTC CAG ACG CAG G-3′) and were displayed as -fold change relative to samples of unstimulated cells used as calibrator. PCR products were quantified on an ABI 7300 cycler.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed according to standard protocols (24). An equal amount of chromatin (50–100 mg) was used for each precipitation. The following antibodies used were obtained from Santa Cruz Biotechnology: anti-RNA polymerase II (N-20), anti-NF-κB p65 (A), anti-ATF-2 (C-19), anti-CREB (H-74), anti-CREB (C-21), and control rabbit IgG. An ICER-specific polyclonal rabbit antiserum was generated using the murine sequence NH2-Ala-Val-Thr-Gly-Asp-Glu-Thr-Gly-Gly-Cys-COOH for generating an antigenic peptide (Eurogentec). Amino acids encoded by the ICER-specific exon are underlined. For chromatin immunoprecipitations, affinity-purified antibody was used. Immunoprecipitated Tnfa promoter DNA was quantified by real-time PCR analysis using primers for the proximal Tnfa promoter (sense, 5′-GGA GAT TCC TTG ATG CCT GG-3′; and antisense, 5′-GCT CTC ATT CAA CCC TCG GA-3′) and, as a control, primers spanning a region located ∼5.1 kb upstream of the Tnfa transcriptional start site (sense, 5′-ACT GGC TTT ACC TAA TGG-3′; and antisense, 5′-ACA TAC AAG TGC CAC AGG-3′). For each amplification product, DNA amounts in immunoprecipitates were normalized against those present in input chromatin. Results are displayed as -fold change relative to precipitations with control rabbit IgG used as calibrator. PCR products were quantified on an ABI 7300 cycler.
Statistical Analysis
Statistical analysis of the data was performed using Student's t test. Data are presented as means ± S.E. with the number of independent experiments indicated in the figure legends. Differences between groups were considered significant for p < 0.05.
RESULTS
Inhibition of TNFα Production by CGRP Is Dependent on the Induction of Endogenous ICER
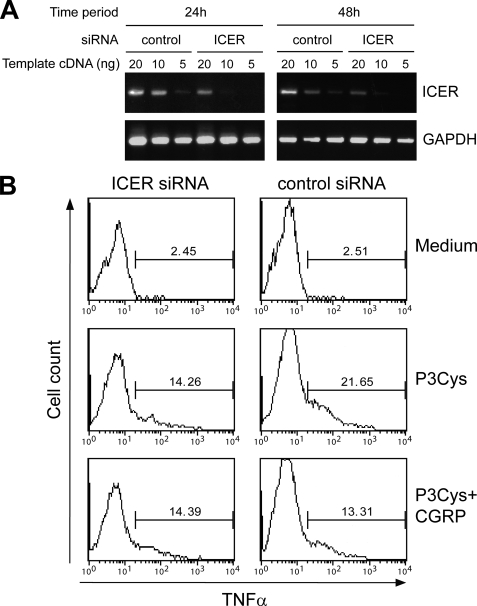
This study was designed to further delineate the molecular mechanisms underlying the immunosuppressive function of the neuropeptide CGRP. Previous work has shown that CGRP inhibits TNFα production by TLR-stimulated dendritic cells and reduces systemic TNFα levels in endotoxemic mice (12). The inhibitory activity of CGRP correlates with the up-regulation of the repressor protein ICER, and the ectopic overexpression of ICER ameliorates TNFα production, suggesting that the effect of CGRP may be mediated by ICER (12). To directly explore the role of endogenous ICER in the suppression of TNFα production by CGRP, BMDC were transfected with control or ICER-specific siRNA, and 24 or 48 h later, cells were stimulated with CGRP to induced ICER expression. ICER mRNA levels were determined by semiquantitative RT-PCR using titrated amounts of cDNA as template. As shown in Fig. 1A, induction of ICER mRNA by CGRP was markedly reduced both 24 and 48 h after transfection of BMDC with ICER-specific siRNA compared with control siRNA, indicating a sustained and specific knockdown of ICER. In subsequent experiments, BMDC were transfected with siRNAs together with a plasmid directing expression of GFP and stimulated with P3Cys. TNFα levels were determined in GFP-positive BMDC by flow cytometry. We found that, in BMDC transfected with control siRNA, the P3Cys-stimulated expression of TNFα was strongly reduced by CGRP treatment (Fig. 1B). In contrast, transfection of BMDC with ICER-specific siRNA almost completely prevented the inhibitory effect of CGRP (Fig. 1B). Comparable results were obtained when BMDC were stimulated with LPS instead of P3Cys to induce TNFα production (data not shown). These results indicate that inhibition of dendritic cell TNFα production by CGRP is dependent on the expression of endogenous ICER.
FIGURE 1.
Inhibition of dendritic cell TNFα production by CGRP is dependent on ICER. A, BMDC were transfected with control or ICER-specific siRNA. Cells were allowed to recover for 24 or 48 h and stimulated with CGRP for 16 h. Expression of ICER was determined by semiquantitative RT-PCR using titrated amounts of cDNA as template. B, BMDC were transfected with siRNAs together with an expression construct for GFP. After 24 h, cells were treated with culture medium, P3Cys, or a combination of P3Cys and CGRP for 16 h, and expression of TNFα by GFP-positive cells was determined by flow cytometry analysis of fixed and permeabilized cells. The data depicted are representative of three independent experiments yielding comparable results.
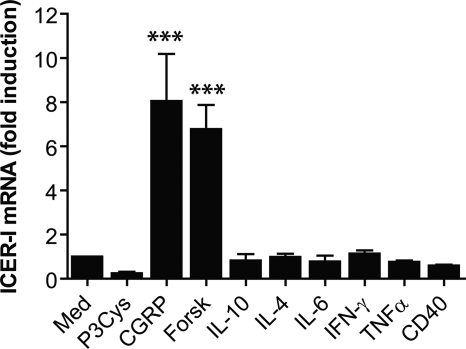
To address the question as to whether ICER may also be involved in the regulation of dendritic cell functions by other immune mediators, the expression of ICER was examined in more detail. BMDC were treated with the indicated agonists, and ICER mRNA levels were quantified by real-time RT-PCR. The results in Fig. 2 demonstrate that ICER mRNA was significantly induced by CGRP and forskolin, both of which lead to increased cAMP levels in BMDC (12). In contrast, exposure of BMDC to IL-10, IL-4, IL-6, TNFα, or IFN-γ or cross-linking of CD40 did not alter ICER mRNA levels (Fig. 2). Stimulation of BMDC with P3Cys revealed a trend for reduced amounts of ICER mRNA (Fig. 2). These results indicate that, in dendritic cells, ICER is selectively induced by CGRP and elevation of cellular cAMP levels.
FIGURE 2.
ICER is selectively induced by CGRP and elevation of cAMP. BMDC were treated with the indicated mediators, and expression of ICER mRNA was determined by quantitative real-time RT-PCR (n = 3). ***, p < 0.001. Med, medium; Forsk, forskolin.
CGRP Inhibits TNFα Biosynthesis at the Transcriptional Level
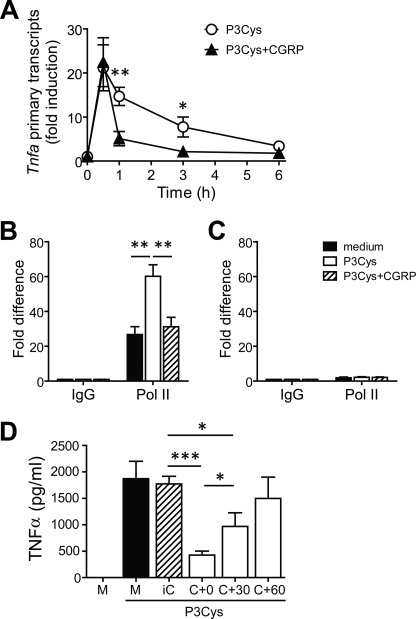
Having established that the inhibitory effect of CGRP on dendritic cell TNFα production is ICER-dependent, we next examined the influence of CGRP on the TLR-stimulated transcriptional activity of the Tnfa gene. Quantification of primary Tnfa transcripts revealed that the transcriptional activity of the Tnfa gene peaked between 0.5 and 1 h after stimulation of BMDC with the TLR2 agonist P3Cys, declining thereafter and returning to near base-line levels after 6 h (Fig. 3A). Importantly, treatment of BMDC with CGRP in addition to P3Cys substantially reduced the amount of primary Tnfa transcripts at 1 and 3 h after initiation of transcription by P3Cys (Fig. 3A). However, 0.5 h after the addition of P3Cys, we did not observe an inhibitory effect of CGRP treatment. Thus, treatment of BMDC with CGRP results in a delayed repression of TLR2-stimulated Tnfa transcription. The delayed effect of CGRP is consistent with the kinetics of induction of ICER protein expression reported previously (12).
FIGURE 3.
CGRP causes delayed transcriptional repression of the Tnfa gene. A, BMDC were stimulated for the indicated time periods with P3Cys alone or in combination with CGRP. The transcriptional activity of the Tnfa gene was determined by real-time primary transcript RT-PCR. Induction of Tnfa primary transcripts is given relative to unstimulated cells (n = 3–4). B and C, chromatin samples of BMDC that were untreated or stimulated for 1 h with P3Cys or a combination of P3Cys and CGRP were immunoprecipitated with an antibody against RNA polymerase II (Pol II) or control rabbit IgG. DNA isolated from immunoprecipitates was analyzed by real-time PCR using primers spanning the Tnfa proximal promoter region (B) or a region located ∼5.1 kb upstream of the Tnfa transcriptional start site (C). Values for RNA polymerase II are given as -fold differences relative to the IgG controls (n = 7). D, BMDC were left untreated in medium (M) or were stimulated with P3Cys alone or together with CGRP (C). The addition of CGRP either was concomitant with P3Cys (C+0) or was delayed for 30 (C+30) or 60 (C+60) min. As a control, BMDC were also incubated with P3Cys and inhibitory CGRP (iC), which is an inactive mutant of CGRP lacking the N-terminal seven amino acids (n = 8). *, p < 0.05; ***, p < 0.001.
To further corroborate these findings, chromatin immunoprecipitation experiments with antibodies against RNA polymerase II were performed. Immunoprecipitated Tnfa promoter fragments were quantified by real-time PCR analysis. The results in Fig. 3B demonstrate that the amount of RNA polymerase II bound to the proximal Tnfa promoter region was significantly increased 1 h after stimulation of BMDC with P3Cys. However, P3Cys-induced recruitment of RNA polymerase II was almost completely prevented when BMDC were treated with CGRP in addition to P3Cys (Fig. 3B). The specificity of the chromatin immunoprecipitation experiments was demonstrated by the lack of specific amplification products derived from a region located ∼5.1 kb upstream of the transcriptional start site of the Tnfa gene (Fig. 3C).
A transcriptional mechanism for the inhibition of TNFα biosynthesis by CGRP that is mediated by the up-regulation of ICER would predict CGRP not to be effective when administered after the peak transcriptional activity of the Tnfa gene. To test this hypothesis, the addition of CGRP to P3Cys-stimulated BMDC was delayed for different time periods, and the effects on TNFα production were determined. The results in Fig. 3C show that the inhibitory effect of CGRP on TNFα production was significantly attenuated when it was added 0.5 h after P3Cys compared with concomitant administration of P3Cys and CGRP. Moreover, when CGRP was added to BMDC 1 h after P3Cys, TNFα production was not affected (Fig. 3C).
CGRP Prevents TLR2-induced Recruitment of ATF-2 to the Tnfa Promoter
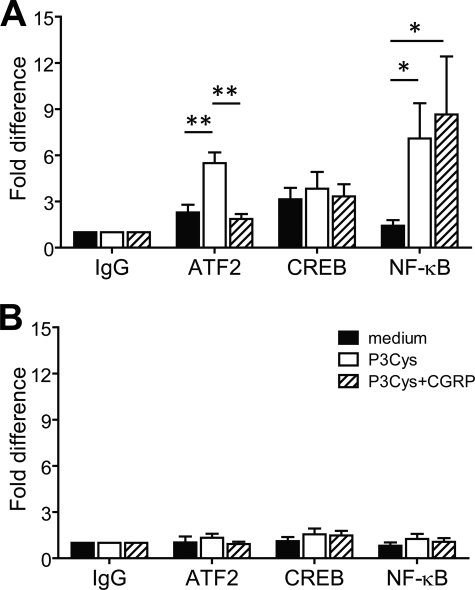
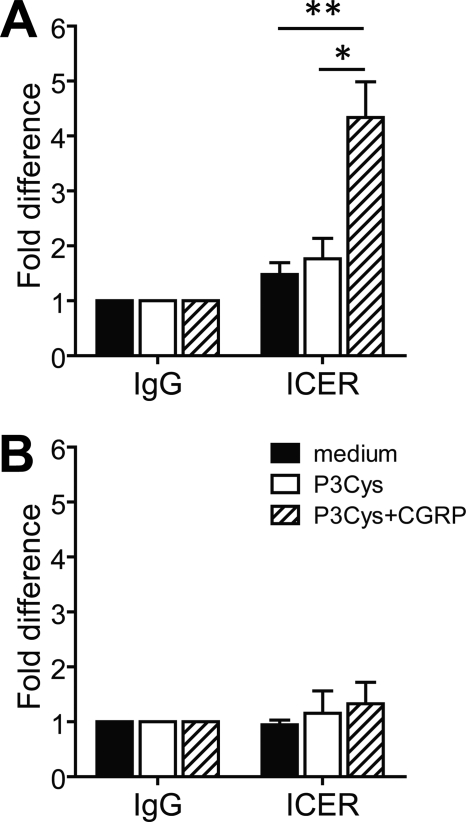
The promoter region of the Tnfa gene contains a CRE that is essential for LPS-induced Tnfa transcription in macrophages (25). Because ICER has been reported to repress gene transcription by binding to CREs (23), we addressed the question as to whether transcriptional repression of Tnfa in CGRP-treated BMDC may be mediated by alterations of transcription factor recruitment to the Tnfa promoter. Chromatin immunoprecipitation experiments showed that stimulation of BMDC with P3Cys significantly increased binding of the transcription factor ATF-2 to the Tnfa promoter, whereas inducible recruitment of CREB was not detectable (Fig. 4A). The lack of inducible binding of CREB was confirmed with independent antibodies (data not shown). Importantly, when BMDC were treated with CGRP, the P3Cys-induced recruitment of ATF-2 to the Tnfa promoter was completely abrogated (Fig. 4A).
FIGURE 4.
CGRP inhibits recruitment of ATF-2 to the Tnfa promoter in BMDC. BMDC were untreated or stimulated with P3Cys or a combination of P3Cys and CGRP for 1 h. Chromatin samples of BMDC were immunoprecipitated with antibody against ATF-2, CREB, or NF-κB p65 or with control rabbit IgG. DNA isolated from immunoprecipitates was analyzed by real-time PCR using primers spanning the Tnfa proximal promoter region (A) or a region located ∼5.1 kb upstream of the Tnfa transcriptional start site (B). Values are given as -fold differences relative to the IgG controls (n = 5). *, p < 0.05; **, p < 0.01.
The CRE site in the Tnfa promoter is in close proximity to a κB element, which is also required for LPS-stimulated Tnfa transcription (25). We therefore tested whether binding of NF-κB to the Tnfa promoter may be altered by CGRP treatment of BMDC. Stimulation of BMDC with P3Cys caused a marked increase in the binding of NF-κB p65 to the Tnfa promoter, but in contrast to ATF-2, recruitment was not significantly altered by treatment of BMDC with CGRP (Fig. 4A). Control PCR analyses of ATF-2, CREB, and NF-κB p65 chromatin immunoprecipitates with primers spanning a genomic region far upstream of the Tnfa promoter did not reveal significant amounts of amplification products (Fig. 4B), confirming the specificity of the assay. Together, these results indicate that treatment of BMDC with CGRP prevents TLR2-induced recruitment of ATF-2, but not CREB or NF-κB, to the Tnfa promoter.
CGRP Stimulates Recruitment of ICER to the Tnfa Promoter
The ICER protein consists of a unique N-terminal sequence that is encoded by an ICER-specific exon of the CREM gene and a bZIP DNA-binding domain that is shared between ICER and various CREM isoforms (21). To specifically examine the role of ICER in the regulation of Tnfa gene expression, an affinity-purified polyclonal antiserum was generated against the ICER-specific sequence. The antiserum readily detected ICER in control Western blot analyses of transiently transfected HEK293 cells (data not shown). Chromatin immunoprecipitation assays directly demonstrated that binding of ICER to the Tnfa promoter was markedly induced by concomitant treatment of BMDC with P3Cys and CGRP (Fig. 5A). In contrast, stimulation of BMDC with P3Cys alone did not induce promoter recruitment of ICER (Fig. 5A). Control PCR analyses of ICER chromatin immunoprecipitates detecting a genomic region far upstream of the Tnfa transcriptional start site did not reveal significant amounts of amplification products (Fig. 5B). Thus, these results support the concept that CGRP represses Tnfa gene expression through ICER-mediated inhibition of ATF-2 promoter recruitment.
FIGURE 5.
CGRP promotes recruitment of ICER to the Tnfa promoter. BMDC were untreated or stimulated with P3Cys or a combination of P3Cys and CGRP for 1 h. Chromatin samples of BMDC were immunoprecipitated with an affinity-purified polyclonal rabbit antibody against ICER or control rabbit IgG. DNA isolated from immunoprecipitates was analyzed by real-time PCR using primers spanning the Tnfa proximal promoter region (A) or a region located ∼5.1 kb upstream of the Tnfa transcriptional start site (B). Values are given as -fold differences relative to the IgG controls (n = 5). *, p < 0.05; **, p < 0.01.
DISCUSSION
The neuropeptide CGRP exerts potent immunosuppressive activities by inhibiting the production of proinflammatory cytokines (1, 2). CGRP suppresses TLR-stimulated dendritic cell TNFα production by a mechanism that is independent of IL-10. Instead, it stimulates the rapid up-regulation of the transcriptional repressor ICER (12). Moreover, ectopic expression of ICER in transfected macrophage-like cells causes reduced TNFα production (12). In this study, we have extended these findings and have shown by RNA interference experiments in primary dendritic cells that inhibition of TNFα production by CGRP is dependent on the up-regulation of endogenous ICER. Consistent with the function of ICER as a transcriptional repressor, we have further shown by primary transcript analyses and quantitative chromatin immunoprecipitation analysis of RNA polymerase II-bound promoter fragments that CGRP suppresses the transcriptional activity of the Tnfa promoter in TLR2-stimulated dendritic cells. Kinetic analysis of primary transcript levels revealed that CGRP did not interfere with the induction of Tnfa gene expression but rather repressed the established transcriptional activity of the Tfna gene. Consistent with these findings, we have previously shown that CGRP does not alter TLR signaling pathways leading to the activation of MAPKs and NF-κB (12). Considered together, these results provide strong evidence that CGRP mediates crucial immunosuppressive activities in dendritic cells by inducing the expression of ICER, resulting in repression of inflammatory gene expression.
The proximal promoter region of the Tnfa gene contains a CRE site that is essential for full transcriptional activity in macrophages and T cells (25–27). Because we found inhibition of TNFα production by CGRP to be dependent on ICER and because ICER is known to repress gene transcription by competing with transactivating factors for binding to CRE sites (23), the influence of CGRP on the recruitment of CRE-binding transcription factors to the Tnfa promoter was examined. We found that stimulation of BMDC through TLR2 markedly elevated binding of ATF-2, whereas moderate levels of constitutive CREB binding were observed, which were not altered in stimulated cells. These observations suggest that CRE-dependent transcription of Tnfa is driven by ATF-2 rather than CREB. This conclusion is also supported by independent studies showing that, in LPS-stimulated macrophages, the recruitment of CREB to the Tnfa promoter is not increased (25) and that expression of an activation-deficient mutant of CREB does not impair TNFα mRNA induction (28). Importantly, we found that treatment of TLR2-stimulated BMDC with CGRP reduced Tnfa promoter binding of ATF-2 to base-line levels, whereas recruitment of ICER to the Tnfa promoter was markedly increased. In contrast, CGRP did not influence recruitment of NF-κB p65. These findings strongly suggest that the inhibitory effect of CGRP is due to specific replacement of ATF-2 by ICER.
Previous studies in T lymphocytes are in accordance with this conclusion. It was shown that, in human T cells, treatment with forskolin or prostaglandin E2 leads to the induction of ICER, interaction of ICER with the composite NF-AT/AP-1 site of the IL-2 promoter, and inhibition of IL-2 production (29, 30). Similarly, ICER may bind to a CRE site in the Ccl4 promoter, thereby attenuating production of CCL4 in activated T cells (31). In conclusion, our results are consistent with a mechanistic model proposing that the CGRP-induced repressor ICER competes with the transactivating factor ATF-2 for binding to the CRE site of the Tnfa promoter, thereby inhibiting TLR-stimulated transcriptional activity and gene expression.
ATF-2 is activated by dual phosphorylation at Thr69 and Thr71 in response to cellular stress or upon exposure of cells to TNFα (32–35). However, TLR signaling pathways resulting in the activation of ATF-2 have not been described. Therefore, we have conducted additional experiments showing that stimulation of BMDC through TLR2 resulted in a marked and rapid phosphorylation of ATF-2 that was partially dependent on JNK (c-Jun N-terminal kinase) (data not shown). Thus, these results are consistent with a role of ATF-2 in the transcriptional regulation of TLR-stimulated TNFα production in dendritic cells.
The results of this study provide evidence that ICER mediates crucial immunosuppressive activities of the neuropeptide CGRP. In addition to CGRP, the expression of ICER may be induced by a diverse array of extracellular mediators, including thyroid-stimulating hormone, follicle-stimulating hormone, noradrenaline, glucagon, nerve growth factor, gastrin, cholecystokinin, and IFN-γ (36–42). In dendritic cells, however, we found the up-regulation of ICER to be restricted to CGRP and pharmacological elevation of cAMP levels. Numerous pro- and anti-inflammatory cytokines or ligation of CD40, which engages distinct signaling pathways that do not increase cellular cAMP levels, did not influence expression of ICER. These results suggest that, in dendritic cells, up-regulation of ICER is dependent on the elevation of cellular levels of cAMP, which is known to lead to the activation of protein kinase A and CREB. Thus, our findings differ from those of a previous report indicating that IFN-γ may induce ICER through casein kinase II-dependent activation of CREB (36). It should be noted, however, that cell type-specific activities of IFN-γ may provide a possible explanation for this discrepancy because up-regulation of ICER by IFN-γ was demonstrated in a macrophage-like cell line (36), whereas our studies were performed with primary dendritic cells.
Although TLRs are considered crucial for the efficient immune defense against invading pathogens, uncontrolled TLR signaling may contribute to the pathogenesis of inflammatory disorders (43). Multiple negative feedback mechanisms have been described that dampen canonical TLR signaling in a cell-autonomous manner (44–46). In addition, intricate interactions between the immune and nervous systems may contribute to a favorable balance between pathogen-directed protective responses and autoaggressive immune reactions (1, 2, 47–49). The results of this study support the concept that the neuropeptide CGRP plays an important role in the negative regulation of immune responses induced by engagement of TLRs and provide a mechanistic explanation for this function.
Acknowledgments
We thank Dr. Günther Schneider (II. Medical Department, Technical University Munich) for help with the chromatin immunoprecipitation experiments and Dr. Peter Murray (St. Jude Children's Research Hospital, Memphis, TN) for advice with primary transcript RT-PCR analysis of Tnfa transcription.
This article was selected as a Paper of the Week.
- CGRP
- calcitonin gene-related peptide
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- CREM
- CRE modulator
- bZIP
- basic leucine zipper
- MAPK
- mitogen-activated protein kinase
- ICER
- inducible cAMP early repressor
- IL
- interleukin
- TNFα
- tumor necrosis factor-α
- TLR
- Toll-like receptor
- BMDC
- bone marrow-derived dendritic cell(s)
- LPS
- lipopolysaccharide
- P3Cys
- (S)-[2,3-bis(palmitoyloxy)-(2-R,S)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-(S)-Lys4-OH
- IFN-γ
- interferon-γ
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein.
REFERENCES
- 1.Steinman L. (2004) Nat. Immunol. 5, 575–581 [DOI] [PubMed] [Google Scholar]
- 2.Sternberg E. M. (2006) Nat. Rev. Immunol. 6, 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naukkarinen A., Järvikallio A., Lakkakorpi J., Harvima I. T., Harvima R. J., Horsmanheimo M. (1996) J. Pathol. 180, 200–205 [DOI] [PubMed] [Google Scholar]
- 4.Stead R. H., Tomioka M., Quinonez G., Simon G. T., Felten S. Y., Bienenstock J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2975–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunzendorfer S., Kaser A., Meierhofer C., Tilg H., Wiedermann C. J. (2001) J. Immunol. 166, 2167–2172 [DOI] [PubMed] [Google Scholar]
- 6.Numao T., Agrawal D. K. (1992) J. Immunol. 149, 3309–3315 [PubMed] [Google Scholar]
- 7.Levite M., Cahalon L., Hershkoviz R., Steinman L., Lider O. (1998) J. Immunol. 160, 993–1000 [PubMed] [Google Scholar]
- 8.Carucci J. A., Ignatius R., Wei Y., Cypess A. M., Schaer D. A., Pope M., Steinman R. M., Mojsov S. (2000) J. Immunol. 164, 3494–3499 [DOI] [PubMed] [Google Scholar]
- 9.Hosoi J., Murphy G. F., Egan C. L., Lerner E. A., Grabbe S., Asahina A., Granstein R. D. (1993) Nature 363, 159–163 [DOI] [PubMed] [Google Scholar]
- 10.Fox F. E., Kubin M., Cassin M., Niu Z., Hosoi J., Torii H., Granstein R. D., Trinchieri G., Rook A. H. (1997) J. Invest. Dermatol. 108, 43–48 [DOI] [PubMed] [Google Scholar]
- 11.Torii H., Hosoi J., Beissert S., Xu S., Fox F. E., Asahina A., Takashima A., Rook A. H., Granstein R. D. (1997) J. Leukocyte Biol. 61, 216–223 [DOI] [PubMed] [Google Scholar]
- 12.Harzenetter M. D., Novotny A. R., Gais P., Molina C. A., Altmayr F., Holzmann B. (2007) J. Immunol. 179, 607–615 [DOI] [PubMed] [Google Scholar]
- 13.Asahina A., Hosoi J., Beissert S., Stratigos A., Granstein R. D. (1995) J. Immunol. 154, 3056–3061 [PubMed] [Google Scholar]
- 14.Gomes R. N., Castro-Faria-Neto H. C., Bozza P. T., Soares M. B., Shoemaker C. B., David J. R., Bozza M. T. (2005) Shock 24, 590–594 [DOI] [PubMed] [Google Scholar]
- 15.Reinshagen M., Flämig G., Ernst S., Geerling I., Wong H., Walsh J. H., Eysselein V. E., Adler G. (1998) J. Pharmacol. Exp. Ther. 286, 657–661 [PubMed] [Google Scholar]
- 16.Reinshagen M., Rohm H., Steinkamp M., Lieb K., Geerling I., Von Herbay A., Flämig G., Eysselein V. E., Adler G. (2000) Gastroenterology 119, 368–376 [DOI] [PubMed] [Google Scholar]
- 17.McLatchie L. M., Fraser N. J., Main M. J., Wise A., Brown J., Thompson N., Solari R., Lee M. G., Foord S. M. (1998) Nature 393, 333–339 [DOI] [PubMed] [Google Scholar]
- 18.Wimalawansa S. J. (1997) Crit. Rev. Neurobiol. 11, 167–239 [DOI] [PubMed] [Google Scholar]
- 19.Evans B. N., Rosenblatt M. I., Mnayer L. O., Oliver K. R., Dickerson I. M. (2000) J. Biol. Chem. 275, 31438–31443 [DOI] [PubMed] [Google Scholar]
- 20.Drissi H., Lasmolest F., Le Mellay V., Marie P. J., Lieberherr M. (1998) J. Biol. Chem. 273, 20168–20174 [DOI] [PubMed] [Google Scholar]
- 21.De Cesare D., Sassone-Corsi P. (2000) Prog. Nucleic Acid Res. Mol. Biol. 64, 343–369 [DOI] [PubMed] [Google Scholar]
- 22.Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 23.Molina C. A., Foulkes N. S., Lalli E., Sassone-Corsi P. (1993) Cell 75, 875–886 [DOI] [PubMed] [Google Scholar]
- 24.Saccani S., Pantano S., Natoli G. (2002) Nat. Immunol. 3, 69–75 [DOI] [PubMed] [Google Scholar]
- 25.Yao J., Mackman N., Edgington T. S., Fan S. T. (1997) J. Biol. Chem. 272, 17795–17801 [DOI] [PubMed] [Google Scholar]
- 26.Tsai E. Y., Falvo J. V., Tsytsykova A. V., Barczak A. K., Reimold A. M., Glimcher L. H., Fenton M. J., Gordon D. C., Dunn I. F., Goldfeld A. E. (2000) Mol. Cell. Biol. 20, 6084–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai E. Y., Jain J., Pesavento P. A., Rao A., Goldfeld A. E. (1996) Mol. Biol. Cell 16, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ananieva O., Darragh J., Johansen C., Carr J. M., McIlrath J., Park J. M., Wingate A., Monk C. E., Toth R., Santos S. G., Iversen L., Arthur J. S. (2008) Nat. Immunol. 9, 1028–1036 [DOI] [PubMed] [Google Scholar]
- 29.Bodor J., Habener J. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 273, 9544–9551 [Google Scholar]
- 30.Bodor J., Spetz A. L., Strominger J. L., Habener J. F. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3536–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barabitskaja O., Foulke J. S., Jr., Pati S., Bodor J., Reitz M. S., Jr. (2006) J. Leukocyte Biol. 79, 378–387 [DOI] [PubMed] [Google Scholar]
- 32.Morton S., Davis R. J., Cohen P. (2004) FEBS Lett. 572, 177–183 [DOI] [PubMed] [Google Scholar]
- 33.Gupta S., Campbell D., Dérijard B., Davis R. J. (1995) Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 34.Livingstone C., Patel G., Jones N. (1995) EMBO J. 14, 1785–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., Angel P. (1995) EMBO J. 14, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead J. R., Hughes T. R., Irvine S. A., Singh N. N., Ramji D. P. (2003) J. Biol. Chem. 278, 17741–17751 [DOI] [PubMed] [Google Scholar]
- 37.Uyttersprot N., Costagliola S., Dumont J. E., Miot F. (1999) Eur. J. Biochem. 259, 370–378 [DOI] [PubMed] [Google Scholar]
- 38.Kameda T., Mizutani T., Minegishi T., Ibuki Y., Miyamoto K. (1999) Biochim. Biophys. Acta 1445, 31–38 [DOI] [PubMed] [Google Scholar]
- 39.Thonberg H., Lindgren E. M., Nedergaard J., Cannon B. (2001) Biochem. J. 354, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain M. A., Daniel P. B., Habener J. F. (2000) Diabetes 49, 1681–1690 [DOI] [PubMed] [Google Scholar]
- 41.Monaco L., Sassone-Corsi P. (1997) Oncogene 15, 2493–2500 [DOI] [PubMed] [Google Scholar]
- 42.Thommesen L., Nørsett K., Sandvik A. K., Hofsli E., Laegreid A. (2000) J. Biol. Chem. 275, 4244–4250 [DOI] [PubMed] [Google Scholar]
- 43.Cook D. N., Pisetsky D. S., Schwartz D. A. (2004) Nat. Immunol. 5, 975–979 [DOI] [PubMed] [Google Scholar]
- 44.Han J., Ulevitch R. J. (2005) Nat. Immunol. 6, 1198–1205 [DOI] [PubMed] [Google Scholar]
- 45.Kawai T., Akira S. (2007) Semin. Immunol. 19, 24–32 [DOI] [PubMed] [Google Scholar]
- 46.Liew F. Y., Xu D., Brint E. K., O‘Neill L. A. (2005) Nature 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 47.Brogden K. A., Guthmiller J. M., Salzet M., Zasloff M. (2005) Nat. Immunol. 6, 558–564 [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez-Rey E., Chorny A., Delgado M. (2007) Nat. Rev. Immunol. 7, 52–63 [DOI] [PubMed] [Google Scholar]
- 49.Ulloa L. (2005) Nat. Rev. Drug Discov. 4, 673–684 [DOI] [PubMed] [Google Scholar]