Abstract
The scaffold attachment factors SAFB1 and SAFB2 are paralogs, which are involved in cell cycle regulation, apoptosis, differentiation, and stress response. They have been shown to function as estrogen receptor corepressors, and there is evidence for a role in breast tumorigenesis. To identify their endogenous target genes in MCF-7 breast cancer cells, we utilized a combined approach of chromatin immunoprecipitation (ChIP)-on-chip and gene expression array studies. By performing ChIP-on-chip on microarrays containing 24,000 promoters, we identified 541 SAFB1/SAFB2-binding sites in promoters of known genes, with significant enrichment on chromosomes 1 and 6. Gene expression analysis revealed that the majority of target genes were induced in the absence of SAFB1 or SAFB2 and less were repressed. Interestingly, there was no significant overlap between the genes identified by ChIP-on-chip and gene expression array analysis, suggesting regulation through regions outside the proximal promoters. In contrast to SAFB2, which shared most of its target genes with SAFB1, SAFB1 had many unique target genes, most of them involved in the regulation of the immune system. A subsequent analysis of the estrogen treatment group revealed that 12% of estrogen-regulated genes were dependent on SAFB1, with the majority being estrogen-repressed genes. These were primarily genes involved in apoptosis, such as BBC3, NEDD9, and OPG. Thus, this study confirms the primary role of SAFB1/SAFB2 as corepressors and also uncovers a previously unknown role for SAFB1 in the regulation of immune genes and in estrogen-mediated repression of genes.
Keywords: Cancer/Breast, Chromatin/Immunoprecipitation/ChIP, Receptors/Nuclear, Transcription/Repressor, Transcription/Target Genes, Scaffold Attachment
Introduction
The estrogen receptor α (ERα)3 plays a pivotal role in both normal and pathophysiological processes, such as osteoporosis and breast cancer (1). The transcriptional activity of ERα is not only regulated by its ligand estrogen but also through its interaction with cofactors, which can either enhance (coactivators) or repress (corepressors) its activity (2). Briefly, upon binding of its ligand, ERα undergoes major structural rearrangements leading to exposure of binding surfaces that subsequently results in facilitated recruitment of different coactivators, including histone acetyltransferases or histone methyltransferases, and proteins from the SWI/SNF chromatin remodeling complex (3). In the absence of ligand, recruitment of corepressors results in the creation of locally repressed chromatin, subsequently resulting in inhibition of transcription (4).
Our group has identified the scaffold attachment factors SAFB1 and SAFB2 as ERα corepressors (5, 6). The two proteins are paralogs that share 74% similarity at the amino acid levels, with up to 98% in some functional domains (7). The genes reside in close proximity on chromosome 19p13.3, oriented in a head-to-head orientation, separated by an ∼500-bp GC-rich promoter (6). The C termini of the proteins harbor a repression domain that can refer repression when transferred to a heterologous DNA-binding protein. This repression domain can be further defined into two independent repression domains, termed RD1 and RD2 (8). The interaction between SAFB1 and ERα was mapped to the DNA binding domain hinge domain, a region of high similarity among the nuclear receptor family members. In fact, SAFB1 has been shown to bind to multiple nuclear receptors like farnesoid X receptor α, peroxisome proliferator-activated receptor γ, and the vitamin D receptor (9).
SAFB1 also plays a role in RNA processing; it can bind RNA through its central RNA recognition motif and was shown to effect splice site selection (8, 10). On the N terminus, SAFB1 has a highly conserved region, termed SAF-box, which is a homeodomain-like DNA-binding motif that interacts with scaffold/matrix attachment regions (11). Scaffold/matrix attachment regions are postulated to anchor chromatin onto the nuclear matrix, thereby organizing genomic DNA into topologically distinct loop domains that are important in replication and transcription (12, 13).
Although details have yet to be studied, SAFB1 has already been implicated in a number of cellular processes, including stress response, regulation of cell cycle, apoptosis, and differentiation. Briefly, upon stress, SAFB1 moves to stress-induced nuclear bodies, also called SNB (Src-activated during mitosis nuclear body), where it can colocalize with heat shock factor HSF1 (14). Another group showed that stress also resulted in movement of the zona occludens ZO-2 protein into the nucleus, where it binds to and colocalizes with SAFB1 (15). Lee et al. (16) have recently shown that SAFB1 nuclear distribution changes after induction of apoptosis and that it is cleaved in a caspase 3-dependent manner. Overexpression of SAFB1 caused growth arrest and multinuclearity, suggesting a role for cell proliferation and division (17). Finally, Debril et al. (9) showed that SAFB1 levels changed dramatically during early phases of adipocyte and enterocyte maturation, suggesting a role for SAFB1 in differentiation.
A number of observations collectively provide strong support for SAFB1/SAFB2 playing a role in breast tumorigenesis. First, the SAFB genes map to a chromosomal locus that displays frequent loss of heterozygosity in breast cancer, and second, mutations in SAFB1/2 have been identified (18). A meta-analysis of 151 published loss of heterozygosity studies, including >15,000 breast tumors, identified nine regions of consistent and high loss of heterozygosity, with the SAFB1/SAFB2 locus being one of them (19). The same region was recently linked to hereditary breast cancer in Swedish families; however, this study did not identify germ line mutations in the coding regions of SAFB1 and SAFB2 (20). We have recently reported that SAFB1/SAFB2 protein expression was frequently lost in breast tumors and that this was associated with worse overall survival (21). Mouse embryonic fibroblasts deficient in SAFB1 showed a lack of contact inhibition, increased foci formation, and increased oncogene-induced anchorage-independent growth. Interestingly, they also displayed significantly decreased senescence (22).
To further our understanding how SAFB1 and SAFB2 exert their effects on numerous cellular processes, to identify which estrogen-regulated genes are controlled by SAFB1/SAFB2, and finally to begin to understand how SAFB1/SAFB2 might contribute to tumorigenesis, we performed a ChIP-on-chip study, coupled with gene expression array analysis, and the data are reported here.
MATERIALS AND METHODS
Cell Culture
MCF-7 cells were grown in fetal bovine serum (Invitrogen) supplemented with 5% fetal bovine serum and 1% penicillin and streptomycin in a 5% CO2 atmosphere at 37 °C. Before treatments or transfections, the cells were maintained in phenol red-free minimal essential medium containing 5% charcoal-stripped calf serum for a minimum of 2 days.
Immunoblotting
Cells were lysed with 5% SDS; lysates were sonicated, and proteins were subjected to 8% SDS-PAGE and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with monoclonal anti-SAFB1 (1:1000, see below) and polyclonal anti-SAFB2 (1:1000) (6) primary antibodies. Subsequently, the nitrocellulose membranes were incubated with secondary antibodies (GE Healthcare) conjugated with peroxidase. Densitometry of protein bands was acquired using an AlphaImager EC gel documentation system (Alpha Innotec), and bands were analyzed with the spot densitometry analysis tool (Alpha Ease FC software, version 4.1.0).
Generation of SAFB1-specific Monoclonal Antibody
The SAFB1 peptide AEIDNGSVADCVEDDDADNLQESLSDSRELVEGEMKELPEQLQEHAIEDKETINNLDTSSSDFTILQEIEEPS, corresponding to amino acids 137–209, maps to a region of low similarity between SAFB1 and SAFB2. It was generated as a glutathione S-transferase fusion protein and purified, and a mouse monoclonal antibody was generated in the Baylor College of Medicine Dan L. Duncan Cancer Center Protein Core following standard procedures. The supernatant was subsequently purified using peptide columns, and the final monoclonal antibody is highly specific for SAFB1.
siRNA Transfection, Estrogen (E2) Treatment, and RNA Isolation
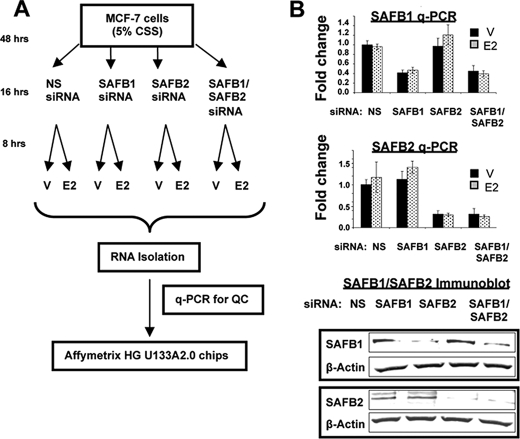
The MCF-7 cells were grown in charcoal-stripped serum for at least 2 days. The cells were then transfected with either control siRNA (NS) (Dharmacon, siGenome nontargeting siRNA 2, catalogue no. D-00121002-20), SAFB1 (siGENOME SMARTpool, catalogue no. L-005150-00), and/or SAFB2 (siGENOME SMARTpool, catalogue no. L-020373-01) siRNA, using Lipofectamine 2000 (Invitrogen). The overall study design is shown in Fig. 1A. Forty eight hours post-transfection, the cells were treated with 19 nm E2 or ethanol vehicle for 8 h prior to RNA extraction. Total RNA was isolated using the RNeasy RNA isolation kit from Qiagen (Valencia, CA) according to the manufacturer's protocols, and RNA quality was analyzed by the 260:280 ratio. To exclude off-target effects, some studies were repeated with a second independent SAFB1 siRNA (Invitrogen, catalogue no. 1299001; SAFBHSS109450).
FIGURE 1.
Design of gene expression array study, and quality control of SAFB1/SAFB2 siRNA in MCF-7 cells. A, model of experimental setup of gene expression array study. V, vehicle. B, specific and efficient down-regulation of SAFB1 and SAFB2 by siRNA. MCF-7 cells were transiently transfected with NS siRNA, SAFB1 siRNA, SAFB2 siRNA, or SAFB1/SAFB2 siRNA. mRNA levels of SAFB1 and SAFB2 were measured by q-PCR (top panel), and protein expression by immunoblotting (bottom panel) using antibodies as indicated.
Quantitative Real Time PCR (q-PCR)
RNA was treated with DNase (Roche Applied Science) as described before (6). The RNA was reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen). For q-PCR analysis, we used either TaqMan, or SYBR Green chemistry (Applied Biosystems, Foster City, CA), and the details, including primer sequences, are provided in supplemental Table S1. The assay was carried out in a ABI PRISM 7700 sequence detector system (Applied Biosystems, Foster City, CA). Relative mRNA expression was calculated by ΔΔCt method (23), using β-actin as the reference gene.
Chromatin Immunoprecipitation (ChIP) and ChIP-on-chip Assay
The ChIP assays were essentially performed as we described previously (24). Immunoprecipitations were performed with polyclonal ERα (H-184, Santa Cruz Biotechnology), a pan-SAFB monoclonal (5, 24) or polyclonal SAFB1 (25) or hemagglutinin antibodies (Covance). The primer sequences for the regulatory regions tested in ChIP assays are provided in supplemental Table S1. The relative enrichment of ChIP DNAs is presented as percent input.
For the ChIP-on-chip study, we used a pan-SAFB antibody that recognizes both paralogs SAFB1 and SAFB2 (Upstate, now Millipore). Specificity of the antibody was previously tested using several approaches, including enzyme-linked immunosorbent assay, immunohistochemistry, and immunoblotting, and use of the antibody for SAFB studies, including ChIP, has previously been published (5, 24). An initial validation by q-PCR was performed to confirm that there was enrichment of the known SAFB target genes hsp27/HSBP1 and pS2/TIFF (7, 24). ChIP products and their respective input genomic fragments were amplified by ligation-mediated PCR. qChIP was repeated after amplifications to verify that enrichment of pS2 and hsp27 was retained. The genomic products of three biological ChIP replicates were labeled with Cy5 (for ChIP products) and Cy3 (for input) and cohybridized according to standard NimbleGen protocols (NimbleGen Systems, Inc., Madison, WI). A NimbleGen human promoter array representing 1.5 kb of promoter sequence (15 probes of 50 bp each) from 24,275 genes (human genome from May, 2004) was used. As described previously (26), for the data analysis, the log-ratio was calculated between the probe intensities of the ChIP product and input chromatin, which were cohybridized on the same assay. For each of the 24,275 promoter regions, the moving average of log-ratio along its 15 probes with window size 3 was computed to find the maximum value of the averages of three neighboring probes. Subsequently, the log-ratio of all probes across the entire array was randomly permutated, and the maximum values of moving average for each gene was re-computed to find the 90th, 95th, and 99th percentile of these 24,275 values from the random permutation. The positive threshold was set up using the signal of positive control genes pS2 and hsp27. This threshold was greater than the 90th percentile in the three replicates. To identify candidate SAFB-binding sites, we took promoter regions whose maximum moving average was above the cutoffs in the three experiments.
Gene Expression Arrays and Analysis
RNA was isolated as mentioned above and was subjected to DNase treatment before reverse transcription. We studied gene expressions using Affymetrix Human Genome HG-U133A2.0 Gene Chip (Santa Clara, CA) expression arrays (22,215 probe sets). Five micrograms of total RNA from each sample were labeled using the bioarray high yield transcript kit according to the manufacturer's conditions (ENZO, Farmingdale, NY). Labeled RNAs were hybridized and washed according to the manufacturer's recommendations (Affymetrix, Inc., Santa Clara, CA). All expression array data have been deposited into the Gene Expression Omnibus, accession number GSE15548.
The data were analyzed using the dChip software for gene expression estimation (with PM Perfect Match only model) and clustering (27). Analysis of variance with contrast analysis with Insightful ArrayAnalyzer was used for group comparisons. To find differentially expressed genes, we used Benjamini-Hockberg correction for multiple comparisons (28) with false discovery rate = 0.1. Experiments were performed in quadruplicates; however, out of 32 samples, 3 samples displayed very low 5′–3′ ratios (1× NS siRNA vehicle, 1× NS siRNA E2, and 1× SAFB1 siRNA vehicle) and were thus not included in the final analysis. For gene ontology analysis and chromosomal enrichment, we used Positional Enrichment Analysis software (29), Ingenuity Pathway Assist, DAVID (30), and EASE.
Finally, the calculation of the number of common genes between the significant genes in expression array and ChIP analyses that would be expected by chance was found based on the following: 1) the number of common genes on the entire chips used in those assays (5827); 2) the number of significant genes from the microarray analysis in the 5827 genes (472); and 3) the number of binding sites in the 5827 genes (363). This gives the number of common genes expected by chance equal to ∼29 (472 × 363/5827).
RESULTS
Identification of SAFB-binding Sites
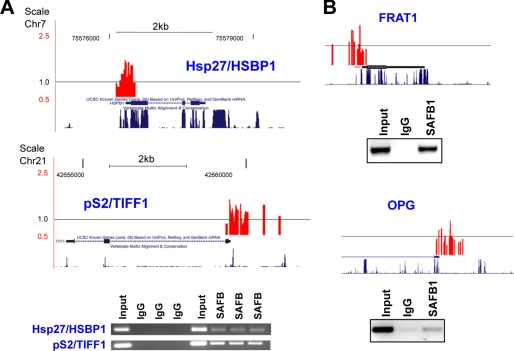
To identify SAFB1 and SAFB2 target genes, we performed a ChIP-on-chip in MCF-7 cells, an estrogen receptor-positive breast cancer cell line. Chromatin fragments were immunoprecipitated with a pan-SAFB antibody or a control IgG antibody, and the products were amplified by ligation-mediated PCR. Specific enrichment of the known SAFB target promoters hsp27/HSBP1 (7) and pS2/TIFF1 (24) was validated by quantitative PCR ChIP before and after ligation-mediated PCR. Subsequently, we labeled the amplicons and hybridized them onto high density oligonucleotide arrays containing 1.5-kb sequences of 24,275 genes. Using a stringent analysis, in which only overlapping peak enrichment in all three replicates was considered positive, we identified 818 SAFB-binding sites (supplemental Table S2, A and B). 541 of those sites map to promoters of known genes (supplemental Table S2A), whereas 277 are upstream of unknown genes, containing high protein probability-coding sequences (KIAA/FLJ/DKFZ/CnORF) (supplemental Table S2B). Fig. 2A shows graphically the presentation of SAFB-binding peaks and ChIP results for hsp27/HSBP1 and pS2/TIFF1 (our controls). Fig. 2B shows the recruitment of SAFB1 to two additional genes identified in the ChIP-on-chip study (FRAT1 and osteoprotegerin (OPG)).
FIGURE 2.
Confirmation of SAFB binding to candidate genes identified in ChIP-chip study. A, confirmation of SAFB binding to control genes hsp27/HSPB1 and pS2/TIFF1. Top panel, graphical presentation of SAFB1-binding peaks in hsp27/HSPB1 and pS2/TIFF1 genes, as identified in the ChIP-chip experiment. The black line around 1 represent the average binding (signal) across the entire array. The gene structure and the mammalian conservation of sequence are shown. Bottom panel, ChIP was performed using pan-SAFB antibodies and triplicate biological samples (primer sequences are provided in supplemental Table S1). B, same as in A, but with FRAT1 and OPG genes. ChIP assay, using SAFB1-specific antibodies, is representative of three independent experiments.
We next asked the question whether there was an enrichment of SAFB target-binding sites on specific chromosomes and/or chromosomal location, which was done by using EASE and positional gene enrichment software packages, respectively. We identified enrichment of 6p arm, with an EASE score of 0.0013. Positional gene enrichment analysis revealed clustering of SAFB-binding sites on chromosome 6, region 26266328 to 28411890 (p = 5.4E-10), and on chromosome 1, region 144218966 to 148167146 (p = 1.11E-7). Interestingly, both of these regions contain histone gene clusters, and as shown in the supplemental Table S2A, SAFB recruitment was identified in a large number of histone genes. This is of interest because genes in the histone cluster have been proposed to be regulated in specific nuclear territories (31) and might therefore suggest that SAFB proteins might also play a role in higher order chromatin regulation.
In summary, the ChIP-on-chip analysis identified 541 SAFB-binding sites in promoters of known genes, and the analysis revealed enrichment of SAFB recruitment to two chromosomal regions that contain histone gene clusters.
SAFB1 Represses More Target Genes than SAFB2
To identify transcripts that were altered upon decrease of SAFB1 or SAFB2 expression, we performed gene expression array analysis in MCF-7 cells. A secondary goal was to identify ERα target genes for which estrogen regulation was dependent upon SAFB expression. We also tested the impact of cotransfecting siRNAs targeting both SAFB1 and SAFB2, which allowed us to address the question of interactions between the two paralogs. We therefore set up an experiment reflecting a 2 × 4 design, with a total of eight experimental groups (Fig. 1A). q-PCR and immunoblot analysis confirmed that SAFB1 and SAFB2 were decreased by ∼60 and ∼75% at the RNA level and by ∼80 to ∼90% at the protein level, respectively (Fig. 1B). The siRNA targeting both SAFB1 and SAFB2 resulted in ∼65 and 75% knockdown at the RNA and protein level, respectively. The RNA from biological quadruplicates was subsequently used for cDNA synthesis and cRNA labeling. Three samples displayed very low 5′–3′ ratios (1× NS siRNA vehicle, 1× NS siRNA E2, and 1× SAFB1 siRNA vehicle) and were thus not used for array hybridization. A total of 29 samples were hybridized to Affymetrix U133A2.0 chips, and data were analyzed as described under “Material and Methods.”
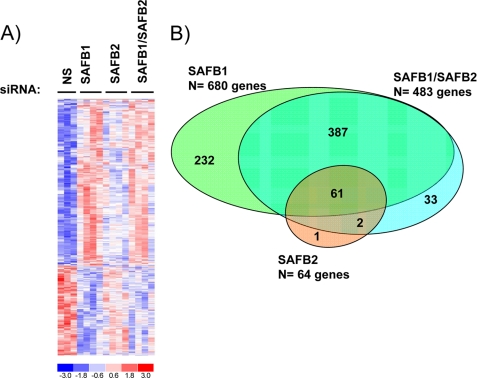
Analyzing the expression of genes across four experimental groups (NS, SAFB1, SAFB2, or SAFB1/SAFB2) in the presence of vehicle allowed us to identify the global SAFB1/SAFB2-regulated gene set. Using a Benjamin-Hochberg adjusted to p = 0.1, we identified 716 genes (Fig. 3A and supplemental Table S3). There were more genes induced (n = 457) than repressed (n = 259), reflecting the known role of SAFB1 and SAFB2 in transcriptional repression (5, 6).
FIGURE 3.
Identification of SAFB1, SAFB2, and SAFB1/SAFB2 target genes in MCF-7 cells. A, heat map representing 716 genes that were significantly regulated by SAFB1, SAFB2, or the combination of SAFB1/SAFB2 siRNA. The cutoff for this analysis was based on a Benjamin-Hochberg adjusted p = 0.1. Expression levels are shown in red and blue (levels are above and below the median, respectively). B, SAFB1 regulates more target genes than SAFB2. The Venn diagram represents genes that are significantly regulated by SAFB1, SAFB2, and SAFB1/SAFB2.
We next asked how many of the 716 target genes identified by gene expression array analysis contained SAFB1/SAFB2-binding sites in their promoters as identified by our ChIP-on-chip study. Overlapping of the lists revealed only 24 genes (Table 1). The number of common genes expected by chance is 29, which suggests that the majority of the SAFB1/SAFB2 target genes are either not regulated through recruitment of SAFB1/SAFB2 to the promoter regions (used in the ChIP-on-chip studies) or are regulated through other mechanisms. Alternatively, it is feasible that at least some of the target genes identified by gene expression array analysis represent indirect targets of SAFB1/SAFB2. Given that Jun and Fos were identified among the SAFB1/SAFB2 target genes, we asked the question whether the regulation was simply a result of changing AP1 activity. A comparison of the AP1 target genes (n = 116) from the study by DeNardo et al. (32) with the 716 SAFB1/SAFB2 target genes (Fig. 3A and supplemental Table S3) revealed only 14 common target genes, which is not significantly different from the number of overlapping genes one would expect by chance. These data would suggest that SAFB1 does not modify its target genes indirectly, through regulating AP1 (fos/jun) activity.
TABLE 1.
SAFB1/SAFB2 target genes that also contained SAFB-bindings sites in their promoter regions
The table contains 24 genes listed in alphabetical order that were identified in two independent analyses, in the ChIP-on-chip study (n = 818) and in the gene expression array analysis (n = 716).
| Accession no. | Official gene symbol | Gene name | -Fold change (siSAFB) | |
|---|---|---|---|---|
| 1 | NM_000425 | L1CAM | L1 cell adhesion molecule isoform 1 precursor | 1.89 |
| 2 | NM_005252 | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 1.63 |
| 3 | NM_002800 | PSMB9 | Proteasome β9 subunit isoform 1 proprotein | 1.58 |
| 4 | X56841 | HLA-E | HLA-E | 1.52 |
| 5 | NM_002228 | JUN | v-jun avian sarcoma virus 17 oncogene homolog | 1.49 |
| 6 | NM_004529 | MLLT3 | Myeloid/lymphoid or mixed lineage leukemia (trithorax homolog) | 1.37 |
| 7 | NM_003337 | UBE2B | Ubiquitin-conjugating enzyme E2B | 1.36 |
| 8 | NM_014417 | BBC3 | BCL2-binding component 3 | 1.33 |
| 9 | NM_002117 | HLA-C | Major histocompatibility complex | 1.31 |
| 10 | AF458589 | PPP1R12A | Myosin phosphatase target subunit 1 variant | 1.24 |
| 11 | NM_002419 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | 1.22 |
| 12 | NM_002121 | HLA-DPB1 | Major histocompatibility complex | 1.21 |
| 13 | BC017093 | RPL35A | RPL35A protein | 1.18 |
| 14 | NM_004048 | B2M | β2-Microglobulin precursor | 1.18 |
| 15 | NM_000520 | HEXA | Hexosaminidase A preproprotein | 1.11 |
| 16 | NM_002623 | PFDN4 | Prefoldin 4 | 1.09 |
| 17 | NM_017582 | UBE2Q | Ubiquitin-conjugating enzyme E2Q | 1.05 |
| 18 | NM_014141 | CNTNAP2 | Cell recognition molecule Caspr2 precursor | 0.81 |
| 19 | NM_003366 | UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | 0.79 |
| 20 | NM_078469 | BCCIP | BRCA2 and CDKN1A-interacting protein isoform C | 0.79 |
| 21 | NM_001417 | EIF4B | Eukaryotic translation initiation factor 4B | 0.77 |
| 22 | NM_001005273 | CHD3 | Chromodomain helicase DNA-binding protein 3 isoform 1 | 0.77 |
| 23 | NM_014649 | SAFB2 | Scaffold attachment factor B2 | 0.75 |
| 24 | NM_005105 | RBM8A | RNA-binding motif protein 8A | 0.64 |
To identify individual contributions of SAFB1 and SAFB2 in gene regulation, and potential interactions, we next investigated the overlap between the following three experimental groups: genes regulated by SAFB1 siRNA, SAFB2 siRNA, or SAFB1/SAFB2 siRNA (Fig. 3B). The most striking result was that out of the 716 SAFB1/SAFB2-regulated genes, 680 genes were significantly regulated by SAFB1 (supplemental Table 4A) and only 64 genes by SAFB2 (supplemental Table S4B). Furthermore, lack of SAFB1 results in altered expression of 163 target genes that were not affected by SAFB2 siRNA (supplemental Table S5), whereas SAFB2 only has one unique target gene (islet cell autoantigen 1, ICA1). To exclude siRNA off-target effects, we utilized a second independent SAFB1 siRNA and performed q-PCR studies on six SAFB1 target genes. We were able to confirm a significant effect of SAFB1 loss in five of those, which strongly suggests that the majority of the targets do not represent off-target effects (data not shown). Finally, the combined knockdown of SAFB1 and SAFB2 did not result in the regulation of additional genes (n = 483, supplemental Table S4C) n.s., not significant), suggesting a lack of interaction between SAFB1 and SAFB2 in gene regulation.
In summary, the gene expression array data support the previously described roles of SAFB1 and SAFB2 in transcriptional repression. Also, our data strongly indicate that SAFB1 plays a more critical role in regulation of gene expression in MCF-7 cells, compared with SAFB2. We therefore focused our subsequent more detailed studies on SAFB1 target genes.
Majority of SAFB1 Targets Are Immune Regulatory Genes
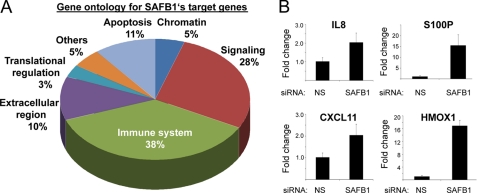
To gain insights into the functions of the identified SAFB1 target genes, we evaluated their associated Gene Ontology Biological Process terms. This analysis revealed that the majority of SAFB1 target genes are involved in immune regulation (38%), followed by genes critical in signaling (28%) and in apoptosis (11%) (Fig. 4A). The effect of SAFB1 on immune regulatory genes was of interest, considering the increasing evidence that immune cells play a pivotal role in tumorigenesis (33). Therefore, we sought to confirm the effect of SAFB1 siRNA on the expression of interesting candidate genes previously assigned to immune regulatory functions and tumorigenesis. We selected four genes from the top candidate gene list (>4-fold change with SAFB1 siRNA) (Table 2) and were able to confirm significant loss of repression for all of them (IL8, S100P, CXCl11, and HMOX1) (Fig. 4B). Thus, the gene expression array analysis uncovered a previously unknown role for SAFB1 in the regulation of genes related to the immune system.
FIGURE 4.
SAFB1 regulates immune genes. A, gene ontology analysis for SAFB1-regulated genes (n = 680) revealed that the majority represent immune regulatory, signaling, and apoptotic genes. B, SAFB1 is necessary for the repression of immune regulatory genes. The SAFB1-mediated repression of four immune regulatory SAFB1 candidate genes was tested (IL8, S100P, CXCL11, and HMOX1). q-PCR was performed using RNA from MCF-7 cells transfected with NS siRNA or SAFB1 siRNA. The data are an average of three replicates ± S.D.
TABLE 2.
SAFB1 target genes identified in gene expression array analysis
Target genes were prioritized based on a ≥4-fold change in the presence of SAFB1siRNA.
| Accession no. | Official gene symbol | Gene name | -Fold change (siSAFB1) | |
|---|---|---|---|---|
| 1 | 218541_s_at | C8orf4 | Chromosome 8 open reading frame 4 | 11.091 |
| 2 | 205890_s_at | GABBR1///UBD | γ-Aminobutyric acid (GABA) B receptor, 1///ubiquitin D | 10.772 |
| 3 | 204351_at | S100P | S100 calcium-binding protein P | 9.666 |
| 4 | 203665_at | HMOX1 | Heme oxygenase (decycling) 1 | 9.06 |
| 5 | 209619_at | CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | 9.024 |
| 6 | 211990_at | HLA-DPA1 | Major histocompatibility complex, class II, DPα1 | 8.116 |
| 7 | 207339_s_at | LTB | Lymphotoxin β (tumor necrosis factor superfamily, member 3) | 6.435 |
| 8 | 204932_at | TNFRSF11B | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | 6.365 |
| 9 | 211122_s_at | CXCL11 | Chemokine (CXC motif) ligand 11 | 5.777 |
| 10 | 202859_x_at | IL8 | Interleukin 8 | 5.539 |
| 11 | 823_at | CX3CL1 | Chemokine (CX3C motif) ligand 1 | 5.485 |
| 12 | 204698_at | ISG20 | Interferon-stimulated exonuclease gene 20 kDa | 5.34 |
| 13 | 209230_s_at | P8 | p8 protein (candidate of metastasis 1) | 5.311 |
| 14 | 208894_at | HLA-DRA | Major histocompatibility complex, class II, DRα | 5.202 |
| 15 | 203060_s_at | PAPSS2 | 3′-Phosphoadenosine 5′-phosphosulfate synthase 2 | 4.971 |
| 16 | 33304_at | ISG20 | Interferon-stimulated exonuclease gene 20 kDa | 4.9 |
| 17 | 1405_i_at | CCL5 | Chemokine (CC motif) ligand 5 | 4.779 |
| 18 | 204655_at | CCL5 | Chemokine (CC motif) ligand 5 | 4.63 |
| 19 | 210163_at | CXCL11 | Chemokine (CXC motif) ligand 11 | 4.623 |
| 20 | 204237_at | GULP1 | GULP, engulfment adaptor PTB domain containing 1 | 4.619 |
| 21 | 202917_s_at | S100A8 | S100 calcium-binding protein A8 (calgranulin A) | 4.606 |
| 22 | 204533_at | CXCL10 | Chemokine (CXC motif) ligand 10 | 4.466 |
| 23 | 214329_x_at | TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 4.167 |
| 24 | 205027_s_at | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 4.16 |
| 25 | 210982_s_at | HLA-DRA | Major histocompatibility complex, class II, DRα | 4.071 |
| 26 | 214321_at | NOV | Nephroblastoma overexpressed gene | 4.045 |
| 27 | 208173_at | IFNB1 | Interferon, β1, fibroblast | 4.022 |
Loss of SAFB1 Leads to De-repression of Estrogen-regulated Genes, Many of Which Are Known Regulators of Apoptosis
Because SAFB1 is a known ERα corepressor, we next asked whether estrogen-regulated genes were affected by loss of SAFB1. Therefore, we performed a subset analysis using the data from the 2 × 2 experimental design, control, and SAFB1 siRNA and vehicle and estrogen treatment. Using adjusted p value = 0.1, we identified a total of 585 genes that were significantly altered by estrogen in control siRNA-transfected cells (supplemental Table S6). We determined that 12% (68 genes) of these estrogen-regulated genes were dependent on SAFB1 (supplemental Table S7), i.e. were altered in the presence of SAFB1 siRNA (p < 0.1). We also asked the following question: How many of the E2-regulated genes have SAFB on their promoters in the ChIP-on-chip analysis, regardless of whether they are affected by SAFB siRNA? Overlap of ChIP-chip (supplemental Table S2) and E2-regulated gene lists (supplemental Table S6) identified a total of 16 E2-regulated genes with SAFB-binding sites in their promoters, suggesting that SAFB has many ER-independent target genes and also supporting the idea that regulation by SAFB occurs outside the proximal promoter region.
The 68 estrogen and SAFB1-regulated genes included 61 repressed genes and 7 induced genes. Even considering the fact that there are more repressed (n = 380) than induced (n = 205) genes following estrogen treatment, there is still a significant effect of SAFB1 on repressed genes compared with induced genes (16 versus 3%, p = 0.01).
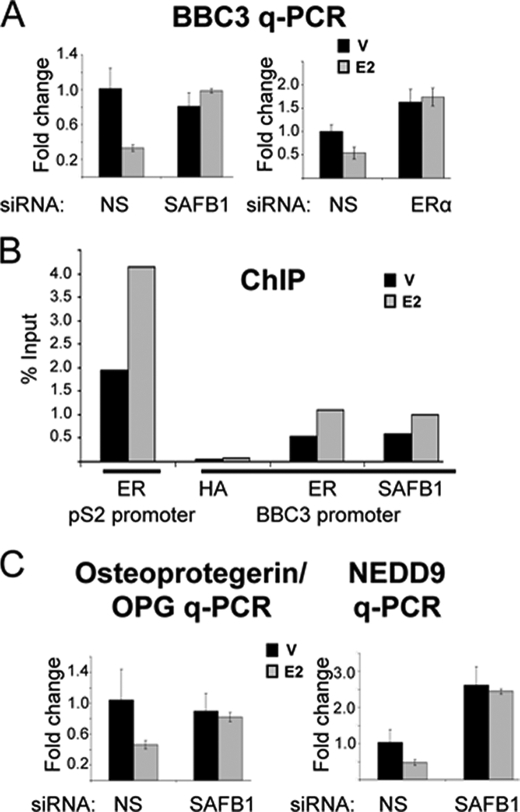
Little is known about estrogen-mediated repression of gene regulation, although recent genome-wide expression studies showed that approximately half of the ERα target genes are repressed upon estrogen treatment (34–36). Because the nature of the repressed target genes is largely unknown, we performed an enrichment analysis for the gene ontology categories of the estrogen-repressed versus induced genes. In contrast to induced genes, which were enriched for cell cycle-related genes, the repressed genes were significantly enriched for apoptosis/programmed cell death (p = 0.008). This observation prompted us to analyze the SAFB1 dependence of apoptotic target gene expression. We focused on the BCL2-binding component 3 (BBC3), previous shown to be critical for apoptosis (37), and which was also identified as a SAFB1/SAFB2 target gene in our ChIP-on-chip study (supplemental Table S2A). We were able to confirm the loss of estrogen repression in the absence of SAFB1 in q-PCR studies (Fig. 5A, left panel). Knockdown of ERα also resulted in loss of repression (Fig. 5A, right panel), confirming that the estrogen-mediated repression is an ERα-dependent effect. As expected, given the ChIP-on-chip results, we were able to detect recruitment of SAFB1 to the BBC3 promoter, along with recruitment of ERα (Fig. 5B).
FIGURE 5.
SAFB1 is involved in estrogen-mediated repression of apoptotic genes. A, E2 repression of BBC3 is SAFB1 and ERα-dependent. BBC3 q-PCR was performed using RNA from MCF-7 cells transfected with NS siRNA, SAFB1 siRNA (left panel), or ERα siRNA (right panel) and treated with vehicle (V) (black) or estrogen (gray) (8 h). The data are an average of three replicates ± S.D. B, ERα and SAFB1 are recruited to the BBC3 proximal promoter. Results of a ChIP analysis on the BBC3 promoter using antibodies against hemagglutinin (negative control), ERα, and SAFB1 are shown. For the ChIP, MCF-7 cells were treated with vehicle (black) or estrogen (gray) for 45 min. Recruitment of ERα to the pS2 promoter was used as positive control. C, loss of estrogen repression of osteoprotegerin and NEDD9 in the absence of SAFB1. The E2-mediated repression of osteoprotegerin and NEDD9 was tested using RNA from MCF-7 cells transfected with NS siRNA and SAFB1 siRNA and treated with vehicle (black) or estrogen (gray) (8 h). q-PCR analysis of mRNA was performed, and the data shown are an average of three replicates ± S.D.
These data prompted the question whether repression by both ER and SAFB1 is a general property of all genes identified in the SAFB1/B2 gene expression array study and the ChIP-on-chip analysis (Table 1). Out of 14 genes tested (B2M, BBC3, CHD3, EIF4B, FOS, HEXA, HLA-C, HLA-DPB1, HLA-E, jun, MLLT3, PFDN4, SAFB2, and UBE2B) 12 were repressed by SAFB1 (B2M, BBC3, fos, HEXA, HLA-C, HLA-DPB1, HLA-E, jun, MLLT3, PFDN4, SAFB2, and UBE2B), but only 4 were repressed by ER (BBC3, fos, HLA-DBP1, and HLA-C) (i.e. induced in the presence of the siRNA) (supplemental Fig. S1). Thus, the analysis of a subset of the genes clearly shows that repression by both ER and SAFB1 is not a general property of all SAFB1/SAFB1 target genes with SAFB-binding sites in their promoter.
Other estrogen-regulated SAFB1 target genes involved in apoptosis are OPG and the neural precursor cell expressed developmentally down-regulated 9 protein (NEDD9), for which we were also able to confirm loss of estrogen-mediated repression in the absence of SAFB1 (Fig. 5C). (The same results were obtained using the second independent SAFB1 siRNA (data not shown), confirming that the observed regulation was not an off-target effect.) Together, these studies suggest that SAFB1 is involved in estrogen-mediated repression of ERα target genes, many of which are critical in apoptotic processes.
DISCUSSION
In this study, we have identified target genes of the transcriptional repressors SAFB1 and SAFB2 in breast cancer cells. Using a combined Chip-on chip and gene expression array analysis approach, we confirmed primary roles of SAFB1/SAFB2 as corepressors, and we discovered previously unknown roles for SAFB1 in the regulation of immune genes and in estrogen-mediated repression of genes.
We found that SAFB1 has many more target genes than its paralog SAFB2. This is not a result of differences in siRNAs efficiencies, because the down-regulation of SAFB1 expression was not stronger compared with SAFB2. Another simple explanation could be that SAFB1 is expressed at higher levels compared with SAFB2 in MCF-7 cells. Although our immunoblotting and q-PCR data do not seem to support this, we cannot exclude this possibility because we currently do not have any data on absolute expression of SAFB1 and SAFB2 RNA and protein. We would therefore like to speculate that the two proteins, although highly conserved, especially in conserved functional domains, have some distinct molecular functions. Sergeant et al. (38) showed that SAFB2 is associated with nuclear protein complexes larger in size than those containing SAFB1 and that SAFB2 but not SAFB1 was detectably associated with nucleic acids, although the integrity of the core SAFB2 protein complexes was micrococcal nuclease-independent. Also, SAFB1 and SAFB2 have distinct subnuclear organizations, although this was cell type-dependent. Finally, the plethora of phenotypes in the SAFB1 knock-out mice suggests that SAFB1 has many functions that cannot be compensated for by SAFB2 (25). Those might involve the regulation of immune target genes we identified in this study.
Intriguingly, SAFB1 regulates the expression of a number of genes critical in the immune system, such as interleukins, chemokines, and members of the major histocompatibility complex class proteins. The in vitro data are supported by in vivo observations in SAFB1−/− mice that have numerous defects in the immune system (25).4 Given the increasing realization that the immune system is an integral part of tumorigenesis, it is interesting to speculate that the regulation of the immune genes might be involved in the role of SAFB1 in breast cancer.
There is a striking similarity in many of the SAFB1-related findings to what has been described for the special AT-rich binding protein 1, SATB1. Like SAFB1, the SATB1 protein binds to scaffold/matrix attachment regions (39, 40), has been shown to play a role in the regulation of the immune system (41), is involved in breast tumorigenesis (42), and has a paralog SATB2, which shares some functions but also has unique properties (43–45). It is of interest that SATB1 orchestrates expression of many genes through its role as a “genome organizer,” in which it directs the formation of chromatin loops through effects on higher order chromatin compaction (12, 46–48). At this point, a similar function has yet to be described for SAFB1; however, some of the data presented here point toward a similar role for SAFB1. First, there is a striking lack of overlap between the genes identified by the ChIP-on-chip study and the gene expression array analysis, suggesting that most SAFB1/SAFB2 target genes do not contain SAFB1/SAFB2 recruitment sites in their proximal promoter region but are regulated through other mechanisms. Second, the ChIP-on-chip study identified an over-representation of two chromosomal loci, chromosome 1 and 6, for binding of SAFB1/SAFB2. These loci contain many members of the histone gene cluster, which has also been shown to be regulated though effects on higher order chromatin structure (31).
There is also strong evidence that SAFB proteins can effect gene regulation through classical action on transcription. It can interact with RNA polymerase II and with a number of nuclear receptors, including ER (5, 6) and peroxisome proliferator-activated receptor γ (9), and it has a strong repression domain that refers repression to heterologous promoters when fused to Gal4-DBD (8). We have previously shown that SAFB1/2 can function as ER corepressor, and we show here that loss of SAFB1 results in loss of estrogen repression of ERα target genes (5, 6). Only a small subset of SAFB1 target genes was modulated after ERα siRNA treatment (including a few target genes known to be estrogen-regulated), which could be due to incomplete knockdown of ERα protein or regulation through ERβ. In any case, this finding highlights the fact that SAFB1 has many ERα-independent target genes.
The estrogen-mediated repression of transcription has only recently come into the spotlight, because in the past ERα has mainly been studied as a classical transcriptional activator, containing two activation domains. It is of interest that many estrogen-repressed genes are involved in apoptosis, and repression of those by estrogen might be a critical process in hormone response, both in normal and in cancer cells. A recent comprehensive global ChIP-on-chip study showed dismissal of RNA polymerase II, decrease of acetylation, and absence of Srcs binding at promoters of repressed genes (49). There is increasing evidence that corepressors such as CtBP1, SMRT, Pax6, and NCoR1 are involved in repression of ERα target genes, and the studies presented here add SAFB1 to this list of corepressors. Together, these findings suggest that SAFB1/SAFB2 belong to a unique class of multifunctional proteins, such as MeCP2 (50), with potentially important roles in chromatin architecture, regulation of RNA splicing, and repression of transcription.
Supplementary Material
Acknowledgments
We thank the Dan L. Duncan Cancer Center Protein Core (supported by National Institutes of Health Grant P30CA125123), Baylor College of Medicine, for generation of monoclonal SAFB1 antibodies. We also thank Dr. Adrian Lee for input and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA9721305 (to S. O.) and P01 CA03019526 (to S. O.). This work was also supported by Dr. Mildred Scheel Stiftung, Deutsche Krebshilfe (German Cancer Aid) (to S. H. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1–7.
B. A. Kaipparettu et al., unpublished results.
- ERα
- estrogen receptor α
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interfering RNA
- NS
- nonspecific
- q-PCR
- quantitative PCR
- E2
- estrogen
- OPG
- osteoprotegerin.
REFERENCES
- 1.Deroo B. J., Korach K. S. (2006) J. Clin. Invest. 116, 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Malley B. W. (2007) Mol. Endocrinol. 21, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 3.Acevedo M. L., Lee K. C., Stender J. D., Katzenellenbogen B. S., Kraus W. L. (2004) Mol. Cell 13, 725–738 [DOI] [PubMed] [Google Scholar]
- 4.Dobrzycka K. M., Townson S. M., Jiang S., Oesterreich S. (2003) Endocr. Relat. Cancer 10, 517–536 [DOI] [PubMed] [Google Scholar]
- 5.Oesterreich S., Zhang Q., Hopp T., Fuqua S. A., Michaelis M., Zhao H. H., Davie J. R., Osborne C. K., Lee A. V. (2000) Mol. Endocrinol. 14, 369–381 [DOI] [PubMed] [Google Scholar]
- 6.Townson S. M., Dobrzycka K. M., Lee A. V., Air M., Deng W., Kang K., Jiang S., Kioka N., Michaelis K., Oesterreich S. (2003) J. Biol. Chem. 278, 20059–20068 [DOI] [PubMed] [Google Scholar]
- 7.Oesterreich S., Lee A. V., Sullivan T. M., Samuel S. K., Davie J. R., Fuqua S. A. (1997) J. Cell Biochem. 67, 275–286 [PubMed] [Google Scholar]
- 8.Townson S. M., Kang K., Lee A. V., Oesterreich S. (2004) J. Biol. Chem. 279, 26074–26081 [DOI] [PubMed] [Google Scholar]
- 9.Debril M. B., Dubuquoy L., Feige J. N., Wahli W., Desvergne B., Auwerx J., Gelman L. (2005) J. Mol. Endocrinol. 35, 503–517 [DOI] [PubMed] [Google Scholar]
- 10.Nayler O., Strätling W., Bourquin J. P., Stagljar I., Lindemann L., Jasper H., Hartmann A. M., Fackelmayer F. O., Ullrich A., Stamm S. (1998) Nucleic Acids Res. 26, 3542–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renz A., Fackelmayer F. O. (1996) Nucleic Acids Res. 24, 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai S., Lee C. C., Kohwi-Shigematsu T. (2006) Nat. Genet. 38, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 13.Horike S., Cai S., Miyano M., Cheng J. F., Kohwi-Shigematsu T. (2005) Nat. Genet. 37, 31–40 [DOI] [PubMed] [Google Scholar]
- 14.Denegri M., Chiodi I., Corioni M., Cobianchi F., Riva S., Biamonti G. (2001) Mol. Biol. Cell 12, 3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traweger A., Fuchs R., Krizbai I. A., Weiger T. M., Bauer H. C., Bauer H. (2003) J. Biol. Chem. 278, 2692–2700 [DOI] [PubMed] [Google Scholar]
- 16.Lee Y. B., Colley S., Norman M., Biamonti G., Uney J. B. (2007) Exp. Cell Res. 313, 3914–3923 [DOI] [PubMed] [Google Scholar]
- 17.Townson S. M., Sullivan T., Zhang Q., Clark G. M., Osborne C. K., Lee A. V., Oesterreich S. (2000) Clin. Cancer Res. 6, 3788–3796 [PubMed] [Google Scholar]
- 18.Oesterreich S., Allredl D. C., Mohsin S. K., Zhang Q., Wong H., Lee A. V., Osborne C. K., O'Connell P. (2001) Br. J. Cancer 84, 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller B. J., Wang D., Krahe R., Wright F. A. (2003) Am. J. Hum. Genet. 73, 748–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergman A., Abel F., Behboudi A., Yhr M., Mattsson J., Svensson J. H., Karlsson P., Nordling M. (2008) BMC Med. Genet. 9, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerich-Hille S., Bardout V. J., Hilsenbeck S. G., Osborne C. K., Oesterreich S. (2009) Breast Cancer Res. Treat., [DOI] [PubMed] [Google Scholar]
- 22.Dobrzycka K. M., Kang K., Jiang S., Meyer R., Rao P. H., Lee A. V., Oesterreich S. (2006) Cancer Res. 66, 7859–7863 [DOI] [PubMed] [Google Scholar]
- 23.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24.Jiang S., Meyer R., Kang K., Osborne C. K., Wong J., Oesterreich S. (2006) Mol. Endocrinol. 20, 311–320 [DOI] [PubMed] [Google Scholar]
- 25.Ivanova M., Dobrzycka K. M., Jiang S., Michaelis K., Meyer R., Kang K., Adkins B., Barski O. A., Zubairy S., Divisova J., Lee A. V., Oesterreich S. (2005) Mol. Cell. Biol. 25, 2995–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo J. M., Juszczynski P., Monti S., Cerchietti L., Ye K., Greally J. M., Shipp M., Melnick A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C., Wong W. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Behav. Brain Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 29.De Preter K., Barriot R., Speleman F., Vandesompele J., Moreau Y. (2008) Nucleic Acids Res. 36, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 31.Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R. (2007) Genes Dev. 21, 2936–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeNardo D. G., Kim H. T., Hilsenbeck S., Cuba V., Tsimelzon A., Brown P. H. (2005) Mol. Endocrinol. 19, 362–378 [DOI] [PubMed] [Google Scholar]
- 33.de Visser K. E., Eichten A., Coussens L. M. (2006) Nat. Rev. Cancer 6, 24–37 [DOI] [PubMed] [Google Scholar]
- 34.Frasor J., Danes J. M., Komm B., Chang K. C., Lyttle C. R., Katzenellenbogen B. S. (2003) Endocrinology 144, 4562–4574 [DOI] [PubMed] [Google Scholar]
- 35.Rae J. M., Johnson M. D., Scheys J. O., Cordero K. E., Larios J. M., Lippman M. E. (2005) Breast Cancer Res. Treat. 92, 141–149 [DOI] [PubMed] [Google Scholar]
- 36.Creighton C. J., Cordero K. E., Larios J. M., Miller R. S., Johnson M. D., Chinnaiyan A. M., Lippman M. E., Rae J. M. (2006) Genome Biol. 7, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., McKinnon P. J., Cleveland J. L., Zambetti G. P. (2003) Cancer Cell 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 38.Sergeant K. A., Bourgeois C. F., Dalgliesh C., Venables J. P., Stevenin J., Elliott D. J. (2007) J. Cell Sci. 120, 309–319 [DOI] [PubMed] [Google Scholar]
- 39.Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T. (1992) Cell 70, 631–645 [DOI] [PubMed] [Google Scholar]
- 40.Dickinson L. A., Dickinson C. D., Kohwi-Shigematsu T. (1997) J. Biol. Chem. 272, 11463–11470 [DOI] [PubMed] [Google Scholar]
- 41.Alvarez J. D., Yasui D. H., Niida H., Joh T., Loh D. Y., Kohwi-Shigematsu T. (2000) Genes Dev. 14, 521–535 [PMC free article] [PubMed] [Google Scholar]
- 42.Han H. J., Russo J., Kohwi Y., Kohwi-Shigematsu T. (2008) Nature 452, 187–193 [DOI] [PubMed] [Google Scholar]
- 43.Szemes M., Gyorgy A., Paweletz C., Dobi A., Agoston D. V. (2006) Neurochem. Res. 31, 237–246 [DOI] [PubMed] [Google Scholar]
- 44.Britanova O., Akopov S., Lukyanov S., Gruss P., Tarabykin V. (2005) Eur. J. Neurosci. 21, 658–668 [DOI] [PubMed] [Google Scholar]
- 45.Dobreva G., Dambacher J., Grosschedl R. (2003) Genes Dev. 17, 3048–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. (2002) Nature 419, 641–645 [DOI] [PubMed] [Google Scholar]
- 47.Galande S., Purbey P. K., Notani D., Kumar P. P. (2007) Curr. Opin. Genet. Dev. 17, 408–414 [DOI] [PubMed] [Google Scholar]
- 48.Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. (2007) Nat. Cell Biol. 9, 45–56 [DOI] [PubMed] [Google Scholar]
- 49.Kininis M., Chen B. S., Diehl A. G., Isaacs G. D., Zhang T., Siepel A. C., Clark A. G., Kraus W. L. (2007) Mol. Cell. Biol. 14, 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hite K. C., Adams V. H., Hansen J. C. (2009) Biochem. Cell Biol. 87, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.