Abstract
Clusterin (CLU) is an extracellular chaperone that is likely to play an important role in protein folding quality control. This study identified three deposition disease-associated proteins as major plasma clients for clusterin by studying CLU-client complexes formed in response to physiologically relevant stress (shear stress, ∼36 dynes/cm2 at 37 °C). Analysis of plasma samples by size exclusion chromatography indicated that (i) relative to control plasma, stressed plasma contained proportionally more soluble protein species of high molecular weight, and (ii) high molecular weight species were far more abundant when proteins purified by anti-CLU immunoaffinity chromatography from stressed plasma were compared with those purified from control plasma. SDS-PAGE and Western blot analyses indicated that a variety of proteins co-purified with CLU from both stressed and control plasma; however, several proteins were uniquely present or much more abundant when plasma was stressed. These proteins were identified by mass spectrometry as ceruloplasmin, fibrinogen, and albumin. Immunodot blot analysis of size exclusion chromatography fractionated plasma suggested that CLU-client complexes generated in situ are very large and may reach ≥4 × 107 Da. Lastly, sandwich enzyme-linked immunosorbent assay detected complexes containing CLU and ceruloplasmin, fibrinogen, or albumin in stressed but not control plasma. We have previously proposed that CLU-client complexes serve as vehicles to dispose of damaged misfolded extracellular proteins in vivo via receptor-mediated endocytosis. A better understanding of these mechanisms is likely to ultimately lead to the identification of new therapies for extracellular protein deposition disorders.
Keywords: Chaperones, Chaperones/Protein folding, Extracellular Matrix/Fibrinogen, Protein/Degradation, Protein/Folding, Receptors/Endocytosis
Introduction
Processes to attain and maintain native protein conformations are vital for organismal viability. Conditions such as thermal and oxidative stress may cause proteins to partially unfold and aggregate, a process thought to underpin the pathology of many so-called protein deposition diseases, including Alzheimer disease, arthritis, type II diabetes, age-related macular degeneration (ARMD),3 and atherosclerosis (1–5). Although intracellular mechanisms to monitor and control the folding state of proteins are well characterized, corresponding extracellular mechanisms have yet to be established. It has been proposed that clusterin (CLU) is one of a small family of abundant extracellular chaperones that form part of a quality control system that acts to stabilize misfolded, aggregating extracellular proteins, and mediate their clearance from the body via receptor-mediated endocytosis and lysosomal degradation (6–8).
CLU can stabilize proteins and prevent their precipitation during exposure to a variety of stresses in vitro (9–14). This action involves the formation of soluble high molecular weight (HMW) complexes incorporating both CLU and the stressed client protein at an approximate mass ratio of 1:2 (CLU:client); when generated in vitro, these complexes have diameters of 50–100 nm (14). CLU is found associated with extracellular protein deposits in many serious diseases including drusen in ARMD (15), renal immunoglobulin deposits in kidney disease (16), prion deposits in Creutzfeldt-Jakob disease (17), amyloid plaques in Alzheimer disease (18, 19), and also atherosclerotic plaques (20). The presence of CLU in these pathological deposits suggests that it associates with unfolding proteins in vivo. Pathological protein deposition may thus result when the chaperone capacity of CLU and other machinery acting to prevent the accumulation of protein aggregates is exceeded by abnormally high levels of protein unfolding.
In addition to misfolded client proteins, CLU also binds to many native ligands; the nature of these latter interactions remains largely uncharacterized; however, it is believed that CLU has discrete binding sites for native ligands and misfolded client proteins (12, 21). Typically, investigations of the chaperone activity of CLU have been carried out using model proteins that can be induced to unfold at experimentally convenient rates. It was previously shown that depletion of CLU from human plasma increased the extent of plasma protein precipitation at both 60 (11) and 37 °C (22). However, the identity of the major chaperone client proteins for CLU in human plasma was previously unknown. The identity of these client proteins may provide important insights into mechanisms underlying the development of extracellular protein deposition diseases. In this study we exposed human plasma to physiologically relevant stress (gentle rotation to produce shear stress ∼36 dynes/cm2 at 37 °C for 10 days) and used electrophoresis, Western blotting, and mass spectrometry to identify proteins that co-purified with CLU from stressed (but not control) plasma by immunoaffinity chromatography. Immunodot blot assays of fractions from size exclusion chromatography (SEC) and sandwich ELISA were used to confirm that these putative endogenous plasma client proteins formed soluble HMW complexes with CLU in stressed human plasma.
EXPERIMENTAL PROCEDURES
Materials
All of the buffer salts were obtained from Ajax Chemical Co. ortho-Phenylenediamine, 2-β-mercaptoethanol, BCA reagent, goat anti-FGN antiserum, goat anti-CERU antiserum, rabbit anti-HSA antiserum, control goat serum, control rabbit serum, and mouse anti-goat Ig-HRP were from Sigma-Aldrich. Sheep anti-rabbit Ig-HRP and control mouse IgG1 were obtained from Millipore. Purified mouse monoclonal anti-CLU IgG1 (G7) was as described in Ref. 23.
Isolation of Plasma Proteins Co-purifying with CLU
Whole blood supplemented with 20 μm sodium citrate was centrifuged at 1,020 × g for 30 min to pellet cells. The plasma was collected and supplemented with CompleteTM protease inhibitor mixture (Roche Applied Science) and 0.1% (w/v) sodium azide (Az). One 50-ml aliquot was immediately filtered through a GF/C microfiber glass filter (Whatman) and passed at 0.5 ml/min over monoclonal anti-CLU immunoaffinity columns (with an approximate total bed volume of 20 ml), as previously described (25). The columns were subsequently washed with several column volumes of phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, 8 mm Na2HPO4, pH 7.4) containing 0.1% (w/v) Az (i.e. PBS/Az) before the bound protein was eluted using 2 m GdnHCl in PBS, pH 7.4. A second 50-ml aliquot of plasma (from the same batch) was “stressed” as follows: plasma was held in a 100-ml Schott bottle in a Bioline 472 incubator shaker (Edwards Instrument Co.) rotating at 200 rpm at 37 °C for 10 days. This is estimated to correspond to an approximate shear stress of 36 dynes/cm2 (24). Subsequently, this sample was processed as above using the same immunoaffinity procedure. In some cases, where it was not possible to use freshly isolated plasma as the control (i.e. in sandwich ELISA and measurements of turbidity where absorbance readings were required to be obtained concurrently), control plasma (from the same batch) was left static at room temperature for 10 days.
Plasma Protein Precipitation Assays
Total plasma protein precipitation was assessed by microprotein assay and by spectrophotometry. For each plasma sample (control or stressed), three 200-μl aliquots of plasma were filtered using separate 0.45-μm Ultrafree®-MC centrifugal filter devices (Millipore). The precipitate collected on the membranes was extensively washed with PBS. The membranes were then covered with 200 μl of 6 m GdnHCl in PBS and incubated at 60 °C with shaking overnight. Parafilm “M” (Pechiney Plastic Packaging) was used to seal the membrane cups and ensure that no liquid volume was lost during heating. The solutions were diluted 1:50 in PBS before a BCA assay was performed (80). In addition, control or stressed plasma was diluted 1:2 in PBS/Az in a quartz cuvette, and the A360 nm was measured using a WPA Biowave S2100 diode array spectrophotometer (Biochrom). Statistical significance was determined using one-way analysis of variance and Tukey Honestly Significant Difference (HSD).
Size Exclusion Chromatography
SEC of 500 μl of whole plasma or anti-CLU immunoaffinity eluate was carried out using a SuperoseTM 6 10/300 column (GE Healthcare) equilibrated in PBS/Az at the recommended flow rate of between 0.3 and 0.5 ml/min, and the A280 nm was continuously monitored using an ÄKTA fast protein liquid chromatography system (GE Healthcare). Mass standards were from a commercial HMW calibration kit (GE Healthcare). All of the buffers and samples were filtered (0.45 μm) before use. For immunodot blot analysis of whole plasma, 0.5-ml fractions were collected.
SDS-PAGE and Identification of Proteins by Mass Spectrometry
Proteins purified by anti-CLU immunoaffinity chromatography from plasma samples (40 mg of total protein/lane) were separated on 8–15% SDS-PAGE gels using a HoeferTM SE 250/260 SDS-PAGE system (GE Healthcare). Mass spectrometry was outsourced commercially and performed by the Australian Proteome Analysis Facility. Selected Coomassie Blue-stained bands were excised from gels using a scalpel blade. Excised bands were treated in-gel with peptide:N-glycanase F followed by tryptic digestion for 16 h. Matrix-assisted laser desorption ionization mass spectrometry was performed with an Applied Biosystems 4800 Proteomics analyzer. The spectra were acquired in reflectron mode in the mass range 700–3500 Da. The instrument was then switched to tandem mass spectrometry (tandem time-of-flight) mode where peptides from the mass spectrometry scan were isolated and fragmented and then reaccelerated to measure their masses and intensities. The spectra were then examined using the data base search program Mascot (Matrix Science Ltd). High Mowse scores (> 69) in the peptide data base search indicated a likely match (p < 0.05).
Western Blot and Immunodot Blot Analyses
For Western blots, following SDS-PAGE performed as described above (loading 10 μg of total protein into each lane), the gels were subsequently equilibrated in transfer buffer (26 mm Tris, 192 mm glycine, 20% (v/v) methanol, pH 8.3), and the separated proteins were transferred to nitrocellulose membrane using a Mini Trans-Blot Cell Western blotting apparatus (Bio-Rad) at 100 V for 1 h at 4 °C. The membrane was subsequently blocked overnight at 4 °C in 1% (w/v) heat-denatured casein in PBS. Primary and appropriate HRP-conjugated secondary antibodies diluted in heat-denatured casein in PBS following the manufacturer's instructions were incubated in turn with the membrane for 1 h at 37 °C. The membrane was then washed in 0.1% (v/v) Triton X-100 in PBS followed by PBS alone. Enhanced chemiluminescence detection was performed using Supersignal Western Pico substrate (Pierce) following the manufacturer's protocols. Amersham Biosciences HyperfilmTM ECL (GE Healthcare) was placed over the membrane in a Kodak X-Omatic cassette to detect chemiluminescence. Once exposed, the film was removed from the cassette and developed using Kodak developer and fixer. Densitometry was performed using a GS 800 calibrated densitometer (Bio-Rad) and Quantity One software (Bio-Rad). The average optical density/mm2 of the bands was used to estimate their relative quantities.
For immunodot blots, plasma (control or stressed) was fractionated over a SuperoseTM 6 10/300 column as described above. Four microliters of each 0.5-ml fraction was spotted onto nitrocellulose membrane and allowed to dry before a second 4-μl aliquot was applied. The dried membranes were then blocked and processed as described above for Western blot.
Sandwich ELISA
The wells of an ELISA plate (Greiner Bio-one) were coated with 10 mg/ml purified G7 anti-CLU antibody (23) or control mouse IgG1 antibody and then blocked with 1% (w/v) bovine serum albumin in PBS or heat-denatured casein in PBS. Control plasma (left static at room temperature) or stressed plasma was next added to the wells. Subsequently, primary antisera reactive with either CERU, FGN, or HSA diluted in the blocking solution (following the manufacturer's instructions) were added to the appropriate wells. Finally, an appropriate HRP-conjugated secondary antibody diluted in blocking solution (following the manufacturer's instructions) was added. All of the incubations were for 1 h at 37 °C with shaking, and extensive washing with PBS was performed between each step. The final wash was performed using 0.1% (v/v) Triton X-100 in PBS followed by PBS alone. ortho-Phenylenediamine (2.5 mg/ml) and 0.03% (v/v) H2O2 in 50 mm citric acid, 100 mm Na2HPO4, pH 5, was then added to the wells of the plate. The reaction was stopped using 1 m HCl before the A490 nm was measured using a SpectraMax Plus384 microplate reader (Molecular Devices). Nonspecific binding was assessed using species-matched anti-sera of irrelevant specificity and the appropriate secondary antibody. The results presented are adjusted for nonspecific binding by calculating the absorbance in wells coated with G7 relative to the absorbance in wells coated with control mouse IgG1 (of irrelevant specificity) that were treated with the same plasma and primary and secondary antibodies. Statistical significance was determined using Student's t test.
RESULTS
Stress-induced Protein Precipitation
After 10 days, compared with control plasma held at room temperature, stressed plasma was visibly more turbid. The turbidity of control plasma remained unchanged over this period; however, after 10 days the turbidity of stressed plasma was significantly greater than at day 0 and that of control plasma at either time point (Fig. 1; F(3,8) = 433, Tukey HSD, p < 0.0001 in all cases). When performed as described, the BCA assay was unable to detect protein in the filtrates of freshly obtained control plasma but detected a mean ± S.E. (n = 3) of 8.47 ± 0.99 mg of protein in the corresponding filtrates from 200-μl aliquots of stressed plasma.
FIGURE 1.
Turbidity of control and stressed plasma at day 0 and day 10. The plasma samples were diluted 1:2 in PBS on days 0 and 10, and the A360 nm was measured. The figure shows the average A360 nm (n = 3 ± standard error) for each sample. *, significantly increased turbidity relative to the three other sample types (Tukey HSD, p < 0.0001 in all cases). These results are representative of three independent experiments.
Bias toward HMW Species as Detected by SEC of Stressed Plasma and Anti-CLU Co-purifying Proteins
There were differences in the SEC profiles of freshly isolated control and stressed plasma samples (Fig. 2A). Most notably, the respective areas underneath the traces of A280 nm suggest that there was less soluble protein in stressed plasma compared with an equal volume of control plasma. Although there appeared to be less soluble protein in the stressed sample, the amount of protein eluting at the exclusion limit of the column (4 × 107 Da) was similar for both plasma samples (Fig. 2A), indicating that overall a greater proportion of stressed plasma proteins migrated as very large species. SEC profiles of proteins purified by anti-CLU immunoaffinity chromatography from freshly isolated control versus stressed plasma were very different (Fig. 2B). The A280 nm peak eluting at the column exclusion limit was approximately four times greater for proteins immunoaffinity-purified from stressed plasma versus control plasma. Moreover, the majority of species in the former sample were larger than ∼600 kDa, whereas those in the latter sample were predominately smaller in mass (Fig. 2B).
FIGURE 2.
SEC of stressed or control plasma (A) and anti-CLU immunoaffinity-purified proteins (B) from stressed or control plasma. The samples analyzed were: whole plasma exposed to shear stress for 10 days or freshly isolated control plasma (A) and proteins purified by anti-CLU immunoaffinity chromatography from the plasma samples described in A (B). The positions of molecular mass markers are indicated by labeled arrows; the exclusion limit (Vo) ≥ 4 × 107 Da. The results shown are representative of two independent experiments.
Identification of Major Plasma Clients for Clusterin
When equivalent amounts of total protein purified by anti-CLU immunoaffinity chromatography from control and stressed plasma were analyzed side-by-side by reducing SDS-PAGE, many bands were detected in both samples (Fig. 3). A major band at ∼36 kDa, corresponding to the co-migrating α and β subunits of CLU, was detected in both samples; however, this was considerably more prominent in the control sample (Fig. 3). In the sample prepared from control plasma, other protein bands detected probably include those representing ApoA-I (∼28 kDa) and IgG (light chain, ∼23 kDa; heavy chain, ∼50 kDa), which are known to co-purify with CLU from normal human plasma (23, 25), as well as other minor contaminants that can be removed from CLU by subsequent ion exchange chromatography (26). The additional bands that were unique or more prominent in the sample prepared from stressed plasma were also detected (Fig. 3). Several of these bands (Fig. 3, open arrows) were excised and subjected to mass spectrometry analysis (multiple gels of various percentage acrylamide were used to adequately resolve bands for mass spectrometry; however, a single representative gel is shown). These analyses identified CERU, FGN (β chain), and HSA as putative chaperone client proteins for CLU (Table 1).
FIGURE 3.
SDS-PAGE of proteins purified from stressed or control human plasma by anti-CLU immunoaffinity chromatography. The proteins were separated using 12% SDS-PAGE under reducing conditions. The figure shows molecular mass markers (left lane, masses indicated in kDa) and proteins immunoaffinity-purified from freshly isolated control plasma (labeled C) or stressed plasma (labeled S). The open arrows indicate bands that were selected for mass spectrometry analysis. The solid black arrow indicates the position of CLU. The results shown are representative of many independent experiments.
TABLE 1.
Identification of proteins by mass spectrometry
Proteins co-purifying with CLU from stressed plasma (indicated by open arrows in Fig. 3) were analyzed by matrix-assisted laser desorption ionization tandem time-of-flight. The spectra were examined using the data base search program Mascot (Matrix Science Ltd., London, UK). Mowse scores (>69) in the peptide data base search indicated a likely match (p < 0.05).
| Band | Mass | Identity | Mowse score | Coverage | Significance |
|---|---|---|---|---|---|
| kDa | % | ||||
| 1 | 17 | CERU | 175 | 35 | p < 0.05 |
| 2 | 43 | FGN (β chain) | 157 | 54 | p < 0.05 |
| 3 | 67 | HSA and CERU | 398 | 54 | p < 0.05 |
| 97 | 34 | p < 0.05 |
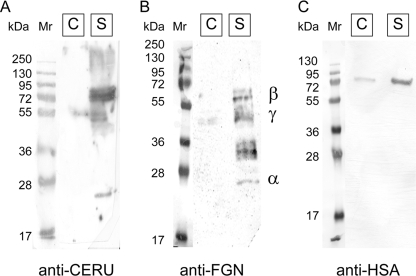
Western blot analysis was used to confirm that CERU, FGN, and HSA co-purified with CLU from stressed plasma but not (or to a lesser extent) from control plasma (Fig. 4). At most, only traces of CERU and FGN were detected in the corresponding protein fractions purified in the same way from control plasma (Fig. 4, A and B). There was measurable HSA in the fraction prepared from control plasma; however, this was reproducibly substantially less than in the sample prepared from stressed plasma (Fig. 4C). A band representing HSA was detected at about the position expected for the intact molecule (69 kDa, Fig. 4C), but the same was not true for CERU. Intact CERU has a mass of 122 kDa; however, it is prone to autolysis in plasma to yield fragments of 67, 53, and 20 kDa (27) that corresponded to the approximate position of the major bands detected on the blot (Fig. 4A). Bands corresponding to the α, β, and γ chains of FGN (25, 56, and 48 kDa, respectively) were detected in the lane containing proteins from stressed plasma. In plasma, FGN is highly susceptible to proteolysis and is present in many fragmented and cross-linked forms (28, 29). Therefore, FGN bands detected between ∼29 and 36 kDa are likely to represent β and γ chain degradation products (Fig. 4B).
FIGURE 4.
Western blot analysis of proteins purified by anti-CLU immunoaffinity chromatography from control or stressed human plasma. The proteins separated by SDS-PAGE (10 μg of total protein loaded per lane) were transferred to nitrocellulose membrane and probed with anti-CERU (A), anti-FGN (B), or anti-HSA antisera (C). Each panel shows the position of molecular mass markers (left lanes, masses in kDa indicated), and proteins immunoaffinity-purified from freshly isolated control plasma (labeled C) or stressed plasma (labeled S). In B, the position of the α, β, and γ subunits of FGN are indicated. The results shown are representative of three independent experiments.
CLU forms HMW complexes with chaperone client proteins in vitro (9, 10, 12, 14). If CLU-client protein complexes formed in situ in plasma are also large, then relative to unstressed control plasma, a greater proportion of CLU and the client proteins would be detected as HMW species in stressed plasma. This was examined by using SEC to fractionate both control and stressed plasma on the basis of molecular size and then probing the fractions with specific antibodies. Fig. 5 shows immunodot blot results for SEC fractions representing species corresponding to between 460 kDa and the exclusion limit of the column (≥4 × 107 Da) in mass probed with specific antibodies for CLU, CERU, FGN, and HSA. Strikingly, CLU and all three client proteins were detected much more strongly in the HMW fractions (≥4 × 107 Da) prepared from stressed plasma compared with the corresponding fractions of control plasma (Fig. 5). Most of the corresponding fractions from control plasma showed little reactivity with antibodies specific for CLU, CERU, FGN, and HSA. This is consistent with the known masses of these individual proteins (CLU, 61 kDa; CERU, 122 kDa; FGN, 340 kDa; HAS, 69 kDa); the bulk of these proteins would elute in later fractions not represented in this figure unless they were present as aggregates or complexes. The limited reactivity detected for the client proteins in the control plasma lanes probably represents low level associations of these proteins with other unidentified plasma components or self-association (see “Discussion”).
FIGURE 5.
Immunodot blot analyses of SEC-fractionated control or stressed human plasma. Freshly isolated control plasma (C) or stressed plasma (S) were fractionated using a SuperoseTM 6 column (V0 ≥ 4 × 107 Da). Aliquots from each fraction were spotted onto a nitrocellulose membrane that was then incubated with antibodies against CLU (A), CERU (B), FGN (C), or HSA (D) followed by the appropriate HRP-conjugated secondary antibody prior to development by ECL. The approximate molecular mass of the SEC fractions is indicated at the top of the figure. The results shown are representative of three independent experiments.
The results thus far indicated a preferential association of CERU, FGN, and HSA with CLU in stressed human plasma. If this association was the result of the chaperone action of CLU, then it would be expected that complexes incorporating both CLU and one or more of these three putative client proteins would be present in stressed plasma but not in control plasma. This was verified by sandwich ELISA. The wells of an ELISA plate were coated with an anti-CLU antibody (or an isotype-matched control antibody) and subsequently incubated with control or stressed plasma. Bound CERU, FGN, or HSA were then detected with specific antibodies. In this assay, absorbance at 490 nm significantly above that of the controls will only occur if species containing both CLU and a putative client protein are specifically bound to the anti-CLU antibody on the wells. For all three putative client proteins tested, significantly greater absorbance was measured in wells incubated with stressed plasma compared with those incubated with control plasma (Fig. 6; CERU, FGN, and HSA t(4) = 9.06, t(4) = 4.85, and t(4) = 4.99, respectively; p ≤ 0.02 in all cases). When irrelevant control antibodies matched by isotype and species to the primary detection antibody were used in the same assay, there was no significant difference between the wells incubated with stressed or control plasma (control antisera results; Fig. 6).
FIGURE 6.
Detection of CLU-client protein complexes in stressed plasma by sandwich ELISA. The results of sandwich ELISA detecting CLU-client protein complexes containing CERU (A), FGN (B), or HSA (C) in human plasma. The results for “control antisera” were obtained using species-matched control antisera as the primary detection antibody in each case. The results shown are the average A490 nm (n = 3, ± standard error) relative to the nonspecific binding generated in wells coated with mouse IgG1 control antibody. *, increased A490 nm relative to wells incubated with control plasma (stored static at 4 °C; Student's t test, p ≤ 0.02). The results shown are representative of three independent experiments.
DISCUSSION
Exposure to physical and chemical stresses, such as elevated temperature, shear stress, oxidative stress, and UV irradiation, challenge all biological systems. One potential impact of these stresses is damage to proteins, inducing misfolding, loss of function, and aggregation. Although much is known about intracellular mechanisms that act to repair or dispose of stress-damaged proteins, little is known about corresponding extracellular mechanisms. When comparing the intracellular and extracellular environments, some stresses that can contribute to protein unfolding are greater in the latter. This includes hydrodynamic shear stress resulting from the hydraulic force of plasma being pumped around the body (30, 31); normal arterial shear stress is between 10 and 70 dynes/cm2 (32). The extracellular environment is also more oxidizing than the cytosol (33). In this study, we used a constant temperature of 37 °C and an estimated shear stress (generated by orbital shaking) of ∼36 dynes/cm2 (calculated using the formula of Ref. 24) to simulate physiologically relevant extracellular conditions. Relative to plasma left stationary at room temperature, plasma stressed in this way for 10 days showed increased turbidity and protein precipitation (Fig. 1). This result indicates that plasma proteins are prone to unfolding and aggregation under conditions of temperature and shear stress that are likely to occur in vivo. If this is the case, then intuitively, the human body must have systems in place to control this problem.
SEC indicated that the level of soluble protein remaining in stressed plasma was less than in batch-matched control plasma and that proportionately more proteins were present as HMW species in stressed plasma (Fig. 2). These findings may be explained by the stress-induced partial unfolding of plasma proteins, which subsequently aggregate to form increasingly large aggregates, some of which eventually become too large to stay in solution and form insoluble precipitate. Superimposed on this process, we suggest that under these conditions CLU forms large soluble complexes with probably many different misfolded plasma client proteins. This has the effect of ameliorating the extent of protein precipitation measured. We have previously shown that the immunoaffinity depletion of CLU from human plasma significantly increased total protein precipitation in this fluid when it was subsequently incubated at either 60 °C (11) or 37 °C (22). Species ≥4 × 107 Da in stressed plasma may be aggregates on their way toward becoming insoluble or may be aggregates stabilized by extracellular chaperones such as CLU. CLU is itself physically very stable and does not aggregate or precipitate even in response to sustained heating at 60 °C (9, 12–14). Thus the apparent depletion of CLU from the pool of soluble proteins in stressed plasma in the current study (Fig. 3) probably results from its incorporation into growing aggregates; under the conditions tested, CLU is unable to maintain the solubility of all plasma proteins. Supporting the involvement of CLU in forming HMW chaperone-client complexes, SEC of proteins purified by anti-CLU immunoaffinity chromatography from stressed and control plasma indicated that, relative to the latter, the former was dominated by HMW protein species. If it had been possible to elute proteins from the anti-CLU columns using nondenaturing conditions (instead of the 2 m GdnHCl used in this study), the difference in mass profile for the two samples might have been even greater. Our recent work showed that when CLU-client protein complexes were generated from purified proteins in vitro, the mass ratio was, for each of three different client proteins, ∼1:2, respectively (14). The complexes in the current study were purified from stressed human plasma by immunoaffinity chromatography, which (unavoidably) involved eluting bound complexes with denaturing conditions. The harsh elution conditions are likely to have led to partial disruption of the complexes, thus making measurements of stoichiometry rather meaningless. For this reason we did not measure the apparent stoichiometry of the complexes in the current study.
SDS-PAGE analyses of proteins bound from control plasma to anti-CLU columns were consistent with our experience in routine purifications of plasma CLU (26). The protein bands detected (additional to CLU) are likely to represent known CLU ligands such as complement components (34), IgG (23) and Apo-A1 (25). However, other bands were detected as uniquely present (or more abundant) in samples prepared from stressed plasma; these bands were regarded as corresponding to putative endogenous plasma client proteins for the chaperone action of CLU. In this study, three bands were selected from one-dimensional SDS-PAGE for further analysis. It is expected that in the future, by applying a protein separation technique with greater resolution such as two-dimensional SDS-PAGE, further putative client proteins will be identified. Previous studies indicate that the chaperone action of CLU is promiscuous (6, 9–14, 35); thus it is likely that CLU will interact with most misfolding proteins, regardless of their identity, and that endogenous CLU-client complexes will contain a heterogeneous mix of client proteins. However, using the approaches described, identification of specific plasma proteins as clients for CLU will depend on their individual relative abundances and stabilities. In the current study, mass spectrometric analysis of the three bands resolved by one-dimensional SDS-PAGE identified them as corresponding to CERU, FGN, and HSA (Fig. 3 and Table 1). Western blot analyses showed that these three putative client proteins were preferentially detected in protein fractions prepared by anti-CLU immunoaffinity chromatography of stressed plasma versus control plasma (Fig. 4). The small amounts of the putative client proteins co-purifying with CLU from control plasma (Fig. 4, control, C lanes) may be the result of low levels of CLU-client complexes present in freshly isolated plasma. However, low level nonspecific binding of client proteins to the anti-CLU immunoaffinity columns may also contribute in this regard, particularly in the case of HSA, which is known to bind to many surfaces.
Species much larger than expected for monomeric CLU, CERU, FGN, and HSA were detected by immunodot blot analysis of SEC-fractionated samples of both control and stressed plasma. CLU is well known to oligomerize in solution to form a variety of HMW aggregates (9, 14) and also to associate with HDL particles in plasma (25); this would account for the large (460 to >737 kDa) CLU-containing species detected in control plasma (Fig. 5A). In the case of CERU, FGN, and HSA, the large (>460 kDa) species containing these proteins detected in control plasma probably result from interactions between them and other plasma proteins. Many examples of such interactions are known; for example, lactoferrin (36), protein C (37), and myeloperoxidase (38) interact with CERU; at least 11 proteins are known to interact with FGN including vitronectin (39), histidine-rich glycoprotein (40), and apolipoprotein(a) (41); and over 60 different proteins are believed to interact with HSA (42). Self-aggregation, which has been reported for both FGN (43) and HSA (44), may also account for the detection of these proteins as larger species. Corresponding analyses of stressed plasma strongly detected CLU and each of the three client proteins CERU, FGN, and HSA in fractions representing still larger species approaching (or at) the exclusion limit of the SEC column (4 × 107 Da; Fig. 5). This observation is consistent with CLU forming large HMW complexes with client proteins in stressed plasma, as has been described for purified proteins in buffered solutions (9, 10, 12, 14). Finally, using sandwich ELISA, we were able to confirm that CLU-CERU, CLU-FGN, and CLU-HSA complexes were present in stressed but not control plasma (Fig. 6). Collectively, these results indicate that under conditions of mild, physiologically relevant stress, CLU forms soluble chaperone-client complexes in plasma containing one or more of CERU, FGN, and HSA.
The observation that CLU is found associated with insoluble protein aggregates in all protein deposition diseases in which this has been examined (45) suggests that CLU (i) binds to unfolding proteins in vivo and (ii) becomes incorporated into protein deposits when its chaperone action is overwhelmed by an excess of misfolded protein. All three CLU client proteins identified in this study are involved in protein deposition diseases. CERU, FGN, and HSA are found in ARMD deposits known as drusen (15, 46–49). Additionally, plasma concentrations of CERU and FGN are higher in ARMD compared with normal patients (50, 51). Co-localization of CLU with drusen proteins is very common in ARMD (15); furthermore, in this same disease, non-drusen FGN deposition is implicated in the atrophy of the retinal pigment epithelium and choroidal neovascularization (52, 53). The progression of both atrophic and neovascular ARMD is supported by platelet activation, which results in secretion of growth factors and monocyte chemoattractants. Platelet activation is enhanced by unfolding of FGN (54) and results in the release of CLU from the platelets by degranulation (55). The effects of the interaction between CLU and stressed FGN on platelet activation are currently unknown; this may have significance not only to ARMD but also to the many ischemic and atherosclerotic vascular conditions where FGN deposition is known to occur (5, 56–58). As in ARMD, FGN deposition in such diseases is associated with the recruitment and activation of platelets and has been proposed as a mechanism for vascular injury (59). Deposition of FGN has been reported in breast cancer (60), mesothelioma (61), colon cancer (62), and lymphoma (63). Although the reasons for this are unknown, one potential implication of this observation is that FGN deposition promotes angiogenesis and cancer progression. FGN deposits are also found in renal disease (64, 65), hereditary renal amyloidosis (66), and systemic lupus erythematosus (67). In Type 1 (insulin-dependent) diabetes mellitus, extracellular deposition of HSA is observed around dermal capillaries (68), kidney (69, 70), skeletal muscle (71), and the thyroid gland (72).
Given the large number of diseases in which extracellular protein deposition occurs (1–5, 8), characterization of mechanisms that clear damaged proteins in healthy individuals is likely to shed light on how protein deposition pathologies arise. In this study we have identified three plasma client proteins that bind to CLU during physiologically relevant stress. Their deposition in numerous conditions suggests that overwhelming or disruption of normal activities that prevent their accumulation in healthy individuals is important in the progression of disease. The in vivo interaction of CLU with these client proteins (and others) in vivo is likely to be an important mechanism to prevent the pathological deposition of misfolded extracellular proteins. Significantly, it has been shown that CLU knock-out mice develop progressive glomerulopathy, which is characterized by the accumulation of insoluble protein deposits in the kidneys (73). This directly implicates CLU in the clearance of potentially pathological aggregating proteins, although the precise mechanism underlying this has yet to be described. It has been proposed to occur via the receptor-mediated endocytosis and subsequent lysosomal degradation of extracellular chaperone-client protein complexes (6). Evidence is rapidly accumulating that CLU and other abundant extracellular chaperones are key elements in a quality control system for extracellular protein folding (6–14, 21, 22, 35, 45, 74–79). This report is an important step toward a more complete understanding of the vital mechanisms involved in extracellular protein folding quality control.
Acknowledgment
Wollongong Hospital kindly donated human blood for use in this study.
This work was supported by Australian Research Council Grant DP0773555.
- ARMD
- age-related macular degeneration
- Az
- azide
- CERU
- ceruloplasmin
- CLU
- clusterin
- FGN
- fibrinogen
- HMW
- high molecular weight
- HSA
- human serum albumin
- PBS
- phosphate-buffered saline
- SEC
- size exclusion chromatography
- ELISA
- enzyme-linked immunosorbent assay
- HRP
- horseradish peroxidase
- GdnHCl
- guanadine hydrochloride.
REFERENCES
- 1.Carrell R. W., Lomas D. A. (1997) Lancet 350, 134–138 [DOI] [PubMed] [Google Scholar]
- 2.Carrell R. W., Gooptu B. (1998) Curr. Opin. Struct. Biol. 8, 799–809 [DOI] [PubMed] [Google Scholar]
- 3.Thomas P. J., Qu B. H., Pedersen P. L. (1995) Trends Biochem. Sci. 20, 456–459 [DOI] [PubMed] [Google Scholar]
- 4.Soto C. (2001) FEBS Lett. 498, 204–207 [DOI] [PubMed] [Google Scholar]
- 5.Mullins R. F., Russell S. R., Anderson D. H., Hageman G. S. (2000) FASEB J. 14, 835–846 [PubMed] [Google Scholar]
- 6.Yerbury J. J., Stewart E. M., Wyatt A. R., Wilson M. R. (2005) EMBO Rep. 6, 1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson M. R., Yerbury J. J., Poon S. (2008) Mol. BioSyst. 4, 42–52 [DOI] [PubMed] [Google Scholar]
- 8.Wyatt A. R., Yerbury J. J., Poon S., Wilson M. R. (2009) Curr. Med. Chem. 16, 2855–2866 [DOI] [PubMed] [Google Scholar]
- 9.Humphreys D. T., Carver J. A., Easterbrook-Smith S. B., Wilson M. R. (1999) J. Biol. Chem. 274, 6875–6881 [DOI] [PubMed] [Google Scholar]
- 10.Wilson M. R., Easterbrook-Smith S. B. (2000) Trends Biochem. Sci. 25, 95–98 [DOI] [PubMed] [Google Scholar]
- 11.Poon S., Rybchyn M. S., Easterbrook-Smith S. B., Carver J. A., Pankhurst G. J., Wilson M. R. (2002) J. Biol. Chem. 277, 39532–39540 [DOI] [PubMed] [Google Scholar]
- 12.Poon S., Easterbrook-Smith S. B., Rybchyn M. S., Carver J. A., Wilson M. R. (2000) Biochemistry 39, 15953–15960 [DOI] [PubMed] [Google Scholar]
- 13.Poon S., Treweek T. M., Wilson M. R., Easterbrook-Smith S. B., Carver J. A. (2002) FEBS Lett. 513, 259–266 [DOI] [PubMed] [Google Scholar]
- 14.Wyatt A. R., Yerbury J. J., Wilson M. R. (2009) J. Biol. Chem. 284, 21920–21927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabb J. W., Miyagi M., Gu X., Shadrach K., West K. A., Sakaguchi H., Kamei M., Hasan A., Yan L., Rayborn M. E., Salomon R. G., Hollyfield J. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14682–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French L. E., Tschopp J., Schifferli J. A. (1992) Clin. Exp. Immunol. 88, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freixes M., Puig B., Rodríguez A., Torrejón-Escribano B., Blanco R., Ferrer I. (2004) Acta Neuropathol. 108, 295–301 [DOI] [PubMed] [Google Scholar]
- 18.Ghiso J., Matsubara E., Koudinov A., Choi-Miura N. H., Tomita M., Wisniewski T., Frangione B. (1993) Biochem. J. 293, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calero M., Rostagno A., Matsubara E., Zlokovic B., Frangione B., Ghiso J. (2000) Microsc. Res. Tech. 50, 305–315 [DOI] [PubMed] [Google Scholar]
- 20.Jordan-Starck T. C., Lund S. D., Witte D. P., Aronow B. J., Ley C. A., Stuart W. D., Swertfeger D. K., Clayton L. R., Sells S. F., Paigen B., Harmony J. A. (1994) J. Lipid Res. 35, 194–210 [PubMed] [Google Scholar]
- 21.Lakins J. N., Poon S., Easterbrook-Smith S. B., Carver J. A., Tenniswood M. P., Wilson M. R. (2002) Biochemistry 41, 282–291 [DOI] [PubMed] [Google Scholar]
- 22.French K., Yerbury J. J., Wilson M. R. (2008) Biochemistry 47, 1176–1185 [DOI] [PubMed] [Google Scholar]
- 23.Wilson M. R., Easterbrook-Smith S. B. (1992) Biochim. Biophys. Acta 1159, 319–326 [DOI] [PubMed] [Google Scholar]
- 24.Dardik A., Chen L., Frattini J., Asada H., Aziz F., Kudo F. A., Sumpio B. E. (2005) J. Vasc. Surg. 41, 869–880 [DOI] [PubMed] [Google Scholar]
- 25.Jenne D. E., Lowin B., Peitsch M. C., Böttcher A., Schmitz G., Tschopp J. (1991) J. Biol. Chem. 266, 11030–11036 [PubMed] [Google Scholar]
- 26.Park D. C., Yeo S. G., Wilson M. R., Yerbury J. J., Kwong J., Welch W. R., Choi Y. K., Birrer M. J., Mok S. C., Wong K. K. (2008) Neoplasia 10, 964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingston I. B., Kingston B. L., Putnam F. W. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5377–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connaghan D. G., Francis C. W., Lane D. A., Marder V. J. (1985) Blood 65, 589–597 [PubMed] [Google Scholar]
- 29.Gorinstein S., Caspi A., Goshev I., Aksu S., Salnikow J., Scheler C., Delgado-Licon E., Rosen A., Weisz M., Libman I., Trakhtenberg S. (2003) J. Agr. Food Chem. 51, 822–827 [DOI] [PubMed] [Google Scholar]
- 30.Ker Y. C., Chen R. H. (1998) Lebenson Wiss Technol. 31, 107–113 [Google Scholar]
- 31.Schneider S. W., Nuschele S., Wixforth A., Gorzelanny C., Alexander-Katz A., Netz R. R., Schneider M. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7899–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malek A. M., Alper S. L., Izumo S. (1999) JAMA 282, 2035–2042 [DOI] [PubMed] [Google Scholar]
- 33.Sitia R., Braakman I. (2003) Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 34.Tschopp J., Chonn A., Hertig S., French L. E. (1993) J. Immunol. 151, 2159–2165 [PubMed] [Google Scholar]
- 35.Yerbury J. J., Poon S., Meehan S., Thompson B., Kumita J. R., Dobson C. M., Wilson M. R. (2007) FASEB J. 21, 2312–2322 [DOI] [PubMed] [Google Scholar]
- 36.Zakharova E. T., Shavlovski M. M., Bass M. G., Gridasova A. A., Pulina M. O., De Filippis V., Beltramini M., Di Muro P., Salvato B., Fontana A., Vasilyev V. B., Gaitskhoki V. S. (2000) Arch. Biochem. Biophys. 374, 222–228 [DOI] [PubMed] [Google Scholar]
- 37.Walker F. J., Fay P. J. (1990) J. Biol. Chem. 265, 1834–1836 [PubMed] [Google Scholar]
- 38.Segelmark M., Persson B., Hellmark T., Wieslander J. (1997) Clin. Exp. Immunol. 108, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podor T. J., Campbell S., Chindemi P., Foulon D. M., Farrell D. H., Walton P. D., Weitz J. I., Peterson C. B. (2002) J. Biol. Chem. 277, 7520–7528 [DOI] [PubMed] [Google Scholar]
- 40.Schinke T., Koide T., Jahnen-Dechent W. (1997) FEBS Lett. 412, 559–562 [DOI] [PubMed] [Google Scholar]
- 41.Klose R., Fresser F., Kochl S., Parson W., Kapetanopoulos A., Fruchart-Najib J., Baier G., Utermann G. (2000) J. Biol. Chem. 275, 38206–38212 [DOI] [PubMed] [Google Scholar]
- 42.Zhou M., Lucas D. A., Chan K. C., Issaq H. J., Petricoin E. F., 3rd, Liotta L. A., Veenstra T. D., Conrads T. P. (2004) Electrophoresis 25, 1289–1298 [DOI] [PubMed] [Google Scholar]
- 43.Kim S. H., Haimovich-Caspi L., Omer L., Yu C. M., Talmon Y., Wang N. H., Franses E. I. (2007) Langmuir 23, 5657–5664 [DOI] [PubMed] [Google Scholar]
- 44.Atmeh R. F., Shabsoug B. (1997) Electrophoresis 18, 2055–2058 [DOI] [PubMed] [Google Scholar]
- 45.Wilson M. R., Yerbury J., Poon S. (2008) in Heat Shock Proteins and the Brain: Implications for Neurodegenerative Diseases and Neuroprotection (Asea A., Brown I. eds) Springer Science, Berlin, Germany [Google Scholar]
- 46.Sakaguchi H., Miyagi M., Shadrach K. G., Rayborn M. E., Crabb J. W., Hollyfield J. G. (2002) Exp. Eye Res. 74, 547–549 [DOI] [PubMed] [Google Scholar]
- 47.Umeda S., Suzuki M. T., Okamoto H., Ono F., Mizota A., Terao K., Yoshikawa Y., Tanaka Y., Iwata T. (2005) FASEB J. 19, 1683–1685 [DOI] [PubMed] [Google Scholar]
- 48.Hageman G. S., Luthert P. J., Victor Chong N. H., Johnson L. V., Anderson D. H., Mullins R. F. (2001) Prog. Retin. Eye Res. 20, 705–732 [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues E. B. (2007) Ophthalmologica 221, 143–152 [DOI] [PubMed] [Google Scholar]
- 50.Newsome D. A., Swartz M., Leone N. C., Hewitt A. T., Wolford F., Miller E. D. (1986) Invest. Ophthalmol. Vis. Sci. 27, 1675–1680 [PubMed] [Google Scholar]
- 51.Smith W., Mitchell P., Leeder S. R., Wang J. J. (1998) Arch. Ophthalmol. 116, 583–587 [DOI] [PubMed] [Google Scholar]
- 52.Shiose S., Hata Y., Noda Y., Sassa Y., Takeda A., Yoshikawa H., Fujisawa K., Kubota T., Ishibashi T. (2004) Graefes Arch. Clin. Exp. Ophthalmol. 242, 777–783 [DOI] [PubMed] [Google Scholar]
- 53.van der Schaft T. L., Mooy C. M., de Bruijn W. C., de Jong P. T. (1993) Br. J. Ophthalmol. 77, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbucci R., Lamponi S., Magnani A. (2003) Biomacromolecules 4, 1506–1513 [DOI] [PubMed] [Google Scholar]
- 55.Witte D. P., Aronow B. J., Stauderman M. L., Stuart W. D., Clay M. A., Gruppo R. A., Jenkins S. H., Harmony J. A. (1993) Am. J. Pathol. 143, 763–773 [PMC free article] [PubMed] [Google Scholar]
- 56.Badimon J. J., Lettino M., Toschi V., Fuster V., Berrozpe M., Chesebro J. H., Badimon L. (1999) Circulation 99, 1780–1787 [DOI] [PubMed] [Google Scholar]
- 57.Mailhac A., Badimon J. J., Fallon I. T., Fernández-Ortiz A., Meyer B., Chesebro J. H., Fuster V., Badimon L. (1994) Circulation 90, 988–996 [DOI] [PubMed] [Google Scholar]
- 58.Ernst E., Resch K. L. (1993) Ann. Intern. Med. 118, 956–963 [DOI] [PubMed] [Google Scholar]
- 59.Massberg S., Enders G., Matos F. C., Tomic L. I., Leiderer R., Eisenmenger S., Messmer K., Krombach F. (1999) Blood 94, 3829–3838 [PubMed] [Google Scholar]
- 60.Costantini V., Zacharski L. R., Memoli V. A., Kudryk B. J., Rousseau S. M., Stump D. C. (1991) Cancer Res. 51, 354–358 [PubMed] [Google Scholar]
- 61.Wojtukiewicz M. Z., Zacharski L. R., Memoli V. A., Kisiel W., Kudryk B. J., Rousseau S. M., Stump D. C. (1989) Thromb Res. 55, 279–284 [DOI] [PubMed] [Google Scholar]
- 62.Wojtukiewicz M. Z., Zacharski L. R., Memoli V. A., Kisiel W., Kudryk B. J., Rousseau S. M., Stump D. C. (1989) J. Thromb. Haemost. 62, 1062–1066 [PubMed] [Google Scholar]
- 63.Costantini V., Zacharski L. R., Memoli V. A., Kisiel W., Kudryk B. J., Rousseau S. M., Stump D. C. (1992) J. Lab. Clin. Med. 119, 124–131 [PubMed] [Google Scholar]
- 64.Koffler D., Paronetto F. (1965) J. Clin. Invest. 44, 1665–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koffler D., Paronetto F. (1966) Am. J. Pathol. 49, 383–395 [PMC free article] [PubMed] [Google Scholar]
- 66.Uemichi T., Liepnieks J. J., Benson M. D. (1994) J. Clin. Invest. 93, 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dujovne I., Pollak V. E., Pirani C. L., Dillard M. G. (1972) Kidney Int. 2, 33–50 [DOI] [PubMed] [Google Scholar]
- 68.Chavers B., Etzwiler D., Barbosa J., Bach F. H., Michael A. F. (1984) Diabetalogia 26, 415–419 [DOI] [PubMed] [Google Scholar]
- 69.Westberg N. G., Michael A. F. (1972) Diabetes 21, 163–168 [DOI] [PubMed] [Google Scholar]
- 70.Miller K., Michael A. F. (1976) Diabetes 25, 701–708 [DOI] [PubMed] [Google Scholar]
- 71.Cohn R. A., Mauer S. M., Barbosa J., Michael A. F. (1978) Lab. Invest. 39, 13–16 [PubMed] [Google Scholar]
- 72.Raij J., Michael A. F. (1981) The Lancet 1, 671. [DOI] [PubMed] [Google Scholar]
- 73.Rosenberg M. E., Girton R., Finkel D., Chmielewski D., Barrie A., 3rd, Witte D. P., Zhu G., Bissler J. J., Harmony J. A., Aronow B. J. (2002) Mol. Cell. Biol. 22, 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hatters D. M., Wilson M. R., Easterbrook-Smith S. B., Howlett G. J. (2002) Eur. J. Biochem. 269, 2789–2794 [DOI] [PubMed] [Google Scholar]
- 75.Carver J. A., Rekas A., Thorn D. C., Wilson M. R. (2003) IUBMB Life 55, 661–668 [DOI] [PubMed] [Google Scholar]
- 76.Yerbury J. J., Rybchyn M. S., Easterbrook-Smith S. B., Henriques C., Wilson M. R. (2005) Biochemistry 44, 10914–10925 [DOI] [PubMed] [Google Scholar]
- 77.Stewart E. M., Aquilina J. A., Easterbrook-Smith S. B., Murphy-Durland D., Jacobsen C., Moestrup S., Wilson M. R. (2007) Biochemistry 46, 1412–1422 [DOI] [PubMed] [Google Scholar]
- 78.Kumita J. R., Poon S., Caddy G. L., Hagan C. L., Dumoulin M., Yerbury J. J., Stewart E. M., Robinson C. V., Wilson M. R., Dobson C. M. (2007) J. Mol. Biol. 369, 157–167 [DOI] [PubMed] [Google Scholar]
- 79.Yerbury J. J., Kumita J. R., Meehan S., Dobson C. M., Wilson M. R. (2009) J. Biol. Chem. 284, 4246–4254 [DOI] [PubMed] [Google Scholar]
- 80.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]