Abstract
IIp45 (aka MIIP) is a newly discovered gene whose protein product inhibits cell migration. HDAC6 is a class IIb deacetylase that specifically deacetylates α-tubulin, modulates microtubule dynamics, and promotes cell migration. A yeast two-hybrid assay using IIp45 as bait identified HDAC6 protein as a binding partner of IIp45. This physical interaction of the two functionally antagonistic proteins was confirmed by glutathione S-transferase pulldown assay and co-immunoprecipitation assay in human cells. Serial deletion constructs of HDAC6 were used to characterize the interaction of HDAC6 and IIp45, and this analysis found that the two catalytic domains of HDAC6 protein are required for IIp45 binding. We examined the protein expression patterns of IIp45 and HDAC6 in glioma tissues. Elevated protein levels of HDAC6 were found in high grade glioma samples, in contrast to the decreased protein expression of IIp45. The potential negative regulation of HDAC6 expression by IIp45 was confirmed in cell lines with altered IIp45 expression by constitutive overexpression or small interfering RNA knockdown. Protein turnover study revealed that overexpression of IIp45 significantly reduces the intracellular protein stability of endogenous HDAC6, indicating a possible mechanism for the negative regulation of HDAC6 by IIp45. Results from the HDAC activity assay demonstrated that overexpressed IIp45 effectively decreases HDAC6 activity, increases acetylated α-tubulin, and reduces cell migration. The increased cell migration resulting from siIIp45 knockdown was significantly reversed by co-transfection of siHDAC6. Thus, we report here for the first time a novel mechanism by which IIp45 inhibits cell motility through inhibition of HDAC6.
Keywords: Cell, Cell/Migration, Cell/Motility, Cytoskeleton/Microtubules, Cytoskeleton/Tubulin, Diseases/Cancer, Diseases/Neurological, HDAC6, IIp45, Glioma
Introduction
Cell motility is important for normal tissue development and remodeling as well as for pathological conditions such as cancer invasion and metastasis (1, 2). Cell motility is controlled by well orchestrated programs that modulate the microtubule stabilization and destabilization cycles that permit cells to move efficiently (3). One of the key regulatory mechanisms of microtubule dynamics is acetylation of α-tubulin (4). Acetylation of protein lysine residues is a well recognized post-translational modification that modulates various pathways of cellular signal transduction. Protein acetylation is regulated by a number of acetyltransferases and deacetylases, which include the histone deacetylase (HDAC)2 family (5). Among the 18 HDAC family members that have been reported in humans, HDAC6, classified as a type II histone deacetylase, is the only member that has two functional deacetylase domains and a zinc finger motif (6). Both catalytic domains are required for HDAC6 enzymatic activity (7). HDAC6 is also unique in that it is localized mainly in the cytoplasm and specifically deacetylates α-tubulin, the most abundant microtubule component, rather than histone proteins (8). In addition to α-tubulin, heat shock protein 90 (hsp90) and cortactin are two well recognized substrate proteins for HDAC6-mediated deacetylation (9). Hsp90 is a highly conserved molecular chaperone that interacts with >100 client proteins and regulates various cell signaling and cellular response pathways, including many that are activated in cancers (10–13). Cortactin is a protein that promotes polymerization and rearrangement of the actin cytoskeleton and cell migration (14–16). A number of studies have shown that HDAC6 regulates microtubule dynamics/cytoskeleton structure and increases cell migration through reduction of α-tubulin and cortactin acetylation (7–9, 17). HDAC6 has been proposed as a therapeutic target, and there is a significant ongoing effort to develop specific and effective HDAC6 inhibitors for various diseases, including cancer (for review, see Refs. 18–20).
We previously reported a cell migration inhibitory protein, invasion inhibitory protein 45 (IIp45), that is capable of interacting with insulin-like growth factor-binding protein 2 (IGFBP2), a cell migration-promoting protein, and attenuating IGFBP2-mediated cell migration (21). However, IIp45 also inhibits cell motility in cells that express low levels of IGFBP2, suggesting that IGFBP2 is not the only target for the inhibitory action of IIp45 on cell migration.
To search for other potential targets of IIp45 in the regulation of cell migration, we performed a yeast two-hybrid assay using IIp45 as bait and subsequently identified HDAC6 as an IIp45-binding protein. The physical interaction between IIp45 and HDAC6 was further confirmed by glutathione S-transferase (GST) pulldown and co-immunoprecipitation (co-IP) assays using human cells. We found that the levels of HDAC6 protein were frequently high in advanced gliomas. In contrast, the expression of IIp45 was low in these gliomas, consistent with our previous observation that IIp45 expression was not detectable in many of tested glioblastoma tissue samples (21).
The knowledge that HDAC6 and IIp45 function in an antagonistic fashion in the modulation of cell migration, together with the observation of direct binding and inversed protein expression patterns of IIp45 and HDAC6 in brain tumors, led us to hypothesize that IIp45 utilizes an alternative mechanism for inhibition of cell migration through HDAC6. The inversed expressed pattern was reconstituted in cells with IIp45-siRNA knockdown, as well as in IIp45-overexpressing cells. Protein turnover analysis revealed that overexpression of IIp45 significantly reduces the intracellular protein stability of endogenous HDAC6. Direct binding of IIp45 to the catalytic domains of HDAC6 results in decreased HDAC6 activity and resultant increased levels of acetylated α-tubulin, culminating in reduced cell migration. The increased cell migration produced by siIIp45 knockdown was reversed by co-transfection of siHDAC6. Thus, we report here a novel mechanism by which IIp45 inhibits cell motility through inhibition of HDAC6.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Routine chemicals, protein G-agarose, and antibodies against acetylated tubulin, α- and β-tubulin, HA and FLAG tag, as well as agarose beads conjugated with anti-HA and anti-FLAG antibodies, were purchased from Sigma. Antibody against human IIp45 was generated as described previously (21). Anti-HDAC6 and anti-actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). SuperSignal Chemiluminescent reagents were purchased from Thermo Fisher Scientific (Rockford, IL). Negative control siRNA, siRNA of HDAC6 (ID 120451), and IIp45 (ID 123298) were purchased from Ambion (Austin, TX). [35S]Methionine was from PerkinElmer Life Sciences. The HDAC assay kit was from Abcam (Cambridge, MA), including the HDAC inhibitor trichostatin (TSA). Tubacin was kindly provided by Drs. James E. Bradner and Ralph Mazitschek of Harvard Medical School (Cambridge, MA).
Primary Glioma Tissues
All primary glioma tissues were obtained from the Brain Tumor Tissue Bank of the M. D. Anderson Cancer Center with the approval of the institutional review board.
Cell Culture, Expression Vectors, and IIp45 Stable Cell Lines
IIp45/pcDNA3.1 was described previously (21). FLAG-tagged wild-type HDAC6 and deletion constructs were kindly provided by Dr. T. P. Yao at Duke University (Durham, NC) and described previously (22). The C-terminal HA-tagged wild-type IIp45 construct was customer made by OriGene Technologies, Inc. (Rockville, MD), based on the Precision Shuttle vector pCMV6-AC-HA (PS100004).
The HEK293T cell line and glioma cell lines were continually maintained in Dulbecco's modified Eagle's medium and Dulbecco's modified Eagle's medium/F-12 medium, respectively, supplemented with 10% fetal calf serum and incubated at 37 °C in 5% CO2. The human glioma cell line U87MG was purchased from the American Type Culture Collection. The other two human glioma cell lines, LN229 and SNB19, were obtained from the Brain Tumor Center at the University of Texas M. D. Anderson Cancer Center.
Transfection and Western Blotting
Transfections of plasmid DNA and siRNA were performed using Lipofectamine 2000 reagent and its protocol (Invitrogen). Briefly, cells were subcultured on dishes or multiwell plates at a cell density of 30% for siRNA or 50% for plasmid DNA. Transfections were performed the next day using 24 μg of plasmid DNA/100-mm dish or a final concentration of 20 nm (siIIp45) or 40 nm (siHDAC6) with negative control siRNA. Cells were collected 2 days after transfection for cell migration assay and for cell lysate or RNA preparation for Western blotting, HDAC activity assay and real time RT-PCR, respectively.
Freshly selected positive IIp45 stable-transfected cells of LN229 and U87MG were obtained after 2 weeks selection following the procedure as described previously (21, 23) with minor modifications. Western blotting was performed using the SuperSignal Chemiluminescent method.
Yeast Two-hybrid Screening Analysis
The full-length human IIp45 cDNA was inserted into the pGBKT7 bait vector containing the GAL4 DNA binding domain to form in-frame IIp45-BD fusion protein at the EcoRI site. The recombinant IIp45-BD plasmid was used as bait to screen a human fetal brain cDNA library constructed in the pACT2 vector (Clontech, Mountain View, CA) under the selection of histidine, adenine, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Transformants were screened and evaluated according to the Matchmaker user manual provided by the vendor. Genes of positive clones were identified by DNA sequence analysis. The full-length cDNA of HDAC6 was obtained from human fetal brain total RNA by RT-PCR cloning and confirmed by DNA sequencing.
GST Pulldown Assay
GST pulldown assay was performed as described previously (21) using purified GST-IIp45 fusion protein produced by GST-IIp45/pGEX-2T fusion construct. Cell lysates prepared from LN229- and HDAC6-transfected HEK293T cells.
Co-IP Assays
Cells from untransfected cells and cells transfected with wild-type or each of the deletion constructs were collected 2 days after transfection. IIp45-HDAC6 reciprocal co-IP and binding domain analysis were performed using cell lysates prepared in Nonidet P-40 (1%) lysis buffer containing protease inhibitors with a protein concentration of 250 μg/ml. Samples (80% of each of the total cell lysate) were incubated at 4 °C overnight with 20 μl of anti-HA or anti-FLAG beads or 25 μl of protein G-agarose (1:1 slurry) conjugated with anti-IIp45 polyclonal antibody from rabbit or anti-HDAC6 polyclonal antibody from goat. The immunoprecipitated proteins were resolved on SDS-PAGE and analyzed by Western blotting for detection of HDAC6 or IIp45. Original cell lysates were used for protein expression as input quantification (10% or 20% of total lysate of each sample) using β-actin as loading control.
Real Time RT-PCR
RNA samples for real time RT-PCR were prepared from aliquots of siIIp45-treated SNB19 cells as well as IIp45 stable-transfected LN229 cells and their matched control cells. Real time RT-PCR was performed on the ABI Prism 7900 (Applied Biosystems) using the commercially available TaqMan gene expression assay for human HDAC6 (Assay ID Hs00195869_m1) and housekeeping gene human GAPDH (Assay ID 4326317E) as control. The 7900 Sequence Detection System 2.3 software was used to determine the fold-change for HDAC6 using the δδCt method with 95% confidence.
Cell Migration Assays
2 × 105 cells/well of each sample were seeded in triplicate on a Transwell polycarbonate chamber with a pore size of 8 μm (Becton Dickinson Biosciences,) as described (21) and incubated for 3–15 h at 37 °C in 5% CO2. Migrated cells were then fixed and stained following the protocol provided by the vendor and counted under a microscope. Mean value ± S.E. were calculated based on three independent experiments.
Metabolic Labeling
Metabolic labeling of cultured cells was performed by pulse-chase experiment. Cells were grown in 100-mm-diameter dishes and subcultured into 60-mm dishes at 1:5 ratios upon confluence. On the next day, cells were incubated with 1 ml of methionine-free Dulbecco's modified Eagle's medium (Invitrogen) for 1 h. For metabolic labeling (pulse), 10 μl of [35S]methionine (14 mCi/ml)/dish was added and incubated with cells for 30 min. The labeling medium was then removed, and cells were washed with phosphate-buffered saline. Dishes of the starting time point 0 were collected on ice and later harvested in Nonidet P-40 cell lysis buffer containing 0.05 m Tris-HCI, pH 7.4, 0.15 m NaCl, 1 mm phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40. The rest of dishes were continuously incubated in 1 ml of Dulbecco's modified Eagle's medium containing 0.2 mm excess unlabeled l-methionine (chase). Dishes were collected on ice at desired time points during the chase period of time, and cells were then harvested and lysed in 0.5% Nonidet P-40 lysis buffer for immunoprecipitation and then separated by SDS-PAGE. Metabolically labeled and immunoprecipitated protein was detected by fluorography. Quantitative analysis was performed by densitometry using IMAGE software from the National Institutes of Health.
HDAC Assay
Following the protocol provided by the vender, HDAC activity was measured using a HDAC activity assay kit (colorimetric) from Abcam, in which an established HDAC inhibitor TSA is included as control. Briefly, protein concentration of cell lysates prepared from control and experimental groups was determined by protein assay (Bradford method; Bio-Rad). Cell lysates containing 100 μg of protein with and without HDAC inhibitor (0.5 μm for TSA, 1 μm for tubacin) were used in each of duplicated wells for each sample and incubated for 1 h at 37 °C in reaction buffer with the substrate of a synthetic acetylated lysine. The samples were then incubated with color developer for 30 min. The amount of released deacetylated product was measured by absorption at 405 nm using an enzyme-linked immunosorbent assay plate reader (Dynatech, U.K.). HDAC activity is represented as the mean value of O.D. ± S.E. adjusted by blank control in 100 μg of protein based on three independent assays.
Statistical Analysis
Significant difference in the comparison of two experimental groups was determined by t test (p < 0.05). GraphPad Prism 5 software (GraphPad Software, La Jolla, CA) was used for correlation analysis.
RESULTS
IIp45 Binds Directly to the Catalytic Domains of HDAC6
A yeast two-hybrid assay with IIp45 as bait was used initially to explore the possibility of physical interactions of IIp45 with other established proteins that are involved in the regulation of cell migration. After two rounds of screening, HDAC6 was identified as a binding partner of IIp45.
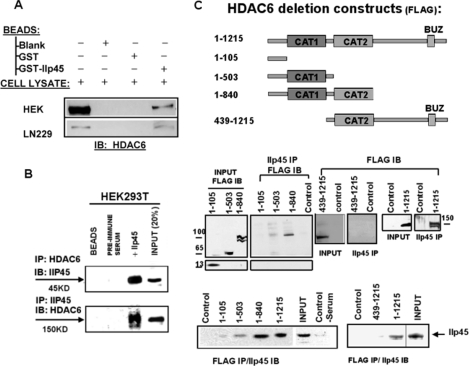
To confirm that IIp45 indeed binds to HDAC6 in human cells, we carried out GST pulldown assays using GST-IIp45 fusion protein with total proteins extracted from HEK293T cells and LN229 glioma cells. The results showed that HDAC6 protein is pulled down by GST-IIp45 as detected by anti-HDAC6 antibody (Fig. 1A). Next, we performed a reciprocal co-IP assay followed by Western blotting with HEK293T cellular extracts. Results showed that IIp45 and HDAC6 form a complex (Fig. 1B).
FIGURE 1.
Interaction between IIp45 and HDAC6. A, GST pulldown assay. Cell lysates of LN229 cells and HDAC6-transfected HEK293T cells were subjected to GST pulldown assay with glutathione-Sepharose beads, GST, and GST-IIp45 fusion protein, followed by SDS/PAGE and Western blot analysis using anti-HDAC6 antibody. B, IIp45-HDAC6 co-IP in HEK293T cells using anti-IIp45 or anti-HDAC6 antibody with preimmune serum or blank protein G beads as negative control. Proteins bound to protein G beads were collected, washed, and subjected to SDS-PAGE/Western blot (IB) analysis. C, binding domain analyses of HDAC6 for interaction with IIp45 in HDAC6-FLAG construct-transfected HEK293T cells. Top panel, schematic description of the constructs of FLAG-tagged wild-type HDAC6 and four truncated HDAC6: HDAC6(1–105), from which two catalytic domains (CAT1 and CAT2) are deleted; HDAC6(1–503), which contains the CAT1 but not the CAT2 domain; HDAC6(1–840), which contains both CAT1 and CAT2; and HDAC6 (493–1215), which contains the CAT2 but not the CAT1 domain. Middle and bottom panels, reciprocal co-IP assay of deletion HDAC6 and endogenous IIp45 in HEK293T cells transfected with wild-type or each of the deletion constructs using monoclonal anti-FLAG immunoprecipitation (IP) and anti-IIp45 serum from rabbit. INPUT, immunoblot of steady level of HDAC6 or IIp45 in cell lysates (10–20% of the amount, as indicated, of the same cell lysate sample used for immunoprecipitation).
To characterize further the binding domains on HDAC6 for interaction with IIp45, expression vectors for wild-type and serial deleted HDAC6 with C-terminal FLAG tag (Fig. 1C) were transfected into HEK293T cells followed by co-IP Western blotting assays to examine binding with endogenous IIp45. HEK293T cells were used because they express detectable IIp45 and relatively low levels of endogenous HDAC6. We used an anti-IIp45 antibody to immunoprecipitate endogenous IIp45 protein and an anti-FLAG antibody to detect HDAC6 protein in the immunoprecipitated complexes. As shown in Fig. 1C, HDAC6(1–105), from which two catalytic domains (CAT1 and CAT2) are deleted, did not bind to IIp45. HDAC6(1–503), which contains the CAT1 domain but not the CAT2 domain, weakly but detectably bound to IIp45. HDAC6(493–1215), which contains the CAT2 domain but not the CAT1 domain, showed no detectable binding to IIp45. HDAC6(1–840), which contains both CAT1 and CAT2 domains, exhibited strong binding to IIp45, comparable with that seen with wild-type HDAC6. These results suggest that the presence of both catalytic domains in HDAC6 is required for efficient HDAC6 binding to IIp45. A reciprocal co-IP assay using a monoclonal anti-FLAG antibody for immunoprecipitation followed by anti-IIp45 antiserum immunoblotting confirmed these findings (Fig. 1C, bottom panel).
Protein Expression of IIp45 and HDAC6 Are Inversely Correlated in Gliomas and Human Cell Lines
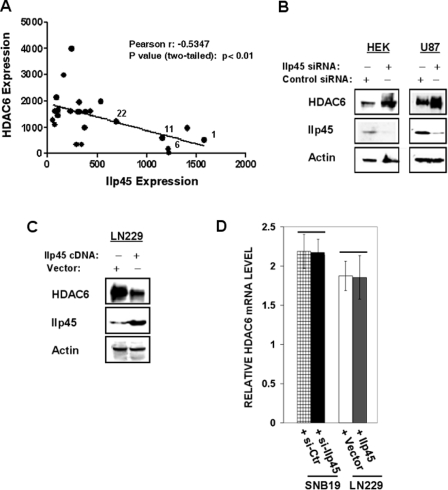
IIp45 and HDAC6 play antagonistic roles in the control of cell migration. Because the role of HDAC6 in gliomas was not known previously, we first examined the expression of HDAC6 and IIp45 in 24 randomly selected human glioma tissues, including both low grade (gliomas 1, 6, 11, and 22) and high grade gliomas (gliomas 2–4, 7–10, 12–21, and 23 and 24). IIp45 expression was decreased in advanced gliomas compared with the lower grade gliomas. In contrast, HDAC6 expression was found to be increased in high grade gliomas. Correlation analysis showed an inverse pattern of IIp45 and HDAC6 protein expression in glioma tissue samples (Fig. 2A), suggesting that there is a negative regulatory relationship between these two proteins.
FIGURE 2.
Inversed protein expression pattern of IIP45 and HDAC6 in gliomas and human cell lines. A, protein extracts of four low grade glioma tissue samples (gliomas 1, 6, 11, and 22) and 20 high grade gliomas (glioblastoma multiforme) tissue samples analyzed for HDAC6 and IIp45. The level of β-actin was measured as a loading control. The results were quantified with densitometry and normalized with β-actin. Statistic correlation analysis was performed using the GraphPad Prism 5 program. B and C, Western blot analysis of HDAC6 steady levels in HEK293T and U87MG cells transfected with siIIp45 (B) or with the IIp45 expression vector (C). D, real time RT-PCR analysis of the HDAC6 mRNA levels in IIp45 knockdown or overexpressed cells. There is no statistical difference.
To test this hypothesis, we first examined the IIp45 and HDAC6 protein levels in cell lines in which the expression of IIp45 was down-regulated by treatment with IIp45-siRNA. Similar to our observation in high grade gliomas, siIIp45 knockdown resulted in increased steady levels of HDAC6 (Fig. 2B). Consistently, overexpression of IIp45 resulted in decreased expression of HDAC6 protein (Fig. 2C). HDAC6 transcript was not altered after modulation of IIp45 in real time RT-PCR assays (Fig. 2D). These results suggest that IIp45 down-regulates HDAC6 at the protein level.
IIp45 Reduces Protein Stability of HDAC6
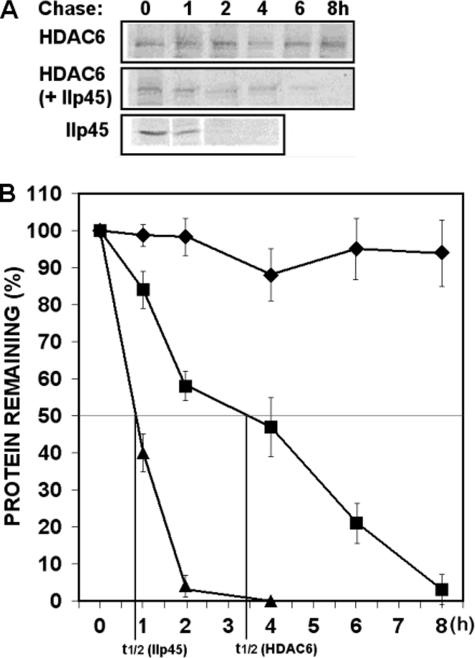
It is possible that IIp45 down-regulates the intracellular steady level of HDAC6 through destabilizing the HDAC6 protein. To determine the effect of IIp45 on intracellular protein stability of HDAC6, we examined the protein half-lives of endogenous HDAC6 protein in IIp45-transfected LN229 cells compared with control cell line. HDAC6 protein has been shown to be stable for over 8 h (24). In a previous cycloheximide treatment experiment, we observed that intracellular IIp45 protein was degraded by >50% within 1 h (21). In this study, we measured the half-life of newly synthesized endogenous HDAC6 and exogenously expressed IIp45 protein by pulse-chase experiments in [35S]methionine-labeled cells (Fig. 3A) followed by densitometry quantification. Consistent with the previous studies, the half-life of IIp45 in transfected cells was 50 min (Fig. 3, A and indicated in B, bottom line, t1/2), and the newly synthesized endogenous HDAC6 was stable over the time of 8 h in LN229 control cells (Fig. 3 A and top line of B). In contrast, under the condition of IIp45 overexpression in IIp45-transfected cells, the half-life of endogenous HDAC6 was reduced to less than 4 h (Fig. 3, A and middle line of B).
FIGURE 3.
IIp45 reduces protein stability of HDAC6. Intracellular protein stability of endogenous HDAC6 was measured by protein turnover assay in IIp45 stable-transfected LN229 cells and control cell line. Cells were split and grown overnight to 50% confluence and radiolabeled with [35S]methionine in a pulse-chase experiment. Cell lysates were collected at time points 0, 1, 2, 4, 6, and 8 h of chase. Clarified cell lysates were used for immunoprecipitation of endogenous HDAC6 or overexpressed IIp45 in parallel samples. Immunoprecipitated proteins were resolved on SDS-PAGE and detected by fluorography. A, increased protein turnover rate of endogenous HDAC6 was shown in IIp45 stable-transfected cell line. B, graphic data of HDAC6 protein turnover was based on quantification of gel densitometry using the National Institutes of Health IMAGE program. Protein degradation curves from the top: endogenous HDAC6 in LN229 (diamonds), endogenous HDAC6 in LN229-IIp45-stable cell line (squares), IIp45 in LN229-IIp45-stable cell line (triangles).
IIp45 Inhibits HDAC6 Activity and Increases Cellular Levels of Acetylated α-Tubulin
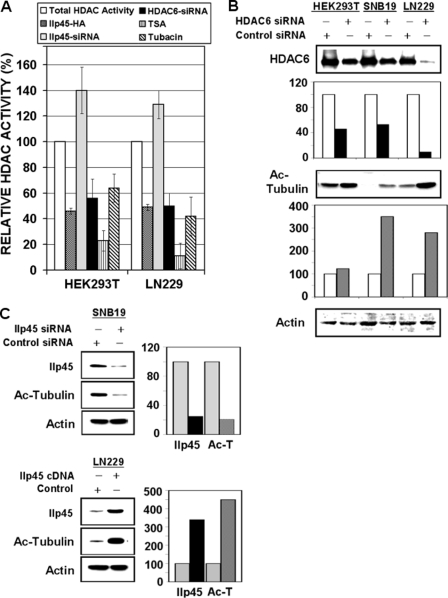
The observation that both catalytic domains of HDAC6 are required for its efficient binding to IIp45 suggests that IIp45 might modulate the deacetylase activity of HDAC6. To test this hypothesis, we performed the colorimetric HDAC activity assay that measured deacetylation of a common substrate. An established HDAC inhibitor, TSA, was used as the positive control. The effect of IIp45 on HDAC activity was compared with that of (i) siRNA-HDAC6, representing an HDAC6-specific inhibition; (ii) TSA, a broad deacetylase inhibitor of most members of the HDAC family, including HDAC6; and (iii) tubacin, an HDAC6-specific chemical inhibitor. Tubacin has been shown to inhibit specifically HDAC6-mediated deacetylation of α-tubulin, resulting in an increased intracellular level of acetylated α-tubulin (25, 26). We measured HDAC activity in cell lysates extracted from HEK293T and LN229 cells transfected with IIp45 expression vector, IIp45 siRNA, HDAC6 siRNA, or control nontargeting siRNA. Data were normalized to control cells (as 100%). The analyses showed average inhibition rates of 54% (p < 0.02) and 51% (p < 0.05) for HEK293T and LN229 cells, respectively, upon overexpression of IIp45, which were comparable with inhibition rates seen in cells treated by siHDAC6 or in tubacin-containing (1 μm) cell lysates (Fig. 4A). Knockdown of IIp45 showed an opposite effect in all tested cell lines. The highest inhibitory rates were shown in assays on HEK293T and LN229 cell lysates combined with 0.5 μm TSA (79% and 83%, respectively).
FIGURE 4.
IIp45 inhibits HDAC6 activity and increases intracellular acetylated α-tubulin level. A, HDAC activity assay was performed on cell lysates (100 μg of protein) of HEK293T and LN229 cells. Cells were transfected with wild-type IIp45-HA or treated by IIp45-siRNA or HDAC6-siRNA. Data were calculated with control reagent-treated cells and are presented as a percentage of HDAC activity compared with the total HDAC activity (as 100%) in untreated parental cells. Bars in each cell line from left: untreated parental cells HDAC activity (as 100%; white bars), percentage of the HDAC activity in IIp45-transiently transfected cells compared with parental (dark gray bars), percentage of the HDAC activity in siIIp45-treated cells (light gray bars), percentage of HDAC activity in siHDAC6-treated cells (black bars), percentage of HDAC activity in parental cell lysates measured by HDAC activity assays containing HDAC inhibitors TSA (0.5 μm) (striped bars) and tubacin (1 μm) (hatched bars). Error bars, mean ± S.E. B and C, Western blotting of acetylated α-tubulin. The intensity of the bands was quantified with densitometry and normalized with that of β-actin loading control. Values are presented as bar graphs in percentage relative to control (100%). B, increased level of acetylated α-tubulin in HDAC6 siRNA (40 nm) transfected HEK293T, SNB19, and LN229. C, effects of altered IIp45 expression on acetylated α-tubulin, showing a decreased level in siIIp45-treated SNB19 (upper panels, compared with control siRNA) and increased level in IIp45-transfected LN229 (lower panels, compared with control vector).
We next examined the status of α-tubulin acetylation in cells after modulation of HDAC6 or IIp45 by Western blotting with an antibody that specifically detects acetylated α-tubulin. HDAC6 knockdown resulted in increased levels of acetylated α-tubulin in HEK293T and glioma cell lines (Fig. 4B). Decreased levels of acetylated α-tubulin were observed in cells transfected by IIp45 siRNA, whereas overexpression of IIp45 resulted in increased α-tubulin acetylation (Fig. 4C).
IIp45 Inhibits HDAC6-mediated Cell Migration
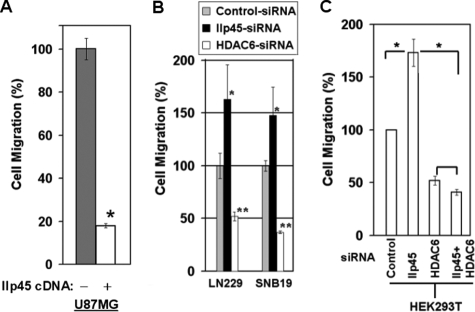
Based on results from the above studies, we speculated that IIp45 inhibits cell migration mediated by HDAC6. We tested this hypothesis using cell migration assays. Over the duration of our cell migration assay from 3 to 15 h after plating cells in Transwells, we observed no significant changes in cell cycle, cell viability, or cell growth in IIp45-transfected cell lines or cells treated with siRNAs (data not shown). Our results demonstrated that the cell migration rate was reduced by 82% (Fig. 5A; p < 0.05) in the IIp45-transfected cell line U87MG compared with its parental cell line, a result similar to that seen previously in LN229 cells (21). Cell motility was enhanced significantly by treatment of siIIp45, increasing 65% (p < 0.05) in LN229 cells and 49% (p < 0.05) in SNB19 cells. In contrast, treatment with siHDAC6 resulted in remarkable reductions in migration of both LN229 (50%; p < 0.001) and SNB19 (63%; p < 0.002) cells (Fig. 5B). These findings provide evidence that IIp45 and HDAC6 play antagonistic roles in cell migration.
FIGURE 5.
IIp45 inhibits cell migration through inhibition of HDAC6. Semiconfluent cells were split into triplicate wells with a pore size of 8 μm. The chambers were incubated at 37 °C in a 5% CO2 incubator for 3 h (U87MG), 6 h (SNB19), or 15 h (for LN229 and HEK293T). Migrated cells were then fixed, stained, and counted. Mean value ± S.E. (error bars) were calculated based on triplicate sample setting and repeated three times. A, IIp45 stable-transfected U87MG cell line compared with the parental cell line (*, p < 0.05). B, siIIp45 (20 nm) and siHDAC6 (40 nm) knockdown showed opposite effects on cell migration in both LN229 (left) and SNB19 (right) cell lines (compared with control siRNA, *, p < 0.05; **, p < 0.002, respectively). C, single and double siRNA knockdown in HEK293T cells (*, p < 0.05).
To test further the hypothesis that IIp45 inhibits cell migration through HDAC6, we performed experiments with single or double siRNA transfection of HEK293T cells in a setting of knockdown expression of IIp45, HDAC6, or both (Fig. 5C). Consistently, compared with control cells, treatment with siIIp45 led to 75% increased migration (p < 0.05), whereas treatment with siHDAC6 led to decreased migration by 50% (p < 0.05). Furthermore, marked attenuation of cell migration by HDAC6 knockdown almost completely abolished the migration-promoting effect of siIIp45 knockdown (p < 0.05). The difference in cell migration between siHDAC6 alone and siHDAC6+siIIp45 double-transfected cells was not statistically significant. These observations demonstrate that the cell migration-promoting effect of siIIp45 is significantly rescued by siHDAC6, supporting the notion that IIp45 is indeed capable of inhibiting HDAC6-mediated cell migration.
DISCUSSION
Cell motility is an important cellular process for both normal tissue remodeling and tumor progression, in which abnormally increased cell motility is associated with cancer invasion. In the present study, we found that IIp45 directly binds to the two catalytic domains of the HDAC6, reduces protein stability of HDAC6, inhibits cellular deacetylase activity, and consequently leads to increased acetylation of α-tubulin and decreased cell motility. These results demonstrate that IIp45 is a molecular inhibitor of HDAC6.
HDAC6 is known as a major regulator of cell migration. However, its role in glioma cell migration was previously unknown. The expression of HDAC6 mRNA was reported to be increased in brain tumor tissues compared with normal brain (5), but the protein levels of HDAC6 were not examined. In this study, we showed that the protein level of HDAC6 is abundant in high grade gliomas, as contrasted with IIp45 protein expression. We further showed that IIp45 negatively regulates the protein stability of HDAC6. These results suggest that IIp45 is an upstream regulator of HDAC6 at the posttranslational level. The exact mechanism by which HDAC6 is degraded through IIp45 binding is not clear at present. Degradation of HDAC6 protein has been reported to occur through the proteasomal degradation pathway (27). In addition, HDAC6 is an hsp90 client protein, and HDAC6 protein degradation is regulated by the chaperone function of hsp90 (27). On the other hand, hsp90 is known as a substrate of HDAC6 activity (28–30). Inhibition of HDAC6 results in hyperacetylation of hsp90, an inactivated form of hsp90 chaperone, which in turn leads to polyubiquitylation and proteasomal degradation of hsp90 client proteins that promote HDAC6 protein degradation (27). Therefore, by inhibiting HDAC6 deacetylation activity, IIp45 may enhance HDAC6 degradation through a regulation loop of hsp90 hyperacetylation-activated proteasomal degradation. Alternatively, HDAC6 is involved in transporting proteins to aggresomes in cells for ubiquitin-mediated degradation (22, 31), and IIp45 may play a role in this process. Further studies are needed to determine whether IIp45 activates an hsp90-mediated HDAC6 proteasomal degradation pathway or promotes degradation of HDAC6 after transportation of protein to aggresomes.
In this study, we have provided evidence that both catalytic domains of HDAC6 protein are important for binding to IIp45. It is known that both catalytic domains of HDAC6 are essential for its deacetylase activity (7). Our analyses of HDAC activity in cells after modulation of IIp45 and HDAC6 reveal a similar HDAC-inhibitory ability between IIp45 and siHDAC6, suggesting that binding of IIp45 to the two catalytic domains on HDAC6 possibly blocks the deacetylase activity of HDAC6. These studies have thus provided strong evidence that IIp45 is a cellular inhibitor of HDAC6 at both the protein stability level and the enzymatic level. This regulation represents a new mechanism through which IIp45 inhibits cancer cell migration.
Acknowledgments
We thank Dr. Kanchana Natarajan Mendes for contributing to this project and Hong Zheng for technical assistance in this work. We thank Don Norwood at the Department of Scientific Publication for editing the manuscript.
This work was supported by a grant from the James S. McDonnell Foundation.
- HDAC
- histone deacetylase
- IIp45
- invasion inhibitory protein 45
- IGFBP
- insulin-like growth factor-binding protein
- GST
- glutathione S-transferase
- co-IP
- co-immunoprecipitation
- siRNA
- small interfering RNA
- HA
- hemagglutinin
- TSA
- trichostatin
- RT
- reverse transcription.
REFERENCES
- 1.Wehrle-Haller B., Imhof B. A. (2003) Int. J. Biochem. Cell Biol. 35, 39–50 [DOI] [PubMed] [Google Scholar]
- 2.Cunha-Ferreira I., Bento I., Bettencourt-Dias M. (2009) Traffic 10, 482–498 [DOI] [PubMed] [Google Scholar]
- 3.Takemura R., Okabe S., Umeyama T., Kanai Y., Cowan N. J., Hirokawa N. (1992) J. Cell Sci. 103, 953–964 [DOI] [PubMed] [Google Scholar]
- 4.Hammond J. W., Cai D., Verhey K. J. (2008) Curr. Opin. Cell Biol. 20, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Ruijter A. J. M., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburgi A. B. P. (2003) Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertos N. R., Wang A. H., Yang X. J. (2001) Biochem. Cell Biol. 79, 243–252 [PubMed] [Google Scholar]
- 7.Zhang Y., Gilquin B., Khochbin S., Matthias P. (2006) J. Biol. Chem. 281, 2401–2404 [DOI] [PubMed] [Google Scholar]
- 8.Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. (2002) Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R., Yao T. P., Lane W. S., Seto E. (2007) Mol. Cell. 27, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch W. J., Feramisco J. R. (1982) J. Biol. Chem. 257, 14949–14959 [PubMed] [Google Scholar]
- 11.Welch W. J. (1991) Curr. Opin. Cell Biol. 3, 1033–1038 [DOI] [PubMed] [Google Scholar]
- 12.Tsutsumi S., Neckers L. (2007) Cancer Sci. 98, 1536–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser C., Lang S. A., Stoeltzing O. (2009) Anticancer Res. 29, 2031–2042 [PubMed] [Google Scholar]
- 14.Wu H., Reynolds A. B., Kanner S. B., Vines R. R., Parsons J. T. (1991) Mol. Cell. Biol. 11, 5113–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohoka Y., Takai Y. (1998) Genes Cells 3, 603–612 [DOI] [PubMed] [Google Scholar]
- 16.Daly R. J. (2004) Biochem. J. 382, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacs J. J., Murphy P. J., Gaillard S., Zhao X., Wu J. T., Nicchitta C. V., Yoshida M., Toft D. O., Pratt W. B., Yao T. P. (2005) Mol. Cell 18, 601–607 [DOI] [PubMed] [Google Scholar]
- 18.Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., Sánchez-Madrid F. (2008) Trends Cell Biol. 18, 291–297 [DOI] [PubMed] [Google Scholar]
- 19.Witt O., Deubzer H. E., Lodrini M., Milde T., Oehme I. (2009) Curr. Pharm. Des. 15, 436–447 [DOI] [PubMed] [Google Scholar]
- 20.Haberland M., Montgomery R. L., Olson E. N. (2009) Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song S. W., Fuller G. N., Khan A., Kong S., Shen W., Taylor E., Ramdas L., Lang F. F., Zhang W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13970–13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 23.Song S. W., Fuller G. N., Zheng H., Zhang W. (2005) Cancer Res. 65, 3562–3567 [DOI] [PubMed] [Google Scholar]
- 24.Hook S. S., Orian A., Cowley S. M., Eisenman R. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hideshima T., Bradner J. E., Wong J., Chauhan D., Richardson P., Schreiber S. L., Anderson K. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8567–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haggarty S. J., Koeller K. M., Wong J. C., Grozinger C. M., Schreiber S. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao R., Fiskus W., Yang Y., Lee P., Joshi R., Fernandez P., Mandawat A., Atadja P., Bradner J. E., Bhalla K. (2008) Blood 112, 1886–1893 [DOI] [PubMed] [Google Scholar]
- 28.Aoyagi S., Archer T. K. (2005) Trends Cell Biol. 15, 565–567 [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Rao R., Shen J., Tang Y., Fiskus W., Nechtman J., Atadja P., Bhalla K. (2008) Cancer Res. 68, 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kekatpure V. D., Dannenberg A. J., Subbaramaiah K. (2009) J. Biol. Chem. 284, 7436–7445 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Rodriguez-Gonzalez A., Lin T., Ikeda A. K., Simms-Waldrip T., Fu C., Sakamoto K. M. (2008) Cancer Res. 68, 2557–2560 [DOI] [PubMed] [Google Scholar]