Abstract
Caveolin-1 (Cav-1) is the main structural protein of caveolae and plays an important role in various cellular processes such as vesicular transport, cholesterol homeostasis, and signal transduction pathways. The expression and functional role of Cav-1 have been reported in liver and in hepatocyte cell lines, in human cirrhotic liver, and in hepatocellular carcinomas. Previous studies demonstrated that Cav-1 was dispensable for liver regeneration, because Cav-1−/− animals survived and fully regenerated liver function and size after partial hepatectomy. In this study, we have investigated the mechanisms by which the lack of Cav-1 accelerates liver regeneration after partial hepatectomy. The data show that transforming growth factor β (TGF-β) signaling is impaired in regenerating liver of Cav-1−/− mice and in hepatocytes derived from these animals. TGF-β receptors I and II do not colocalize in the same membrane fraction in the hepatocytes derived from Cav-1−/− mice, as Smad2/3 signaling decreased in the absence of Cav-1 at the time that the transcriptional corepressor SnoN accumulates. Accordingly, the expression of TGF-β target genes, such as plasminogen activator inhibitor-1, is decreased due to the presence of the high levels of SnoN. Moreover, hepatocyte growth factor inhibited TGF-β signaling in the absence of Cav-1 by increasing SnoN expression. Taken together, these data might help to unravel why Cav-1-deficient mice exhibit an accelerated liver regeneration after partial hepatectomy and add new insights on the molecular mechanisms controlling the initial commitment to hepatocyte proliferation.
Keywords: Cancer/Liver, Cytokines/TGFβ, Lipid/Raft, Membrane/Proteins, Signal Transduction, Tissue/Organ Systems/Hepatocyte, Tissue/Organ Systems/Liver
Introduction
Caveolae are non-clathrin, flask-shaped invaginations of the plasma membrane enriched in sphingolipids, cholesterol, and caveolin, a multifunctional scaffolding protein (1). Three Cavs2 have been identified in differentiated cells with specific patterns of distribution. Cav-1 and -2 are found in adipocytes and endothelial cells, whereas Cav-3 is selectively expressed in muscle cells (2). Although caveolae participate in many cellular processes, including vesicular transport, cholesterol homeostasis, regulation of signal transduction, integrin signaling, and cancer-associated cell growth (3–7), it is rather surprising that all Cav-null mice are viable, including the complete caveolae-less mouse. However, the combined loss of Cav-1 and -3 has profound effects on the cardiovascular function (2). Cav-1 is a potent inhibitor of proliferative pathways, such as the Ras-p42/p44 MAPK cascades (8), and Cav-1 expression negatively regulates cell cycle progression via p53/p21-dependent pathway (9).
In liver and in different hepatocyte cell lines, the internalization of specific ligands through caveolae has been reported (10). Indeed, changes in the expression of Cav-1 in human cirrhotic liver, HCC, and in hepatoma cells and in regenerating liver have been described (11–15). Cav-1 increases in liver after PH. However, regarding the function of Cav-1 in liver regeneration, our data indicate that animals lacking Cav-1 restore liver mass to the same extent as the wild-type counterparts but with an accelerated hepatocyte proliferation after PH (16).
Although many cytokines are involved in the onset of liver regeneration, TGF-β participates in growth arrest once the liver has gained its appropriate functional mass (17). Interestingly, TGF-β is up-regulated during the early regenerative response to PH (18); however, during regeneration, hepatocytes become refractory to TGF-β signals, and the liver grows despite its presence (19). TGF-β signaling occurs when TGF-β binds to and assembles type I and type II serine/threonine kinase receptors (TβRs). TβR-II phosphorylates TβR-I, and this phosphorylation is both essential and sufficient for TGF-β signaling. The activated TβR-I then phosphorylates and activates Smad2/3, which form oligomers with Smad4 that translocate to the nucleus to regulate the expression of target genes (20). The ski/snon family of proto-oncogenes has been shown to inhibit TGF-β/Smad signaling by disrupting heteromeric Smad complexes and blocking recruitment of coactivators. Both Ski and SnoN are highly expressed in cancer cell lines and repress the growth inhibitory function of the Smads (21). In regenerating liver, the induction of SnoN/Ski might explain the hepatocyte resistance to TGF-β signaling during the proliferative phase (22).
It is known that TβRs localize to caveolae; however, the functional consequences differ depending on the receptor and the cell type (23–25). Indeed, it has been recently proposed that TβR endocytosis via clathrin-coated pit-dependent internalization promotes TGF-β signaling; in contrast, the lipid raft-caveolar internalization pathway facilitates the degradation of TGF-β receptors (26). In view of these data, we have investigated the mechanisms by which the lack of Cav-1 accelerated liver regeneration after PH. Our results show that the TGF-β pathway is attenuated in hepatocytes derived from Cav-1−/− mice. We also demonstrated that TβR-I and -II colocalize with Cav-1 in the caveolar fraction, that Smad2/3 signaling decreases in the absence of Cav-1, and that significant up-regulation of the Smad transcriptional corepressor SnoN occurs. Moreover, HGF inhibits TGF-β signaling in the absence of Cav-1 by increasing SnoN expression. These mechanisms might explain why Cav-1−/− mice exhibit an accelerated liver regeneration after PH and provide a model of pathophysiological alteration of the hepatocyte response to TGF-β.
EXPERIMENTAL PROCEDURES
Chemicals
Antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), BD Transduction Laboratories, Cell Signaling (Boston, MA), Upstate (Upstate Biotechnology, Inc., Lake Placid, NY), Abcam (Cambridge, UK), and Invitrogen. TGF-β1 and other reagents were from Roche Applied Science or Sigma. siRNA-SnoN and scrambled sequences were from Santa Cruz Biotechnology. Tissue culture dishes were from Falcon (Lincoln Park, NJ). Tissue culture media were from BioWhittaker (Walkersville, MD).
Animals
C57BL/6J Cav-1+/+ (The Jackson Laboratory, Bar Harbor, ME) and Cav-1 knock-out mice (cav-1−/−, strain Cav-1tm1Mls/J, genetic background 129/Sv, C57BL/6J, and SJL) aged 2 months (20–25 g) were cared for by following Institutional Care Instructions (Bioethical Commission from Consejo Superior de Investigaciones Científicas) and underwent 70% PH as described (16).
Isolation and Immortalization of Neonatal Hepatocytes
Pools of 4–6 livers from Cav-1+/+ and Cav-1−/− neonates (3–5 days old) were submitted to collagenase dispersion for the isolation of hepatocytes and preparation of primary cultures as described previously (27). To obtain immortalized neonatal cell lines (NCLs), viral BOSC 23 packaging cells were transfected at 70% confluence with 3 μg/6-cm dish of the puromycin-resistance retroviral vector pBabe encoding SV40 large T antigen as described previously (28). Primary neonatal hepatocytes were infected at 60% confluence with Polybrene-supplemented virus (4 μg/ml) for 48 h and maintained in culture medium for 72 h before selection with puromycin (0.5–1 μg/ml) for 1 week. Pools of infected cells, rather than individual clones, were selected to avoid potential clone-to-clone variations. Immortalized cell lines were further cultured for at least 2 weeks with arginine-free medium supplemented with 10% FBS and ornithine to avoid growth of nonparenchymal cells.
Cell Culture, Transient and Stable Cell Transfections, and Treatments
Cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FBS and antibiotics (50 μg/ml each of penicillin, streptomycin, and gentamicin) at 37 °C in a humidified 5% CO2 atmosphere. THLE-2 (derived from normal liver), CCL-13 (Chang liver, CHL), and HepG2 cells were purchased from the ATCC, and the hepatoma cell line HuH-7 (JCRB0403) was purchased from Health Science Research Resources Bank (Osaka, Japan). For transient transfections assays, HuH-7 or NCL cells at 60% of confluence were exposed for 12 h to FuGENE-6 (Roche Applied Science) containing pEGF-N1 (encoding Cav-1-WT-GFP or GFP) (16) and 3TP-lux (luciferase reporter gene that contains the activin/TGFβ-responsive promoter element) (29) vectors following the manufacturer's instructions. For stable transfections, HuH-7 cells at 50% of confluence were exposed for 24 h to FuGENE-6 containing Cav-1-GFP expression vector or GFP control vector. The cells that express Cav-1-GFP or GFP were fluorescence-activated cell sorter-subcloned, and subsequent cultures of selected cells were grown in the presence of G418. At 18 h before experiments, the culture medium was replaced with fresh medium containing 1% (v/v) FBS. Luciferase activity was measured using the luciferase assay system (Promega, Madison, WI).
Isolation of Low Density, Triton-insoluble Caveolae-enriched Fractions
Isolation of caveolin-enriched membrane fractions (detergent-resistant membrane, DRMs) was done using a Triton X-100-based method (16). The samples were ultracentrifuged at 200,000 × g for 18–20 h at 4 °C in an SW60Ti rotor. Twelve fractions of 0.375 ml were collected from the top of the gradient after the centrifugation and mixed 1:1 with cold acetone for 4 h at −20 °C. The precipitated proteins were centrifuged at 16,000 × g for 10 min and dried. The resulting pellets were resuspended in 25 mm MES buffer, pH 6.5, and stored at −20 °C.
Protein Analysis
Total extracts were prepared from Cav-1+/+ and Cav-1−/− remnant liver (50 mg) after PH or cells (2–3 × 106) at different times. Samples were homogenized in a medium containing 10 mm Tris/HCl, pH 7.5, 1 mm MgCl2, 1 mm EGTA, 10% (v/v) glycerol, 0.5% (w/v) CHAPS, 1 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, Protease Inhibitor Mixture (Sigma) and Phosphatase Inhibitor Mixture 1 and 2 (Sigma). The extracts were vortexed for 30 min at 4 °C, and after centrifuging for 20 min at 13,000 × g, the supernatants were stored at −20 °C. For cytosolic extracts, NCL Cav-1+/+ and NCL Cav-1−/− cells (2–3 × 106) were harvested into ice-cold phosphate-buffered saline, resuspended in ice-cold buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol), and left to swell on ice for 15 min. Nonidet P-40 was added to 0.5%, and the cells were vortexed and centrifuged at 12,000 × g for 30 s. The supernatants (cytosolic fraction) were collected and maintained at −20 °C. The pellet (nuclear fraction) was resuspended in ice-cold buffer B (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA) and left on ice for 15 min with intermittent mixing, followed by centrifugation at 12,000 × g for 15 min at 4 °C. Equal amounts of protein (10–50 μg) from each extract or 50 μl of the fractions obtained from the gradient were size-fractionated in 8–12% SDS-PAGE, transferred to a HybondP membrane (GE Healthcare), and after blocking with 5% nonfat dry milk were incubated with the corresponding Abs. The amounts of Cav-1, GFP, pSmad2/3, Smad2/3, TβR-I, TβR-II, SnoN (sc-9141), PAI-1, c-Met, GM130, calregulin, annexin VI, 5′-nucleotidase, and p85 were determined using commercial Abs. After incubation with the corresponding anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary Ab, blots were developed by the ECL protocol (GE Healthcare). The blots were revealed, and different exposure times were used for each blot with a CCD camera in a luminescent image analyzer (Gel-Doc, Bio-Rad) to ensure the linearity of the band intensities.
RNA Isolation and RT-PCR Analysis
1 μg of total RNA, extracted with TRIzol reagent (Invitrogen), was reverse-transcribed using SuperScriptTM III first-strand synthesis system following the indications of the manufacturer (Invitrogen). Real time PCR was conducted with SYBR Green (Applied Biosystems) on a MyiQ real time PCR system (Bio-Rad). The primers used were as follows: PAI-1, FP, 5′-CGGCAGATCCAAGATGCTATG-3′, and RP, 5′-GACCAGCTCTAGGTCCCGCT-3′; Cav-1, FP, 5′-AGCCCAACAACAAGGCCAT-3′, and RP, 5′-GCAATCACATCTTCAAAGTCAATCTT-3′; 36B4, FP, 5′-AGATGCAGCAGATCCGCAT-3′, and RP, 5′-GTTCTTGCCCATCAGCACC-3′; and for 18 S rRNA, FP 5′-GCAATTATTCCCCATGAACGA-3′, and RP, 5′-CAAAGGGCAGGGACTTAATCAA-3′. PCR thermocycling parameters were 94 °C for 3 min, 40 cycles of 94 °C for 20 s, 60 °C for 20 s, 72 °C 20 s, and a final cycle of 72 °C 5 min for qPCR and 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min for semiquantitative RT-PCR. Each sample was run in duplicate and was normalized with the expression of 36B4 for qPCR or 18 S for semiquantitative RT-PCR. The fold induction was determined in a ΔΔCt-based fold change calculations (Relative Quantity (RQ) is 2⋀−ΔΔCt).
Data Analysis
Data are expressed as mean ± S.D. Statistical significance was estimated with Student's t test for unpaired observation. The results were considered significant at p < 0.05. Data were analyzed by SPSS for Windows statistical package version 9.0.1.
RESULTS
TGF-β/Smad Signaling Pathway Is Impaired in Cav-1-deficient Mice after PH
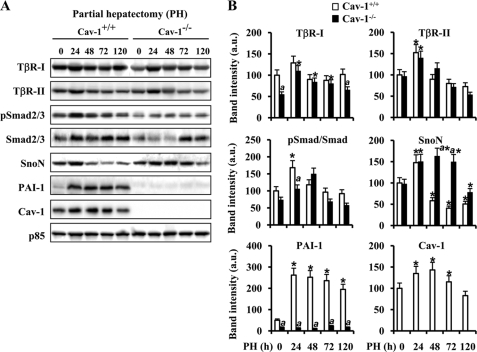
Despite the controversy on the requirement of Cav-1 for liver regeneration after PH, accelerated hepatocyte proliferation was observed in Cav-1-deficient mice, exhibiting a higher liver mass recovery at 80 h after PH (15, 16). To gain insight on this process, we focused on the early TGF-β signaling in regenerating liver. When the protein levels of the TGF-β signalosome, TβR-I, TβR-II, Smad2/3, SnoN, PAI-1, and Cav-1, were analyzed in liver samples from Cav-1+/+ and Cav-1−/− mice after PH, TβR-I and TβR-II exhibited equivalent profiles of changes, except for the consistent observation of a 46% reduction in TβR-I in Cav-1−/− mice before PH (Fig. 1, A and B). However, the phosphorylation of Smad2/3 following TβR-I and TβR-II activation (20), which exhibits a significant increase at 24 h after PH in Cav-1+/+ mice, showed a lesser modification at this time in Cav-1−/− mice. Moreover, the up-regulation of Smad2/3 was delayed up to 72 h after PH in Cav-1−/− animals. Regarding the levels of SnoN and PAI-1, two genes induced by TGF-β (30), SnoN levels increased transiently at 24 h after PH, returning to basal levels at 48 h. However, they remained overexpressed in Cav-1−/− mice livers up to 72 h. The levels of PAI-1, which inhibits the urokinase-type plasminogen activator/plasminogen and blocks the processing of pro-HGF into active HGF, increased significantly at 24 h after PH in Cav-1+/+ mice, but the levels were almost undetectable up to 120 h after PH in Cav-1−/− mice. The changes in Cav-1, with an increase at 24–72 h, after PH were similar to those reported previously (16).
FIGURE 1.
Cav-1 and TGF-β-signaling molecule expression in regenerating liver after PH. Total tissue extracts were prepared from Cav-1+/+ and Cav-1−/− remnant liver after PH at different times. A, protein levels of TβR-I and -II, total and pSmad2/3, Cav-1, SnoN, and PAI-1 were determined by Western blot. Representative blots are shown. B, densitometric analysis of the immunoblots. Data are presented as the means ± S.D. of five independent experiments (animals). *, p < 0.01 versus the corresponding value at 0 time. a, p < 0.01 versus the corresponding value at the indicated time in Cav-1+/+ mice. Blots were normalized with p85 for total extracts.
TβR-I/TβR-II Interaction Is Reduced in the Absence of Caveolae
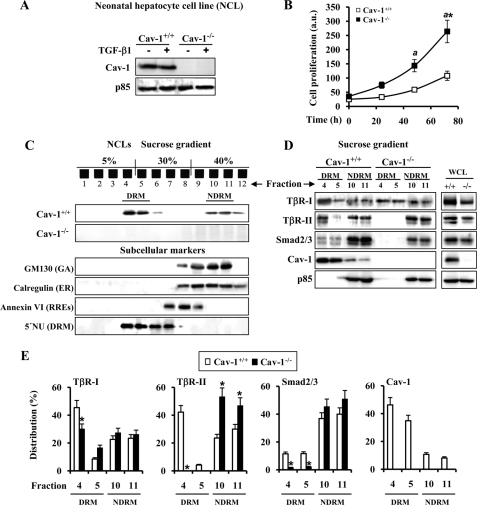
Previous reports described that TβR-I, TβR-II, and Smad-2 cofractionate with Cav-1 in caveolae-enriched microdomains and that Cav-1 directly interacts with TβR-I (23). To better study TβRs-Cav-1 cross-signaling in liver, we generated hepatocyte cell lines (NCL) from neonatal liver of Cav-1+/+ and Cav-1−/− mice. Cav-1 was found only in NCLs from Cav-1+/+ mice (Fig. 2A), and interestingly, cell proliferation of NCL Cav-1−/− clones was always higher (2.5-fold) than the Cav-1+/+ counterparts (Fig. 2B), in agreement with previous work using Cav-1−/− mouse embryo fibroblasts (9). As expected, Cav-1 was present mainly in DRMs and slightly in NDRM fractions of the discontinuous sucrose gradient (Fig. 2C). To identify the subcellular localization in which Cav-1 accumulates, we used different markers as follows: GM130 and calregulin for Golgi and endoplasmic reticulum, respectively, and annexin VI for recycling receptor endosomes and 5′-nucleotidase for the DRM fraction. As shown in Fig. 2C, fractions 4–6, in which Cav-1 is located, correspond mainly to the DRM fraction. Fig. 2, D and E, shows the protein levels and distribution of the TβRs in DRM and NDRM fractions of Cav-1+/+ and Cav-1−/− NCL cells. Whereas the main accumulation of TβR-I in NCL Cav-1+/+ cells was detected in the DRM fraction, this receptor was equally distributed between the DRM and NDRM fractions in NCL Cav-1−/−. Moreover, TβR-II was undetectable in the DRM fraction of NCL Cav-1−/− cells. Smad2/3 were detected in the DRM fraction of NCL Cav-1+/+, accumulating in both cell types in the NDRM fraction.
FIGURE 2.
TβR-I/TβR-II interaction is decreased in the absence of Cav-1. A, protein levels of Cav-1 in NCL lines after treatment with 5 ng/ml TGF-β1 for 30 min. B, time-dependent proliferation of NCL Cav-1+/+ and NCL Cav-1−/− cells. Cells were seeded in medium containing 10% (v/v) FBS and, after the indicated times in culture, were trypsinized, stained with trypan blue, and counted. a.u., arbitrary units. C, sucrose discontinuous (5–30-40%) gradients were performed using a Triton X-100-based method to isolate DRM or NDRM fractions from the NCL cells as described under “Experimental Procedures.” Cav-1 is mainly localized in the fractions 4–5 in NCL Cav-1+/+ cells. GM130 (GA), calregulin (ER), annexin VI (RREs), and 5′-nucleotides (5′NU) (DRM) were used as subcellular markers. D, Western blot detection of TβR-I and II, Smad2/3, and Cav-1 in 50 μl from fractions 4 to 5 for DRM and fractions 10 to 11 for NDRM from NCL cells. Whole cell lysates (WCL) (50 μg) were used as reference for total content and blotted with the same Abs. E, quantification (%) of the distribution of the corresponding protein analyzed in D. The results are expressed as the band intensity of the protein in DRM (fractions 4 to 5) and NDRM (fractions 10 to 11) fractions with respect to the total protein levels. Representative blots are shown. Results are the means ± S.D. of four independent experiments. *, p < 0.01 versus the corresponding value at 0 time (B) or the corresponding value in NCL Cav-1+/+ (E). a, p < 0.01 versus the corresponding value at the indicated time in NCL Cav-1+/+. Blots were normalized with p85 for total extracts.
SnoN Accumulates in NCL Cav-1−/− Cells
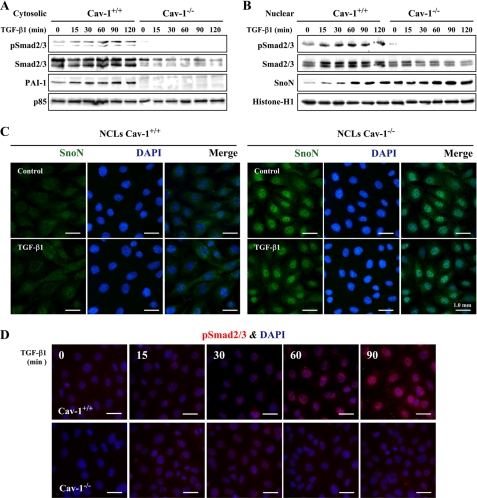
To evaluate the contribution of Cav-1 to TβR signaling, NCL Cav-1+/+ and Cav-1−/− cells were stimulated with TGF-β1, and the activation of Smad2/3 and the expression levels of PAI-1 and SnoN were determined in cytosolic and nuclear extracts. As Fig. 3, A and B, shows, Smad2/3 proteins were detected in both compartments, although the levels were higher in NCL Cav-1+/+ cells. TGF-β1 treatment increased pSmad2/3 in the nucleus, and the phosphorylation was higher in NCL Cav-1+/+ cells (Fig. 3, B and D). The cytosolic levels of PAI-1 increased significantly after 2 h of TGF-β1 treatment in NCL Cav-1+/+ but were lower and TGF-β1-insensitive in Cav-1−/− cells. However, SnoN basal levels were significantly higher in NCL Cav-1−/− and were modulated by TGF-β1 in Cav-1+/+ cells (Fig. 3B). Immunofluorescence analysis of SnoN showed a clear nuclear staining in NCL Cav-1−/−, whereas in NCL Cav-1+/+ the localization of SnoN was mainly cytoplasmic and lesser expressed (Fig. 3C).
FIGURE 3.
SnoN accumulates in NCL Cav-1−/− cells. NCL Cav-1+/+ and NCL Cav-1−/− cells (2–3 × 106) were treated with 5 ng/ml TGF-β1 at the indicated times. Cytosolic (50 μg) (A) and nuclear extracts (10 μg) (B) were obtained, and protein levels of total and pSmad2/3, PAI-1, and SnoN were analyzed. A representative blot is shown. Blots were normalized with p85 for cytosolic extracts and with histone-H1 for nuclear extracts. C, immunofluorescence analysis of SnoN in NCL cells after 1 h of TGF-β1 stimulation. DAPI, 4′,6-diamidino-2-phenylindole. D, immunofluorescence analysis of pSmad2/3 in NCL cells after TGF-β1 stimulation at the indicated times. Neonatal Cav-1+/+ or Cav-1−/− cells (5 × 104) were cultured in 24-multiwell plates on glass coverslips and maintained overnight with 1% FBS. After that period, the cells were fixed for 15 min with 4% paraformaldehyde, pH 7.2, washed with phosphate-buffered saline, and permeabilized with 1% Tween 20 in phosphate-buffered saline for 15 min at room temperature. After blocking with 3% bovine serum albumin for 1 h at room temperature, the cells were incubated 2 h with the corresponding Abs diluted 1:150 in 1% bovine serum albumin, washed several times, and incubated for 2 h with fluorochrome-conjugated Abs (Invitrogen) raised against Fc of primary Abs and treated with 4′,6-diamidino-2-phenylindole for 30 min at room temperature. The glass coverslips were mounting with Vectashield (Vector Laboratories, Burlingame, CA) on microscope slides. The images were acquired in a fluorescence Eclipse E400 microscope (Nikon).
Induction of PAI-1 by TGF-β1 Is Impaired in the Absence of Cav-1
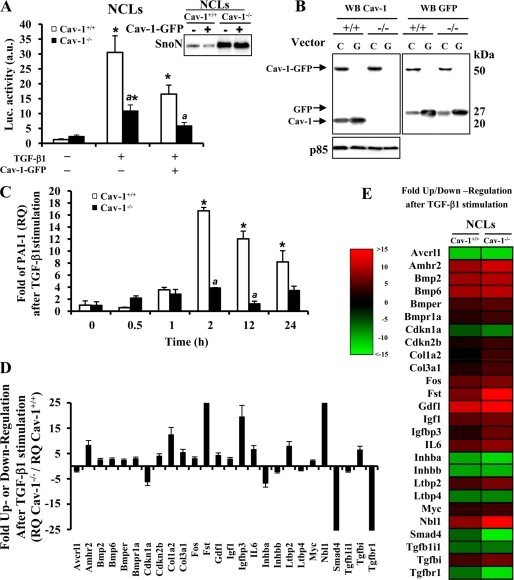
To evaluate further the cross-regulation between Cav-1 and TGF-β signaling, NCL cells were transfected with a TGF-β-responsive promoter (3TP-lux) and with a Cav-1-GFP expression vector (pEGFPN1). Upon TGF-β1 treatment, the activation of the 3TP-lux reporter was 3-fold lower in NCL Cav-1−/− compared with NCL Cav-1+/+. When these cells were cotransfected with 3TP-lux and pEGFPN1-Cav-1 and stimulated with TGF-β1, the activation of 3TP-lux was reduced 50% in both cell lines, suggesting that the transient expression of Cav-1 was unable to restore TGF-β1 responsiveness probably due to the elevated levels of SnoN (Fig. 4A). Fig. 4B shows the expression of Cav-1 (20 kDa), Cav-1-GFP (50 kDa), and GFP (27 kDa) in NCL Cav-1+/+ and Cav-1−/− as a control of the transfection extent. Fig. 4C shows that the induction of PAI-1 by TGF-β1 analyzed by qPCR was significantly lower in NCL Cav-1−/−. From these results, we can suggest that Cav-1 is necessary for the correct signaling of TGF-β; however, and in agreement with previous data reporting Cav-1 as an inhibitor of TGF-β signaling in NIH-3T3 cells (23), the elevated transient expression of Cav-1 down-regulated TGF-β signaling.
FIGURE 4.
TGF-β-responsive promoter and PAI-1 expression are impaired in the absence of Cav-1. A, NCL Cav-1+/+ and NCL Cav-1−/− cells were transfected with pEGFN1 (encoding wild-type Cav-1-GFP or GFP) and with the 3TP-Lux vectors with FuGENE-6 (Roche Applied Science) for 12 h. After 24 h of transfection, cells were treated with TGF-β1 for 12 h, and the luciferase (Luc.) activity was measured by using the luciferase assay system. Inset, SnoN expression in NCL cells after transfection with Cav-1-GFP vector. a.u., arbitrary units. B, Western blot (WB) analysis of the protein, Cav-1-GFP, GFP, or Cav-1, expressed after transfection with the pEGFPN1 vector as a control of the process (C, Cav-1; G, GFP). C, 1 μg of total RNA, extracted with TRIzol reagent (Invitrogen), was reverse-transcribed using SuperScriptTM III first-strand synthesis system for RT-PCR. Real time PCR of PAI-1 mRNA was conducted with SYBR Green on a MyiQ real time PCR system, after treatment of the cells with 5 ng/ml TGF-β1 for the indicated times. Results were normalized with the 36B4 expression and were expressed as relative quantity (RQ) (2⋀−ΔΔCt). D, after the stimulation of the NCL cells with TGF-β1 (5 ng/ml) for 90 min, RNA (2 μg) was used for cDNA synthesis with RT2 first standard kit (SuperArray Bioscience, Frederick, MD). The mouse TGF-β/BMP signaling pathway PCR array was performed according to the manufacturer's protocol, using the Profiler PCR array system and the SYBR Green/fluorescein qPCR master mix (SuperArray Bioscience) on a MyiQ real time PCR system (Bio-Rad). Gene expression was compared with the web-based software package for the PCR array system; this software automatically performs all ΔΔCt-based fold-change calculations from the specific uploaded raw threshold cycle data. E, colorimetric diagram with a selection of the 84 TGF-β signaling related genes analyzed. Genes whose transcription is up/down (red/green) 2-fold in gene expression threshold in NCL Cav-1+/+ and NCL Cav-1−/− cells are shown. All results presented are the means ± S.D. of four independent experiments. *, p < 0.01 versus the corresponding unstimulated condition. a, p < 0.01 versus the corresponding value at the same condition in NCL Cav-1+/+.
To gain further insight into this differential response of the neonatal cells to TGF-β, we used a specific mouse TGF-β-signaling microarray to compare the expression profile in NCL cells after TGF-β1 treatment. Fig. 4, D and E, summarizes the fold up- or down-regulation of the 84 TGF-β-regulated genes analyzed. We sorted the candidate genes whose expression threshold after TGF-β1 treatment was altered more than 2-fold. The down-regulated genes in NCL Cav-1−/− are those implicated in TGF-β signaling (i.e. acvrl1, inhba, inhbb, smad4, tgfbl1l, and tgfbr1) or cell cycle (cdkn1a), and the majority of the up-regulated genes are implicated in proliferation (amhr2, fst, igfbp3, il6, ltbp2, Myc, and nbl1).
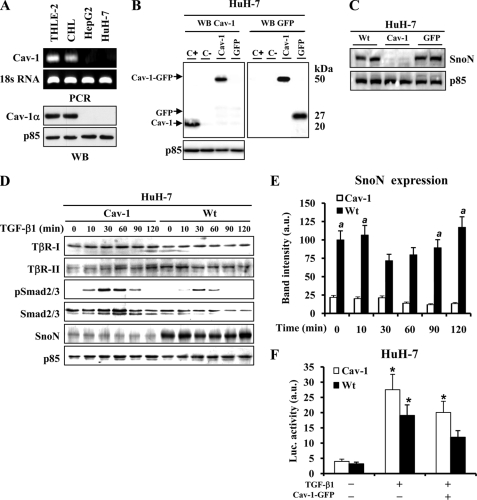
SnoN Protein Levels Are Increased in HCC Human Cells That Lack Cav-1 Expression
The absence of Cav-1 in well differentiated human HCC has been recently reported to be a specific signature of these cells (31). Accordingly, Cav-1 levels were analyzed in various hepatic cell lines, and although the hepatocyte-like THLE-2 or CHL cells showed a significant expression of Cav-1 mRNA and protein, the HCC lines, such as HepG2 or HuH-7, failed to express this gene (Fig. 5A). Moreover, in HuH-7 cells stably transfected with the pEGFPN1-Cav-1 expression vector, the high SnoN levels observed in the parental cells or after transfection with the GFP vector decreased in the presence of Cav-1 (Fig. 5, B and C). Analysis of TβR-I, TβR-II, pSmad2/3, Smad2/3, and SnoN in wild type or Cav-1-expressing HuH-7 cells stimulated with TGF-β1 showed similar levels of TβRs, although the phosphorylation of Smad2/3 was higher in HuH-7-Cav-1-expressing cells compared with HuH-7-WT cells (Fig. 5, D and E). When HuH-7-WT cells were transiently cotransfected with 3TP-lux and pEGFPN1-Cav-1 vectors and stimulated with TGF-β1, an inhibition of the activation of 3TP-lux was observed. Moreover, and in agreement with the data obtained with NCLs in Fig. 4A, HuH-7-Cav-1 cells showed a tendency to up-regulate luciferase activity versus HuH-7 parental cells (Fig. 5F).
FIGURE 5.
SnoN expression is increased in HCC cells that lack Cav-1. A, mRNA and protein levels of Cav-1 were determined by semiquantitative RT-PCR and Western blot (WB) in THLE-2, CHL, HepG2, and HuH-7 cells. B, HuH-7 cells were stably transfected with Cav-1-GFP expression vector or GFP control vector. The cells that express Cav-1-GFP or GFP were fluorescence-activated cell sorter-subcloned and selected in the presence of G418. Western blot analysis of the protein, Cav-1-GFP, GFP, or Cav-1, expressed after transfection with the pEGFPN1 vector as a control of the process (C+, CHL total extract; C−, HuH-7-WT total extract). C, SnoN expression in two different clones from HuH-7-WT, HuH-7-Cav-1, and HuH7-GFP cells. D, TGF-β-signaling molecules expression analyzed by Western blot in HuH-7-WT and HuH-7-Cav-1 cells after stimulation with TGF-β1 at the indicated times. E, densitometric analysis of SnoN protein levels from D. a.u., arbitrary units. F, HuH-7-WT and HuH-7-Cav-1 cells were transfected with pEGF-N1 and 3TP-lux vectors as described in the legend from Fig. 4A. Results are the means ± S.D. of four independent experiments. *, p < 0.01 versus the corresponding unstimulated. a, p < 0.01 versus the corresponding value at the same condition in HuH-7-Cav-1.
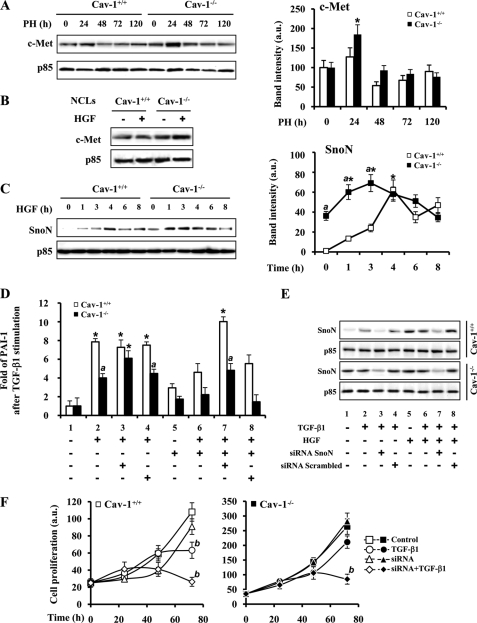
TGF-β and HGF Signaling Cooperate through SnoN Expression to Increase the Proliferation of Cav-1−/− Liver Cells after PH
HGF, a potent inducer of DNA synthesis and cell scatter in cultured hepatocytes, promotes the expression of SnoN (32). In addition to this, the interaction of Cav-1 with c-Met (HGF receptor) has been described (33). After PH, a 75% increase of c-Met at 24 h in Cav-1−/− mice was measured without noticeable changes in Cav-1+/+ mice (Fig. 6A). This situation was partially reproduced in NCLs after HGF challenge (Fig. 6B). Moreover, when the levels of SnoN were determined in NCLs stimulated with HGF, a rise from almost negligible levels to a peak value at 4 h was observed in NCL Cav-1+/+; however, NCL Cav-1−/− cells exhibited significant basal levels, reaching the same peak value at 1 h after HGF administration (Fig. 6C). The preceding data suggest that SnoN overexpression inhibits TGF-β signaling in the absence of Cav-1. As Fig. 6, D and E, shows, pretreatment of cells with a SnoN-specific siRNA increased PAI-1 gene expression in NCL Cav-1−/− cells. When the cells were treated with HGF, the expression of PAI-1 induced by TGF-β was partially abolished, and this was also counteracted after treatment of the cells with SnoN-siRNA. To establish a link between SnoN expression and cell proliferation, a cell counting experiment was performed after transfection of the cells with SnoN-siRNA and stimulation with TGF-β1. As Fig. 6F shows, TGF-β treatment inhibits cell proliferation by 40 and 20% after 72 h in NCL Cav-1+/+ and NCL Cav-1−/− cells, respectively. In the presence of SnoN-siRNA, the inhibition of cell growth promoted by TGF-β1 in NCL Cav-1−/− cells increased up to 70% supporting a role for SnoN in NCL cell proliferation.
FIGURE 6.
TGF-β and HGF signaling cooperate through SnoN expression to increase the proliferation of Cav-1−/− liver cells after PH. A, representative Western blot and the quantification of c-Met protein levels in total liver extracts from Cav-1+/+ and Cav-1−/− mice after PH. B, c-Met protein levels in NCL Cav-1+/+ and NCL Cav-1−/− cells after treatment with 25 ng/ml HGF for 2 h. C, induction of SnoN protein expression by treatment of the NCL cells with HGF for the indicated times. A representative Western blot and a densitometric analysis of the results are shown. D, mRNA PAI-1 levels were measured by qPCR after inhibition of SnoN expression with a specific siRNA or treatment with a scrambled control siRNA. NCL Cav-1+/+ and NCL Cav-1−/− cells were transfected during 6 h with 20 pmol of SnoN siRNA by using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions before TGF-β1 (5 ng/ml) and HGF (25 ng/ml) stimulation for 90 min. E, after transfection of the cells with siRNA and extraction of the mRNA with TRIzol used in D, SnoN protein levels were obtained from organic phase of the TRIzol extraction and analyzed by Western blot. F, 48 h after siRNA transfection, 10 × 103 cells were seeded in a 24-multiwell plate and treated with TGF-β1 (5 ng/ml). The cells were trypsinized, stained with trypan blue, and counted at the indicated times. Results are the means ± S.D. of four independent experiments. *, p < 0.01 versus the corresponding value at 0 time (C) or the corresponding unstimulated data (D). a, p < 0.01 versus the corresponding value at the indicated time (C) or stimulated data in NCL Cav-1+/+(D). b, p < 0.01 versus the corresponding untreated data at 72 h (F). Blots were normalized with p85 for total extracts. a.u., arbitrary units.
DISCUSSION
In this work, we have investigated the contribution of Cav-1 to early TGF-β signaling in regenerating liver after PH, aimed by the observation of an accelerated early commitment to hepatocyte growth in Cav-1−/− mice (16). The role of Cav-1 in the regulation of liver function and regeneration is not clear, and discrepancies exist regarding the requirement of Cav-1 for liver regeneration after PH (15, 16). Our results with hepatoma cell lines show that non-tumor cells and primary cultures of hepatocytes express significant amounts of Cav-1, whereas it was undetectable in the HepG2 and HuH-7 hepatoma cell lines. Besides regeneration and hepatocellular carcinogenesis, recent work proposed a new role for Cav-1 in liver function; in mouse hepatoma cells, Cav-1 expression positively correlated with cell transformation and invasiveness via the activation of the Survivin-mediated pathway (34), and in HepG2 cells Cav-1 negatively regulated the apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand through the internalization of its receptor DR4, which is localized in caveolae (13).
Although many cytokines and growth factors are involved in the onset of liver regeneration, TGF-β has been proposed as an important regulatory factor controlling cell proliferation and death through the transcriptional regulation of cell cycle inhibitors and activators (17); however, sometimes cells become refractory to TGF-β signaling and can proliferate even in the presence of this cytokine. Several groups and our data show an up-regulation of the TβRs during liver regeneration between 24 and 48 h after PH (18). TβRs localize to caveolae although the functional consequences of their activation differ depending on the receptor and the cell type considered, even suppressing TGF-β-mediated signaling in some cases. For instance, Cav-1 interacts with TβR-I and suppresses TGF-β-mediated phosphorylation of Smad2 and subsequent downstream events in NIH-3T3 and in other fibroblast types (23, 35); however, in human endothelial cells, Cav-1 cooperates with ALK1 as a key mediator of the TGF-β pathway in angiogenesis (25). Our data show no differences in TβR-II expression in Cav-1+/+ and Cav-1−/− mice after PH; however, TβR-I levels, although up-regulated after PH, were always reduced in the absence of Cav-1. Moreover, TβR-I and -II do not colocalize in the same membrane fraction in the hepatocytes derived from Cav-1−/− mice. A common model for the TβRs/caveolae interaction proposes that caveolae are involved in receptor degradation, whereas Smad signaling occurs through clathrin-dependent processes (36, 37).
We have used the following three different approaches to analyze the cross-regulation between Cav-1 and the TGF-β signaling pathway: liver regeneration after PH in Cav-1-deficient mice; neonatal hepatocyte cell lines derived from these animals (NCLs); and human hepatoma cell lines lacking Cav-1. In a coherent way, the data support the conclusion that the TGF-β signaling pathway is impaired in the absence of Cav-1 implicating different levels as follows: (a) TβR-I is less abundant in liver from Cav-1−/− mice; (b) as expected, both TβR-I and TβR-II do not colocalize in the DRM fraction in the Cav-1−/− NCL; (c) total and phosphorylated Smad2/3 proteins are decreased in Cav-1−/− liver, Cav-1−/− NCL, and HuH-7 cells; (d) the activity of a Smad2/3-dependent promoter is decreased in Cav-1−/− NCL and in the HuH7 hepatoma cells compared with Cav-1+/+ NCL and the HuH7 cells stably transfected with an expression vector for Cav-1; (e) the expression of TGF-β target genes, such as PAI-1, is decreased both at the mRNA and protein levels, in the absence of Cav-1; (f) good correlation exists between the absence of Cav-1, the decrease in the Smad 2/3 signaling, and the increase of the corepressor SnoN levels in liver from Cav-1−/− mice after PH, in Cav-1−/− NCL cells, and HuH-7 cells. To gain further insight into this differential response of the neonatal cells to TGF-β, we used a specific mouse TGF-β-signaling microarray to compare the expression profile in NCLs cells after TGF-β1 treatment. The down-regulated genes in NCL Cav-1−/− are those implicated in TGF-β signaling or cell cycle.
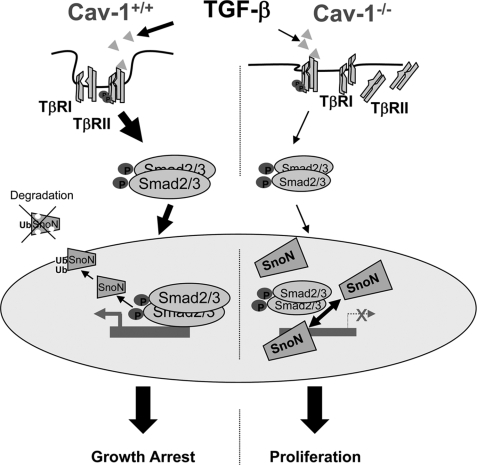
The induction of TGF-β pathway inhibitors, for example the induction of Smad6 and -7, is one of the mechanisms used by tumor cells to overcome the TGF-β restriction (38). In addition to this, other inhibitors, such as the transcriptional repressors TGIF, Ski, and SnoN, allow cell proliferation by blocking Smad activity (39). Ski and SnoN are members of the Ski proto-oncogene family (40). Elevated levels of Ski or SnoN have been detected in many human tumor cell lines, and SnoN has been proposed as a prognostic marker in breast carcinoma (41). In addition to the up-regulation of Ski or SnoN expression, mislocalization may also contribute to malignant progression. SnoN is localized exclusively in the nucleus in cancer tissues, whereas in normal tissues and nontumorigenic cells, SnoN is predominantly cytoplasmic (42). The mechanism of transformation by Ski and SnoN was not defined until diverse studies converged on the conclusion that Ski/SnoN bind directly to the Smad3/4 complex and negatively regulate TGF-β signaling (39). Longerich et al. (43) did not detect mutations or significant pro-tumorigenic expression changes of the snon and ski genes in human hepatocellular carcinomas; however, it was found that both were induced during liver regeneration/proliferation and recruited activated Smad proteins. It is known that SnoN protein levels were increased in the early stages of liver regeneration, whereas Ski was induced at later time points (22). In the absence of Cav-1 in the three models used, Cav-1−/− mice, NCLs, and the HuH7 cells, SnoN is increased, and surprisingly, when Cav-1 is overexpressed SnoN levels did not change, although the TGF-β-dependent luciferase reporter activity is reduced. It has been reported that overexpression of Cav-1 is sufficient to disrupt Smad3 phosphorylation and translocation to the nucleus and consequently TGF-β signaling (23, 35). In this unphysiological situation, SnoN levels are irrelevant because the pathway is impaired at a very upstream level. In addition to this, the subcellular distribution of SnoN between the nucleus and the cytoplasm might be altered and post-translational modifications of SnoN, yet uncovered, cannot be disregarded upon Cav-1 ectopic expression. Our data show that the absence of Cav-1 is related with an increase in SnoN expression and with cell proliferation both in the in vivo model of liver regeneration after PH and in liver cells. According to these data, a schematic representation of TGF-β signaling in Cav-1-deficient hepatocytes is proposed in Fig. 7. In the absence of Cav-1, TβRs, mainly TβRII, delocalize between the membrane and the cytoplasm, leading to a TGF-β-deficient signaling. Lesser activated Smad2/3 translocates to the nucleus, and the higher levels of SnoN impair the transcription of growth-arrest genes and enhance hepatocyte proliferation in the process of liver regeneration. In the presence of Cav-1, TGF-β signaling is sufficient to maintain the balance between cell proliferation and cell growth arrest characteristic of regenerating liver.
FIGURE 7.
TGF-β signaling in Cav-1-deficient hepatocytes. In the absence of Cav-1, TβRs delocalize between the membrane and the cytoplasm, leading to a TGF-β-deficient signaling. Lesser activated Smad2/3 translocates to the nucleus, and the higher levels of SnoN impair the transcription of growth-arrest genes and enhance hepatocyte proliferation in the process of liver regeneration. In the presence of Cav-1, TGF-β signaling is sufficient to maintain the balance between cell proliferation and cell growth arrest characteristic of regenerating liver.
Classic studies in liver regeneration and fibrotic processes have found that HGF specifically counteracts many anti-proliferative and pro-fibrotic actions of TGF-β, suggesting that the balance between HGF and TGF-β might play a pivotal role in the pathogenesis of liver and kidney diseases (17, 44). HGF and TGF-β are induced after PH and tissue injury, suggesting that both are important for the initial wound-healing response and tissue repair. In transient injuries, HGF signaling will predominate resulting in tissue repair and regeneration. In this scenario, TGF-β is important for matrix remodeling, but also for counteracting HGF activity after recovery. In chronic kidney diseases, HGF abolished TGF-β signaling by inducing SnoN in tubular epithelial cells (44). In this regard, the elevated expression of SnoN in the absence of Cav-1 was potentiated by HGF and may be related with the accumulation of c-Met. In summary, in this work we proposed a new cross-talk between TGF-β and HGF signaling and Cav-1 expression during liver regeneration. This cross-talk converges in the Smad transcriptional corepressor, SnoN, which impairs the growth-arrest pathway of TGF-β and thereby might explain why Cav-1−/− mice exhibit an accelerated liver regeneration after PH.
Acknowledgment
We thank Dr. M. A. del Pozo for the vectors encoding WT Cav-1-GFP and GFP.
This work was supported by Grants SAF2007-60551 and BFU2008-02161 from MICINN, S-BIO-0283/2006 from Comunidad de Madrid, and FIS-RECAVA RD06/0014/0025. Red temática Enfermedades Cardiovasculares, Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas, and Centro de Investigación Biomédica en Red de Diabetes y Enfermedades Metabólicas Asociadas are funded by the Instituto de Salud Carlos III.
- Cav
- caveolin
- Cav-1−/−
- caveolin-deficient mice
- TGF-β
- transforming growth factor β
- PH
- partial hepatectomy
- TβR
- TGF-β receptor
- SnoN
- Ski-related novel protein
- HGF
- hepatocyte growth factor
- DRM
- detergent-resistant membrane
- MES
- 4-morpholineethanesulfonic acid
- Ab
- antibody
- NCL
- immortalized neonatal cell line
- PAI-1
- plasminogen activator inhibitor-1
- NDRM
- nondetergent-resistant membrane
- RT
- reverse transcription
- FBS
- fetal bovine serum
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- qPCR
- quantitative PCR
- CHL
- Chang liver
- HCC
- hepatocellular carcinoma.
REFERENCES
- 1.Yamada E. (1955) J. Biophys. Biochem. Cytol. 1, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razani B., Woodman S. E., Lisanti M. P. (2002) Pharmacol. Rev. 54, 431–467 [DOI] [PubMed] [Google Scholar]
- 3.Schnitzer J. E. (2001) Adv. Drug Delivery Rev. 49, 265–280 [DOI] [PubMed] [Google Scholar]
- 4.Fu Y., Hoang A., Escher G., Parton R. G., Krozowski Z., Sviridov D. (2004) J. Biol. Chem. 279, 14140–14146 [DOI] [PubMed] [Google Scholar]
- 5.Okamoto T., Schlegel A., Scherer P. E., Lisanti M. P. (1998) J. Biol. Chem. 273, 5419–5422 [DOI] [PubMed] [Google Scholar]
- 6.del Pozo M. A., Balasubramanian N., Alderson N. B., Kiosses W. B., Grande-García A., Anderson R. G., Schwartz M. A. (2005) Nat. Cell Biol. 7, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams T. M., Lisanti M. P. (2005) Am. J. Physiol. Cell Physiol. 288, C494–C506 [DOI] [PubMed] [Google Scholar]
- 8.Engelman J. A., Zhang X. L., Razani B., Pestell R. G., Lisanti M. P. (1999) J. Biol. Chem. 274, 32333–32341 [DOI] [PubMed] [Google Scholar]
- 9.Galbiati F., Volonté D., Liu J., Capozza F., Frank P. G., Zhu L., Pestell R. G., Lisanti M. P. (2001) Mol. Biol. Cell 12, 2229–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo M., Tebar F., Lopez-Iglesias C., Enrich C. (2001) Hepatology 33, 1259–1269 [DOI] [PubMed] [Google Scholar]
- 11.Yokomori H., Oda M., Ogi M., Sakai K., Ishii H. (2002) Liver 22, 150–158 [DOI] [PubMed] [Google Scholar]
- 12.Yerian L. M., Anders R. A., Tretiakova M., Hart J. (2004) Am. J. Surg. Pathol. 28, 357–364 [DOI] [PubMed] [Google Scholar]
- 13.Zhao X., Liu Y., Ma Q., Wang X., Jin H., Mehrpour M., Chen Q. (2009) Biochem. Biophys. Res. Commun. 378, 21–26 [DOI] [PubMed] [Google Scholar]
- 14.Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. (2004) Mol. Biol. Cell 15, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández M. A., Albor C., Ingelmo-Torres M., Nixon S. J., Ferguson C., Kurzchalia T., Tebar F., Enrich C., Parton R. G., Pol A. (2006) Science 313, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 16.Mayoral R., Fernández-Martínez A., Roy R., Boscá L., Martín-Sanz P. (2007) Hepatology 46, 813–822 [DOI] [PubMed] [Google Scholar]
- 17.Michalopoulos G. K. (2007) J. Cell. Physiol. 213, 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa Y., Wang M., Carr B. I. (1998) J. Cell. Physiol. 176, 612–623 [DOI] [PubMed] [Google Scholar]
- 19.Bouzahzah B., Fu M., Iavarone A., Factor V. M., Thorgeirsson S. S., Pestell R. G. (2000) Cancer Res. 60, 4531–4537 [PubMed] [Google Scholar]
- 20.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 21.Le Scolan E., Zhu Q., Wang L., Bandyopadhyay A., Javelaud D., Mauviel A., Sun L., Luo K. (2008) Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 22.Macias-Silva M., Li W., Leu J. I., Crissey M. A., Taub R. (2002) J. Biol. Chem. 277, 28483–28490 [DOI] [PubMed] [Google Scholar]
- 23.Razani B., Zhang X. L., Bitzer M., von Gersdorff G., Böttinger E. P., Lisanti M. P. (2001) J. Biol. Chem. 276, 6727–6738 [DOI] [PubMed] [Google Scholar]
- 24.Lee E. K., Lee Y. S., Han I. O., Park S. H. (2007) Biochem. Biophys. Res. Commun. 359, 385–390 [DOI] [PubMed] [Google Scholar]
- 25.Santibanez J. F., Blanco F. J., Garrido-Martin E. M., Sanz-Rodriguez F., del Pozo M. A., Bernabeu C. (2008) Cardiovasc. Res. 77, 791–799 [DOI] [PubMed] [Google Scholar]
- 26.Chen Y. G. (2009) Cell Res. 19, 58–70 [DOI] [PubMed] [Google Scholar]
- 27.Martín-Sanz P., Callejas N. A., Casado M., Díaz-Guerra M. J., Boscá L. (1998) Br. J. Pharmacol. 125, 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valverde A. M., Burks D. J., Fabregat I., Fisher T. L., Carretero J., White M. F., Benito M. (2003) Diabetes 52, 2239–2248 [DOI] [PubMed] [Google Scholar]
- 29.Wrana J. L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. (1992) Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q., Pearson-White S., Luo K. (2005) Mol. Cell. Biol. 25, 10731–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cokakli M., Erdal E., Nart D., Yilmaz F., Sagol O., Kilic M., Karademir S., Atabey N. (2009) BMC Cancer 9, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan R., Zhang X., Yang J., Li Y., Liu Y. (2007) J. Am. Soc. Nephrol. 18, 2340–2349 [DOI] [PubMed] [Google Scholar]
- 33.Cantiani L., Manara M. C., Zucchini C., De Sanctis P., Zuntini M., Valvassori L., Serra M., Olivero M., Di Renzo M. F., Colombo M. P., Picci P., Scotlandi K. (2007) Cancer Res. 67, 7675–7685 [DOI] [PubMed] [Google Scholar]
- 34.Wang S., Jia L., Zhou H., Wang X., Zhang J. (2008) IUBMB Life 60, 693–699 [DOI] [PubMed] [Google Scholar]
- 35.Kim S., Lee Y., Seo J. E., Cho K. H., Chung J. H. (2008) Cell. Signal. 20, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 36.Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. (2003) Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 37.Deheuninck J., Luo K. (2009) Cell Res. 19, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heldin C. H., Miyazono K., ten Dijke P. (1997) Nature 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 39.Seoane J. (2006) Carcinogenesis 27, 2148–2156 [DOI] [PubMed] [Google Scholar]
- 40.Stavnezer E., Barkas A. E., Brennan L. A., Brodeur D., Li Y. (1986) J. Virol. 57, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F., Lundin M., Ristimäki A., Heikkilä P., Lundin J., Isola J., Joensuu H., Laiho M. (2003) Cancer Res. 63, 5005–5010 [PubMed] [Google Scholar]
- 42.Krakowski A. R., Laboureau J., Mauviel A., Bissell M. J., Luo K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12437–12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longerich T., Breuhahn K., Odenthal M., Petmecky K., Schirmacher P. (2004) Virchows Arch. 445, 589–596 [DOI] [PubMed] [Google Scholar]
- 44.Liu Y. (2004) Am. J. Physiol. Renal Physiol. 287, F7–F16 [DOI] [PubMed] [Google Scholar]