Abstract
This study investigated the molecular mechanisms underlying the regulatory effect of the newly discovered 45-kDa enzymatically inactive UGT1A spliced polypeptides, named isoform i2, upon UGT1A-mediated glucuronidation. Initially, using an inducible system that mimics the relative abundance of isoforms 1 and 2 of UGT1A1 in human tissues, the rates of formation of glucuronides were significantly reduced. We then used a heterologous system constitutively expressing both isoforms i1 and i2 for an in-depth investigation of the presence of spliced i2 on glucuronidation kinetics. UGT1A1, UGT1A7, and UGT1A8 were selected as candidates for these studies. In all cases, co-expression of i1 and i2 in HEK293 cells leads to a significant reduction of the velocity of the glucuronidation reaction without affecting the affinity (Km app) for all substrates tested and the Km for the co-substrate, UDP-glucuronic acid. The data are consistent with a dominant-negative model of inhibition but do not sustain with an UGT1A_i2-mediated inhibition by competitive binding for substrate or the co-substrate. In contrast, the data from the co-immunoprecipitation experiments are indicative of the existence of a mixture homo-oligomeric (i1-i1 or i2-i2) and hetero-oligomeric (i1-i2) complexes in which the i2-i2 and i1-i2 subunits would be inactive. Thus, protein-protein interactions are likely responsible for the inhibition of active UGT1A_i1 by i2 spliced polypeptides. This new regulatory mechanism may alternatively modulate cellular response to endo/xeno stimulus.
Keywords: Enzymes, Enzymes/Inactivation, Protein/Protein-Protein Interactions, Toxins/Drugs, Glycoconjugate, Glucuronosyltransferase Enzymes, Enzyme Activity
Introduction
Alternative splicing is an important cellular process underlying the transcriptome diversity in eukaryotic cells. Recent transcriptome analyses conducted in several human tissues indicated that the vast majority of human genes undergo alternative splicing and support alternative splicing as one of the most important post-transcriptional mechanisms to regulate gene function and expression (1, 2). In fact, alternative splicing introduces novel mRNA molecules, which may lead to divergent polypeptides, in terms of biological function and/or expression profile. Recent experimental evidence of such alternative splicing variants has been documented in the human UDP-glucuronosyltransferases UGT1A gene (3, 4). This family of proteins represents critical phase II conjugating enzymes in detoxification and elimination processes to avoid accumulation of potentially damaging environmental substances (e.g. pharmaceuticals, dietary carcinogens, and toxins) and in maintaining the homeostasis of several lipophilic endogenous compounds (e.g. bilirubin, bile acids, and hormones).
The UGT1A locus on chromosome 2q37 well illustrates the use of alternative splicing to increase protein diversity from a single genomic locus. Indeed, half of the 19 human functional UGT enzymes are produced from this unique gene. Thirteen different mRNAs are transcribed from this gene, of which nine lead to functional enzymes and four pseudogenes (5, 6). This is driven by alternative usage of the first variable coding exon (encoding for the aglycone-binding domain), which is joined to four constant exons (2–5), encompassing the co-substrate uridine-diphosphate glucuronic acid (UDPGA)-binding4 domain. A new exon (named exon 5b) in the common region of the UGT1A locus, between coding exons 4 and 5, was recently uncovered (3, 4). This new exon 5b might either be used as a terminal exon (leading to the mRNA isoform variant 2 or v2) or be spliced with the common exon 5 (exon 5a) (leading to mRNA isoform variant 3 or v3). Therefore, this splicing event lead to the production of 18 new mRNAs, giving rise to nine new human UGT proteins, named UGT1A isoform 2 (or i2) (3). As a consequence, the novel 45-kDa UGT1A protein isoform 2 (in comparison with the 55-kDa UGT1A isoform 1) lack the 99- amino acid region encoded by the exon 5a, which is substituted by 10 residues encoded by the exon 5b (3, 4).
The presence of endogenous UGT1A_i2 spliced variants is supported by several observations. First, reverse transcriptase-PCR experiments and Western blot analysis using available polyclonal anti-UGT antibodies demonstrated the presence of these new UGT1A spliced in human tissues, including liver, kidney, esophagus, small intestine, and colon (3, 4). An immunohistochemical experiment with a specific polyclonal antibody targeted to exon 5b unique to UGT1A_i2 isoforms clearly demonstrated their existence in human tissues, and they co-localize with the fully active UGT1A enzymes (7). In addition, other immunofluorescence experiments indicated that these shorter proteins co-localize with UGT1A_i1 to the endoplasmic reticulum and perinuclear structures (4). Furthermore, we also revealed interindividual variation in the abundance of the UGT1A_i2 proteins, which has been evidenced in liver microsomes and microsomes derived from extrahepatic tissues (3, 4).
Preliminary functional studies using heterologous UGT1A_i2 expression systems in HEK293 cells clearly indicated that UGT1A_i2 proteins are deprived of glucuronidation activity. Further investigations proposed a negative modulatory effect upon net glucuronidation when stably co-expressed with the fully active UGT1A_i1 (3, 4). However, the clear biological role of 45-kDa proteins still remains to be established. Several mechanisms can be invoked to explain the inhibitory and regulatory effects of UGT1A_i2 upon UGT1A glucuronidation activity, such as competitive binding for substrate and/or co-substrate, allosteric inhibition, post-translational modifications, or even substantial changes in protein quaternary structure. On the other hand, the function and activity of a protein are frequently modulated by the other proteins with which it interacts. Several findings support the possibility that UGTs are highly organized within the endoplasmic reticulum, existing as monomeric proteins as well as oligomers (homodimers and heterodimers) (8–10). Moreover, the cellular co-localization of UGT1A splice forms likely suggests molecular interaction between them. As a secondary objective and to further portray the inhibitory effects of i2 and investigate the mechanisms mediating this regulation, we intended to define whether the action of these spliced forms is mediated by competitive binding for the specific substrate and the co-substrate UDPGA.
According to the strong protein similarity between UGT1A_i1 and i2 isoforms, we first propose to examine whether the action of UGT1A_i2 is mediated by a competitive binding for the specific substrate or the co-substrate UDPGA. In a first step, to clearly establish that UGT1A_i2-mediated inhibition is caused by the presence of spliced i2 forms and not other unknown variables or a clonal effect, we produced an UGT1A1_i2 ecdysone-inducible system in cells stably expressing UGT1A1_i1. To further portray the inhibitory effects of i2, we carried out comparative kinetic analyses between HEK293 clones expressing either isoform 1 of UGT1A1, UGT1A7, or UGT1A8 alone or with their corresponding i2 isoforms. These candidates were retained for subsequent enzyme kinetic profiling based on the previous demonstration of the co-existence of i1 and i2 isoforms in human tissues. Finally, immunoprecipitation assays were conducted to explore the potential close relationship between both UGT1A protein spliced isoforms and to determine whether the UGT1A_i2 negative effect is dependent of protein-protein interaction with the active forms UGT1A_i1. Data from this study support that the inhibitory role of UGT1A_i2 likely occurs through molecular interactions between isoforms 1 and 2 and less likely occurs through competitive binding for specific substrate and co-substrate induced by the presence of i2 forms.
EXPERIMENTAL PROCEDURES
Materials
UDPGA was obtained from Sigma; blasticidin, geneticin (G418), and hygromycin were from Wisent (St. Bruno, Canada); and zeocin and Lipofectin reagent were from Invitrogen. HEK293 cells were obtained from the American Type Culture Collection (Manassas, VA). Protein assay reagents were obtained from Bio-Rad. Bilirubin was purchased from Sigma-Aldrich. Estradiol (E2) was purchased from Steraloids (Newport, RI). Mycophenolate acid was obtained from MP Biomed LLC (Aurora, OH), 4-methylumbelliferone (4-MU) was obtained from Sigma-Aldrich, and SN-38 was prepared by hydrolysis of irinotecan HCl (McKesson) as described (11).
Expression Systems
The inducible system was established as follows. pcDNA3.1/UGT1A1_i1-Myc-His was initially transfected into EcR 293 cells (HEK293 cells that were stably transformed with the regulatory vector pVgRXR) using Lipofectin. UGT1A_i2 cDNA (expressing v5-His tag) was subcloned from pcDNA6 into the pIND ecdysone-inducible mammalian expression vector, and the resulting construct was transfected in clones expressing UGT1A_i1 (HEK RXR+-UGT1A1_i1-Myc-His). Resistance to G418, zeocin, and hygromycin was used to select for potential positive clones. The clonal cell line termed HEKRXR+/UGT1A1_i1-Myc-His+pIND UGT1A_i2-v5-His used for our study displayed undetectable expression of i2 when noninduced with ponasterone A.
The HEK293 clonal cell lines used for the kinetic studies consisted of clones expressing UGT1A_i1 (pcDNA3.1/tagged with Myc-His epitopes), UGT1A_i2 (pcDNA6/tagged with v5-His epitopes), or both pcDNA3.1-UGT1A_i1/Myc-His and pcDNA6-UGT1A_i2/v5-His, as described previously (3).
Western Blot Analysis
Microsomal fractions were prepared as described elsewhere with cells disrupted using 3 × 10 s of sonication (12). To ascertain the level of UGT content in stable UGT1A-HEK293 cell lines, a semiquantitative immunoblot analysis method was performed as described previously (3) using the antibody RC-71 (anti-UGT1A) as the primary antibody and an anti-rabbit IgG horse antibody conjugated with peroxidase as the second antibody (Amersham Biosciences).
Co-immunoprecipitation Assays
HEK293 cells were plated at density of 5.5 × 106 cells/dish in 100-mm dishes and transiently co-transfected with 10 μg of total DNA (10 μg of pUGT1A_i1; 10 μg of pUGT1A_i2; or 5 μg of pUGT1A_i1 and 5 μg of pUGT1A_i2 plasmids) using Lipofectin. After 48 h, the cells were washed twice with phosphate-buffered saline and lysed for 45 min on ice with 1 ml of lysis buffer (0.05 m Tris-HCl, pH 7.4, 0.15 m NaCl, 0.3% deoxycholic acid, 1% Igepal, 1 mm EDTA). The cell lysates were then homogenized by pipetting up and down through fine needles (18 gauge followed by 20 gauge) 10–20 times on ice. The lysates were centrifuged for 15 min at 13,000 × g, and the supernatant was collected. Cell-free lysates were mixed with protein G-Sepharose 4 Fast Flow (50% slurry) (Amersham Biosciences) and stirred for 30 min at 4 °C to preclear nonspecific binding. After centrifugation (13,000 × g for 1 min), 1 mg of supernatant was added with 1 μg of specific monoclonal antibody (Invitrogen) in 1 ml of high salt buffer and incubated at 4 °C with 50 μl of protein G-Sepharose 4 fast flow (50% slurry) for 15 h. The beads were washed three times with 1 ml of lysis buffer and finally with 1 ml of 50 mm Tris, pH 7.5. The beads containing the immunoprecipitated proteins were resuspended with 30 μl of 1× SDS-PAGE solution, heated at 100 °C for 5 min, and centrifuged at 12,000 × g for 20 s. The supernatant was analyzed by SDS-PAGE. The membrane blots were probed with a specific monoclonal antibody linked with horseradish peroxidase (Invitrogen), as specified in the corresponding figure legend.
Functional Assays
The enzymatic assays for the inducible ecdysone system were performed using 20 μg of microsomes in assay conditions described previously (12). Briefly, the reactions were initiated by the addition of 7-ethyl-10-hydroxycamptothecin (SN-38) (5 and 200 μm) for 1 h or estradiol (25 and 200 μm) for 3 h at 37 °C. SN-38 and estradiol assays were stopped with 100 μl (2 n 1% HCl) and 100 μl of ice-cold methanol, respectively. Kinetic assays were further performed in similar conditions and initiated by adding varying concentrations of substrate, whereas those with bilirubin were performed under minimal light conditions. Glucuronide formation was measured by chromatography coupled with mass spectrometry protocols as described previously (4, 11–13). Formation of glucuronides (absolute activity) was corrected by UGT protein content (i1 active enzyme) assessed by Western blot and expressed as relative activity.
Data Analysis and Statistics
Kinetic parameters were calculated using SigmaPlot 8.0 with Enzyme Kinetics 1.1 (SPSS, Chicago, IL). Eadie-Hofstee plots (velocity as a function of (velocity/substrate concentration)) and visual inspection of fitted functions (velocity as a function of substrate concentration) were used to select the best fit enzyme kinetic model (14). The values are expressed as the means of at least two experiments performed in triplicate. The difference in glucuronidation rates and kinetic parameters was evaluated for statistical significance by the paired Student's t test (significant at least at p < 0.05).
RESULTS
Function of UGT1A Splice Isoforms 2 Using an Ecdysone Inducible System
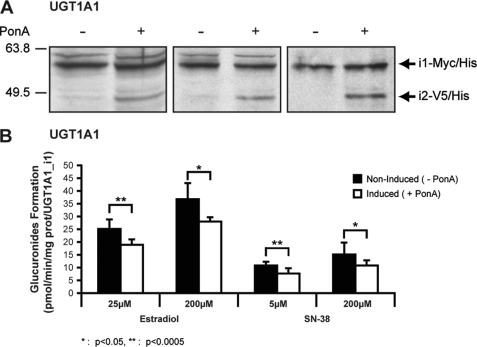
We developed stable UGT1A1_i1-overexpressing HEK293 cells that uniquely express its homolog UGT1A1_i2 when exposed to the ecdysone analog, ponasterone A. This model offers the possibility to test in a single cell line the influence of i2 overexpression on UGT1A_mediated activity, ruling out the possibility that potential differences would arise from interclonal variability. With this model, undetectable expression of i2 was shown in noninduced cells. Upon treatment with ponasterone A, cells demonstrated significant levels of UGT1A1_i2, but no significant changes were noted for UGT1A1_i1 protein expression (Fig. 1A). The i1/i2 expression ratio (3:1) achieved in these conditions corresponds to the physiological levels previously reported in the jejunum microsomal proteins (4). Thus, in a situation that mimics physiological levels of UGT1A1 isoforms in a human tissue, estradiol-G and SN-38-G formation by UGT1A1_i1-expressing cells was significantly decreased by ∼30% upon the induction of UGT1A1_i2 compared with noninduced cells (Fig. 1B). The data clearly support the role of i2 as repressors of UGT1A-mediated glucuronidation, consistent with our previous observations (4) and further validating the use of the heterologous system of stable co-expression of i1+i2 for an in-depth investigation of the presence of i2 on glucuronidation kinetics.
FIGURE 1.
Induction of UGT1A1 protein isoform 2 in UGT1A1_i1-overexpressing cell line. The biological response of i1-expressing cells to i2 expression was investigated by inducing i2 expression by treatment of the cells with ponasterone A (5 μm) at 24 h of induction. A, induction of UGT1A1 isoform 2 proteins following ponasterone A (PonA) treatment for 24 h was visualized by Western blot analysis. Three independent induction experiments are shown. B, induction of UGT1A1_i2 inhibits i1-mediated glucuronidation activity on both substrates tested. Glucuronide formation was quantified by liquid chromatography/mass spectrometric analysis. The data represent the means ± S.D. of three independent experiments performed in triplicate. *, p < 0.5; **, p < 0.005.
The Study of Detailed Enzyme Kinetics
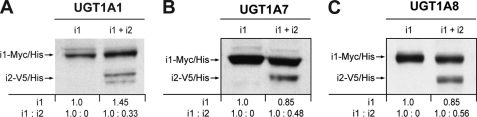
A second series of experiments was designed to test whether and how enzyme kinetics is affected by the presence of the UGT1A_i2 spliced isoforms (Table 1). Although not functional, shorter UGT1A_i2 proteins comprise the substrate and co-substrate-binding domains and might be able to compete for such a binding activity. HEK293 clonal cell lines stably expressing UGT1A_i1 (tagged with Myc-His epitopes) alone or with UGT1A_i2 (tagged with v5-His epitopes) were engineered. The ratios of expression between i1 and i2 proteins are shown in Fig. 2. First, we constructed stably transfected HEK cell clones expressing UGT1A_i1 (tagged with Myc-His epitope) either alone or with UGT1A_i2 (tagged with v5-His epitope). Our study has focused on UGT1A1, UGT1A7, and UGT1A8 as candidates based on our previous work supporting the co-expression of i1 and i2 proteins in human hepatic and extrahepatic tissues (3). The expression data in microsomes and the ratios of expression between splice forms are depicted in Fig. 2. The UGT1A1-overexpressing cell line displayed a 1:0.3 ratio (i1:i2), whereas a slightly higher expression of i2 (relative to i1) was achieved for UGT1A7 (1:0.5) and UGT1A8 (1:0.6) cell lines. Several substrates of the UGT1A1, UGT1A7, and UGT1A8 enzymes were tested and included substrates of high and low affinity for these enzymes, namely, bilirubin, SN-38, and estradiol for UGT1A1; SN-38 and 4-MU for UGT1A7; and finally, mycophenolic acid, estradiol, and 4-MU for the UGT1A8 enzyme.
TABLE 1.
Assay conditions for enzyme kinetics
MeOH, methanol; BHT, butylated hydroxytoluene; MPA, mycophenolic acid; SN-38, ethyl-10-hydroxy-camptothecin.
| Enzymes | Substrates | Concentrationsa | Incubation time | Reactions were stopped with |
|---|---|---|---|---|
| μm | h | |||
| UGT1A1 | Bilirubin | 0–2000 | 0.1 | 100 μl of MeOH + 2% BHT |
| SN-38 | 0–200 | 1 | 100 μl of MeOH + 2% HCl | |
| Estradiol | 0–200 | 3 | 100 μl of MeOH | |
| UDPGA (SN-38 5 μm)b | 0–5000 | 1 | 100 μl of MeOH + 2% HCl | |
| UGT1A7 | SN-38 | 0–200 | 3 | 100 μl of MeOH + 2% HCl |
| 4-MU | 0–2000 | 1 | 100 μl of MeOH | |
| UDPGA (SN-38 5 μm)b | 0–5000 | 1 | 100 μl of MeOH + 2% HCl | |
| UGT1A8 | Estradiol | 0–200 | 1 | 100 μl of MeOH |
| 4-MU | 0–2000 | 1 | 100 μl of MeOH | |
| MPA | 0–1500 | 1 | 200 μl of MeOH + 2% HCl | |
| UDPGA (MPA 100 μm)b | 0–5000 | 1 | 200 μl of MeOH + 2% HCl |
a More than 10 concentrations were used in the specified range.
b Co-substrate.
FIGURE 2.
Western blot analyses of UGT1A1, UGT1A7, and UGT1A8 cell lines stably expressing i1 and co-expressing i1 and i2. The presence of a 58-kDa protein in the HEK293-UGT1A_i1-Myc/His lines and a 48-kDa protein in the HEK293-UGT1A_i1+i2-V5/His lines is confirmed. The relative abundance (OD units) of overexpressed proteins was quantified and is indicated.
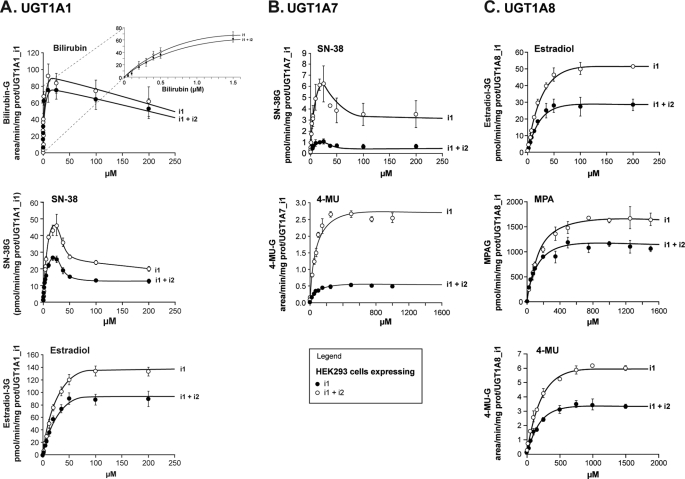
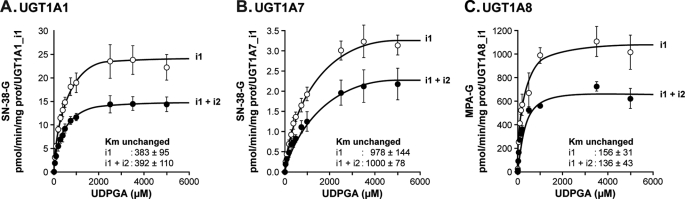
Kinetic plots for selected substrates of high and low affinities are depicted in Fig. 3 and are consistent with kinetic profiles previously reported for these enzymes and substrates (11–13). For each situation tested, the presence of an isoform 2 protein did not modify the kinetic profile compared with the one exhibited by microsomes derived from cells expressing exclusively the i1 protein. However, in all cases, a significant decrease in the rates of formation of glucuronides (corrected for expression relative to active UGT1A_i1) was observed for every substrate and for each combination of UGT1A_i1 and i2. No significant changes in Km or S50 values were observed, supporting that the substrate affinity of i1 proteins is not influenced by the presence of UGT1A_i2 species. The extent of this inhibition varies according to substrate and enzyme and ranged from 18 to 82%. The most striking effect occurred with UGT1A7-derived cell lines, where the percentage of inhibition for glucuronidation of estradiol and 4-MU induced by UGT1A7_i2 reached 82%. The reduction in glucuronidation rates without any influence of Km supports the possibility that the less significant quantity of active enzyme may participate in the reaction when UGT1A_i2 proteins are present. It is noteworthy that that the degree of inhibition did not correlate with i1:i2 protein ratio. For instance, the UGT1A7 protein ratio of 1:0.5 induced ∼80% inhibition of 4-MU glucuronidation, in comparison with ∼30% for a UGT1A8 protein ratio of 1:0.6. We then evaluated whether the presence of UGT1A_i2 proteins influences the enzyme kinetics for the co-substrate UDPGA. In these experiments, the co-substrate concentrations varied from 0 to 5000 μm, whereas the substrate concentration was kept constant (at Km). The presence of UGT1A_i2 proteins does not influence the Km values for UDPGA but significantly reduced the rates of formation of glucuronides for all three enzymes tested (Fig. 4).
FIGURE 3.
Formation of glucuronides for typical UGT1A substrates by cell lines expressing UGT1A1 (A), UGT1A7 (B), and UGT1A8 (C) spliced isoforms. Glucuronide formation was normalized to UGT1A_i1 protein content assessed by Western blot. The data represent the means ± S.D. of two independent experiments performed at least in triplicate.
FIGURE 4.
Formation of glucuronides for typical UGT1A substrates in the presence of varying concentrations of the co-substrate by cell lines expressing UGT1A1 (A), UGT1A7 (B), and UGT1A8 (C) spliced isoforms. Glucuronide formation was normalized to UGT1A_i1 protein content assessed by Western blot. The data represent the means ± S.D. of two independent experiments performed at least in triplicate. Similar Km values for the co-substrate UDPGA were calculated in the presence of i1+i2 compared with cells expressing only i1.
Globally, co-expression of homologous UGT1A_i1 and i2 in HEK293 cells leads to a significant reduction of the velocity of the reaction without altering the affinity for the co-substrate and all of the substrates tested. We then postulated that i2-mediated UGT1A_i1 inhibition of glucuronidation activity of UGT1A_i1-expressing cells would be dependent on protein interaction between i1 and i2 proteins. This was tested by co-immunoprecipitation (co-IP) assays as described below.
Evidence of Protein-Protein Interaction by co-IP Assays
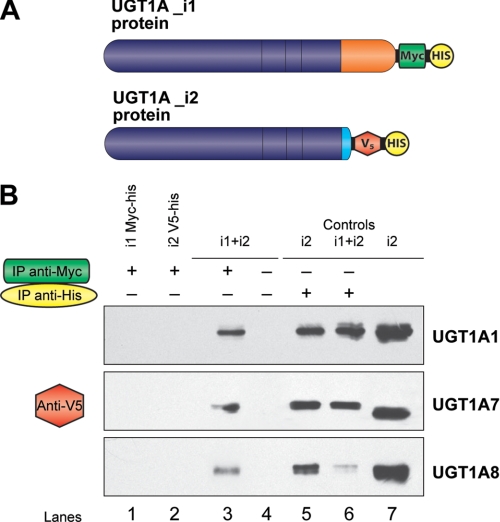
We examined the potential protein-protein interaction between isoforms i1 and i2 of UGT1A1, UGT1A7, and UGT1A8. The total proteins from cell lysates of double-transfected cell lines were used to immunoprecipitate Myc-tagged UGT1A_i1 and associated proteins with the anti-Myc monoclonal antibody. Immunoprecipitated protein complexes were denatured and resolved onto SDS-PAGE, and UGT1A_i2-v5 proteins were revealed with an anti-v5 antibody. The data clearly indicate that v5/His-tagged UGT1A_i2 proteins (48 kDa) are co-precipitated with their homolog UGT1A_i1 for these three enzymes (Fig. 5). To further test the potential homo-oligomerization between either i1 or i2, we established two additional plasmid constructs for UGT1A1, where UGT1A1_i1 was v5-tagged, whereas UGT1A1_i2 was Myc-tagged (Fig. 6A).
FIGURE 5.
Direct interaction between UGT1A spliced forms as demonstrated by co-immunoprecipitation assays. A, UGT1A isoforms 1 were subcloned and tagged with Myc-His epitope, whereas isoforms 2 were tagged with V5-His epitope. B, co-immunoprecipitation assays for UGT1A1, UGT1A7, and UGT1A8. Lanes 1 and 2, negative controls; immunoprecipitation (IP) with anti-Myc (1 μg). Lane 1, UGT1A_i1-Myc/His; lane 2, UGT1A_i2-V5/His; lanes 3 and 4, UGT1A_i1-Myc/His + UGT1A_i2-V5/His; lanes 5 and 6, positive controls immunoprecipitations with anti-His (1 μg); lane 5, visualization of UGT1A_i2-V5/His; lane 6, UGT1A_i2-V5/His (5 μg) was loaded onto the gel as positive control for the Western blot.
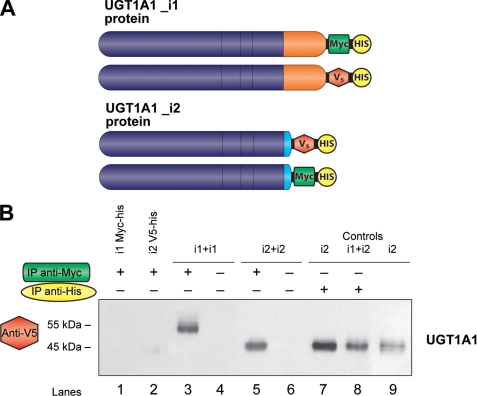
FIGURE 6.
Homo-oligomerization of UGT1A1 spliced forms as demonstrated by co-immunoprecipitation assays. A, UGT1A1_i1 tagged with V5-His epitope and 1A1_i2 tagged with Myc-His epitope constructs were used in these assays. B, co-immunoprecipitation assays for UGT1A1. Lanes 1 and 2, negative controls; immunoprecipitation (IP) with anti-Myc (1 μg). Lane 1, UGT1A1_i1-Myc/His; lane 2, UGT1A1_i2-V5/His; lanes 3 and 4, UGT1A1_i1-Myc/His+UGT1A1_i1-V5/His; lanes 5 and 6, UGT1A1_i2-Myc/His + UGT1A1_i2-V5/His; lanes 7 and 8, positive controls immunoprecipitations with anti-His (1 μg); lane 5, UGT1A1_i2-V5/His; lane 8, UGT1A1_i1-Myc/His_1A1_i2-V5/His; lane 9, UGT1A1_i2-V5/His (5 μg) was loaded onto the gel as positive control for the Western blot.
Thereafter, using both constructs of UGT1A1_i1 (one tagged with Myc-His and another tagged with v5-His) for additional co-IP experiments, it was then showed that isoform 1 of UGT1A1 is able to homo-oligomerize (Fig. 6B, lane 3), which is in line with previous reports (9, 15). When Myc-tagged and v5-tagged UGT1A1_i2 were co-transfected together, the data clearly indicate that UGT1A1_i2 interacts with itself as well (Fig. 6B, lane 5).
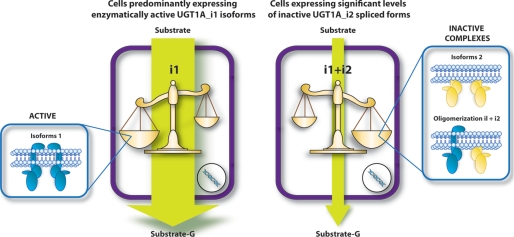
Altogether, these observations reveal the existence of three possibilities of protein complexes: homo-oligomers i1-i1 and i2-i2 and hetero-oligomer i1-i2. We conclude that hetero-oligomer UGT1A_i1/UGT1A_i2, like homo-oligomer i2-i2, would be enzymatically inactive and that the formation of these types of oligomers (dimers of higher oligomers) would be responsible for the overall decrease in activity in the presence of i2 isoforms.
DISCUSSION
The recent discovery of alternatively spliced isoforms of the human UGT1A family, referred to as isoforms 2 or UGT1As_i2, has led us to investigate their biological significance using in vitro models (3, 4, 7). In a first attempt to reveal their function, we previously established HEK293 cell lines that stably express either i1 or i2 or both i1 and i2 simultaneously (3, 4). These experiments demonstrated that although UGT1A_i2 proteins lack transferase activity, they significantly decrease UGT1A_i1-mediated activity. Here, as a first series of experiments, we have used an ecdysone inducible expression system enabling the production of i1+i2 proteins in the same cells to expression levels that mimic our previous observations in human tissues (4). This model clearly demonstrated that the induced expression of the UGT1A1_i2 species in a cell line expressing the UGT1A1_i1 protein lead to inhibition of cellular glucuronidation activity. Furthermore, these results are in line with our previous observations that showed an inhibitory effect caused by i2 species when comparing glucuronidation activity of clonal HEK293 cell lines that separately expressed i1, i2, or both i1+i2. These recent data thus exclude the possibility that the effect is due to interclonal variability or unknown factors. However, the mechanism(s) of such an inhibition was still unclear.
Additional experiments presented here do not support a potential depletion of the substrate or the co-substrate induced by the presence of the i2 protein. Indeed, kinetic plots for substrates conjugated by UGT1A1, UGT1A7, and UGT1A8 enzymes clearly demonstrated that the presence of i2 proteins decreases rates of glucuronide formation without affecting the kinetic model and substrate binding (Km or S50). If UGT1As_i2 were competing for substrate binding, we would have expected to note an effect on Km or S50. At least two substrates/enzyme were tested and displayed similar outcomes. Experiments included substrates of low and high affinities (e.g. previously reported as having a high or low Km, respectively). Similarly, no significant change in the Km for the co-substrate UDPGA was observed, suggesting that the presence of i2 does not compete for the co-substrate either. Overall, this clearly demonstrates that the presence of UGT1A_i2 strongly impairs the function of UGT1A_i1, pointing to a dominant-negative regulatory character.
Based on extensive literature supporting oligomerization of UGTs, we then speculated that inactive UGT1A_i2 proteins might bind active UGT1A_i1 proteins and form inactive heteromeric complexes, thus acting as dominant inhibitors and decreasing only the velocity of the glucuronidation reaction. Our previous observations on UGT1A1 and in human cells support this hypothesis. It was demonstrated that UGT1A_i1 and i2 proteins are co-localized in the endoplasmic reticulum and perinuclear structures, raising the possibility that they may interact together. Indeed, previous experiments exclusively on UGT1A1 suggest a potential interaction between i1 and i2 proteins (4, 7). In the present study, results derived from co-IP experiments support oligomerization between all combinations of i1 and i2 proteins tested, namely for UGT1A1, UGT1A7, and UGT1A8. The binding of isoform 2 protein to active isoform 1 would likely induce a conformational change or disrupt the normal folding process, which could prevent or impair substrate and/or co-substrate binding to the enzymatic complex. The intermolecular interactions between isoform 1 and 2 proteins could thus lead to an enzymatically inactive complex. Still, domain(s) that mediate these interactions need to be identified, and one could possibly expect more than one region involved according to previous reports of defined UGT protein-interaction domains (16–19). Previous observations support the hypothesis of physical interactions between UGT isoforms. Ghosh et al. (9) showed that COS7 cells co-expressing wild-type human UGT1A1 and an inactive mutant form of UGT1A1 (substitution C127Y) displayed a decreased conjugation activity against bilirubin, even though both forms retained the ability to dimerize. Other studies further showed that enzymatic activity of UGT enzymes would be affected by interaction between monomers through homo- and/or heterodimerization (20, 21).
In our system, the degree of inhibition induced by the presence of UGT1A_i2 varies between enzymes and was more pronounced for UGT1A7 compared with UGT1A1 and UGT1A8. For two closely similar proteins such as UGT1A7 and UGT1A8, we observed that even if the presence of i2 proteins for UGT1A8 was higher compared with i1 species (i2 content = 56% of i1) than for UGT1A7 (i2 content = 48% of i1), the inhibitory effect observed on UGT1A8 was not more pronounced than the one obtained for UGT1A7. Because we observed the formation of homo-dimers/oligomers between i1 and i2 by co-IP (i1-i1; i2-i2), the data would be consistent with the existence of a mixture homo-oligomeric (i1-i1 or i2-i2) and hetero-oligomeric (i1-i2) complexes, in which i2-i2 and i1-i2 subunits are inactive. It is thus possible that the ratio of the type of complexes formed depends upon the enzyme. Thus, the inactive complexes (i2-i2 and/or i1-i2) may be more abundant for UGT1A7 than for UGT1A1, but this remains to be demonstrated. Another possibility is that some UGTs may be dimeric, whereas others could form higher oligomers, either trimers or tetramers. We also observed that the UGT1A_i2-mediated inhibition does not seem to be substrate-specific but would rather depend on the UGT1A isoform. A remarkable inhibition of ∼80% for all of the substrates tested was observed for UGT1A7 in the presence of i2, whereas ∼30–40% reduction was observed for UGT1A8, regardless of the affinity of the enzyme for the substrate (Table 2). Altogether, the data suggest a regulation of glucuronidation activity by formation of inactive complexes. The relative abundance of active/inactive complexes, either in the form of dimers or higher oligomers, would be determinant of global transferase activity of the cell (Fig. 7). Whether these molecular interactions might be extended to other UGTs remains to be demonstrated. According to oligomeric interactions between UGT1As (8, 10, 22–25), multiple heterogeneous interactions between UGT1A protein isoforms are potentially expected. Such interactions will require further in-depth investigations and are currently in progress.
TABLE 2.
Functional analysis of co-expressed spliced isoforms demonstrates significant repression of transferase activity
The relative Vmax values are adjusted for the relative level of expression of UGT1A_i1 protein (active isoforms). UGT1A1_i1+i2 = 1.45; UGT1A7_i1+i2 = 0.85; UGT1A8_i1+i2 = 0.85. S, sigmoid profile; SI, substrate inhibition profile; MM, Michaelis-Menten profile.
| Substrates | i2 | Apparent S50 or apparent Km | Relative Vmax | vs. i1 | Kinetic profile |
|---|---|---|---|---|---|
| μm | pmol/min/mg of protein | % | |||
| UGT1A1 | |||||
| Estradiola | − | 17.93 ± 0.2c | 133.8 ± 3.5 | S | |
| + | 15.03 ± 3.4c | 92.9 ± 10.6d | ↓31 | S | |
| SN-38a | − | 9.2 ± 2.7 | 44.0 ± 2.2 | SI | |
| + | 8.9 ± 0.1 | 26.0 ± 0.1d | ↓41 | SI | |
| Bilirubina | − | 0.73 ± 0.21 | 91.4 ± 14.8 | SI | |
| + | 0.70 ± 0.04 | 75.0 ± 11.1d | ↓18 | SI | |
| UGT1A7 | |||||
| SN-38a | − | 6.5 ± 1.1 | 6.3 ± 11.6 | SI | |
| + | 6.8 ± 4.1 | 1.1 ± 0.1d | ↓82 | SI | |
| 4-MUb | − | 82.5 ± 8.9 | 2.54 ± 0.13 | MM | |
| + | 74.6 ± 6.8 | 0.46 ± 0.04d | ↓81 | MM | |
| UGT1A8 | |||||
| Estradiola | − | 22.0 ± 5.6 | 53.1 ± 2.8 | MM | |
| + | 16.1 ± 1.0 | 28.7 ± 0.4d | ↓46 | MM | |
| MPAb | − | 128 ± 40 | 1875 ± 250 | MM | |
| + | 100 ± 27 | 1375 ± 174d | ↓27 | MM | |
| 4-MUb | − | 156 ± 15 | 6.17 ± 0.30 | MM | |
| + | 159 ± 48 | 4.05 ± 0.54d | ↓34 | MM | |
a Referred to as substrates of high affinity.
b Referred to as substrates of low affinity.
c These values are the apparent S50 values. The other values in the column are the apparent Km values.
d Significantly different from UGT1A_i1 alone at p ≤ 0.01.
FIGURE 7.
Schematic representation of the dominant-negative regulatory character of UGT1A_i2 spliced isoform, which is proposed to occur through the formation of inactive complexes with active isoforms 1. In this model, the relative abundance of active (i1-i1)/inactive (i1-i2 and i2-i2) complexes would be determinant of global transferase activity of the cell.
In conclusion, we provide further evidence that the 45-kDa inactive UGT1A_i2 isoforms inhibit the UGT1A-mediated glucuronidation and that these endogenous UGT1A spliced forms act as dominant-negative repressors of glucuronidaton activity. Also the negative function of i2 proteins is supported by recent results of the synthetic small interfering RNA-mediated depletion of UGT1A_i2 in two human colon cancer cell lines that showed that repression of endogenous i2 results in a statistically significant increase of glucuronidation activity (7). According to our findings, one of the molecular mechanisms of the negative activity of the i2 proteins would be through the interaction with active i1 proteins. The potential for i1 and i2 to oligomerize and form inactive complexes drastically increases the mechanisms by which these proteins can be modulated. Additional studies are clearly needed to gain insight into the domains involved in oligomerization and establish the relationship between degrees of inhibition of UGT1A-mediated glucuronidation induced by the presence of i2 and the relative abundance of active/inactive complexes. Finally, because the mRNA and proteins of these naturally occurring UGT1A splice variants are differentially expressed in different human tissues (3, 4, 7), we suggest that under certain conditions, these newly discovered UGT1A splice variants may have a role in the regulation of UGT1A function in vivo. This new regulatory strategy may thus ensure an additional means to modulate cellular response to endo/xeno stimulus. Further studies are required for a more complete understanding of the role of the i2 proteins and their physiological functions.
Acknowledgments
We thank Patrick Caron and Kim Journault for technical support.
This work was supported by funds from the Canadian Institutes of Health Research and the Canada Research Chair Program (to C. G.).
- UDPGA
- UDP-glucuronic acid
- 4-MU
- 4-methylumbelliferone
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1.Stamm S. (2008) J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 2.Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard H., Lévesque E., Bellemare J., Journault K., Caillier B., Guillemette C. (2007) Pharmacogenet. Genomics 17, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 4.Lévesque E., Girard H., Journault K., Lépine J., Guillemette C. (2007) Hepatology 45, 128–138 [DOI] [PubMed] [Google Scholar]
- 5.Gong Q. H., Cho J. W., Huang T., Potter C., Gholami N., Basu N. K., Kubota S., Carvalho S., Pennington M. W., Owens I. S., Popescu N. C. (2001) Pharmacogenetics 11, 357–368 [DOI] [PubMed] [Google Scholar]
- 6.Ritter J. K., Chen F., Sheen Y. Y., Tran H. M., Kimura S., Yeatman M. T., Owens I. S. (1992) J. Biol. Chem. 267, 3257–3261 [PubMed] [Google Scholar]
- 7.Bellemare J., Rouleau M., Harvey M., Têtu B., Guillemette C. (2009) Pharmacogenomics J., in press [DOI] [PubMed] [Google Scholar]
- 8.Finel M., Kurkela M. (2008) Curr. Drug. Metab. 9, 70–76 [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S. S., Sappal B. S., Kalpana G. V., Lee S. W., Chowdhury J. R., Chowdhury N. R. (2001) J. Biol. Chem. 276, 42108–42115 [DOI] [PubMed] [Google Scholar]
- 10.Operaña T. N., Tukey R. H. (2007) J. Biol. Chem. 282, 4821–4829 [DOI] [PubMed] [Google Scholar]
- 11.Gagné J. F., Montminy V., Belanger P., Journault K., Gaucher G., Guillemette C. (2002) Mol. Pharmacol. 62, 608–617 [DOI] [PubMed] [Google Scholar]
- 12.Lépine J., Bernard O., Plante M., Têtu B., Pelletier G., Labrie F., Bélanger A., Guillemette C. (2004) J. Clin. Endocrinol. Metab. 89, 5222–5232 [DOI] [PubMed] [Google Scholar]
- 13.Bernard O., Guillemette C. (2004) Drug. Metab. Dispos. 32, 775–778 [DOI] [PubMed] [Google Scholar]
- 14.Venkatakrishnan K., Von Moltke L. L., Greenblatt D. J. (2001) J. Clin. Pharmacol. 41, 1149–1179 [DOI] [PubMed] [Google Scholar]
- 15.Peters W. H., Jansen P. L., Nauta H. (1984) J. Biol. Chem. 259, 11701–11705 [PubMed] [Google Scholar]
- 16.Ciotti M., Cho J. W., George J., Owens I. S. (1998) Biochemistry 37, 11018–11025 [DOI] [PubMed] [Google Scholar]
- 17.Meech R., Mackenzie P. I. (1997) J. Biol. Chem. 272, 26913–26917 [DOI] [PubMed] [Google Scholar]
- 18.Ouzzine M., Magdalou J., Burchell B., Fournel-Gigleux S. (1999) J. Biol. Chem. 274, 31401–31409 [DOI] [PubMed] [Google Scholar]
- 19.Ouzzine M., Magdalou J., Burchell B., Fournel-Gigleux S. (1999) FEBS Lett. 454, 187–191 [DOI] [PubMed] [Google Scholar]
- 20.Kern A., Hubbard D., Amano A., Bryant-Greenwood G. D. (2008) Endocrinology 149, 1277–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stasiv Y., Regulski M., Kuzin B., Tully T., Enikolopov G. (2001) J. Biol. Chem. 276, 42241–42251 [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara R., Nakajima M., Oda S., Yamanaka H., Ikushiro S., Sakaki T., Yokoi T. (2010) J. Pharm. Sci. 99, 442–454 [DOI] [PubMed] [Google Scholar]
- 23.Fujiwara R., Nakajima M., Yamanaka H., Katoh M., Yokoi T. (2007) Drug Metab. Dispos. 35, 1781–1787 [DOI] [PubMed] [Google Scholar]
- 24.Ikushiro S., Emi Y., Iyanagi T. (1997) Biochemistry 36, 7154–7161 [DOI] [PubMed] [Google Scholar]
- 25.Kurkela M., Patana A. S., Mackenzie P. I., Court M. H., Tate C. G., Hirvonen J., Goldman A., Finel M. (2007) Pharmacogenet. Genomics 17, 115–126 [DOI] [PubMed] [Google Scholar]