Abstract
Capsid proteins that adopt distinct conformations constitute a paradigm of the structural polymorphism of macromolecular assemblies. We show the molecular basis of the flexibility mechanism of VP2, the capsid protein of the double-stranded RNA virus infectious bursal disease virus. The initial assembly, a procapsid-like structure, is built by the protein precursor pVP2 and requires VP3, the other infectious bursal disease virus major structural protein, which acts as a scaffold. The pVP2 C-terminal region, which is proteolyzed during virus maturation, contains an amphipathic α-helix that acts as a molecular switch. In the absence of VP3, efficient virus-like particle assembly occurs when the structural unit is a VP2-based chimeric protein with an N-terminal-fused His6 tag. The His tag has a positively charged N terminus and a negatively charged C terminus, both important for virion-like structure assembly. The charge distributions of the VP3 C terminus and His tag are similar. We tested whether the His tag emulates the role of VP3 and found that the presence of a VP3 C-terminal peptide in VP2-based chimeric proteins resulted in the assembly of virus-like particles. We analyzed the electrostatic interactions between these two charged morphogenetic peptides, in which a single residue was mutated to impede the predicted interaction, followed by a compensatory double mutation to rescue electrostatic interactions. The effects of these mutations were monitored by following the virus-like and/or virus-related assemblies. Our results suggest that the basic face of the pVP2 amphipathic α-helix interacts with the acidic region of the VP3 C terminus and that this interaction is essential for VP2 acquisition of competent conformations for capsid assembly.
Keywords: Methods/Electron Microscopy, Protein/Molecular Dynamics, Protein/Protein-Protein Interactions, Protein/Assembly, Protein/Chimeras, Protein/Multifunctional, Protein/Self-assembly, Viruses/Structure
Introduction
Viruses represent an elegant example of spontaneous self-assembly. There are nonetheless fundamental differences between natural crystals and viruses; a crystal is a continuous three-dimensional network without size limits, whereas viruses build a container with a defined cargo space for nucleic acid packaging in a biologically feasible time frame (1). Based on the basic principles of crystal growth, viruses require additional mechanisms to control their dimensions (2). The tertiary structure of capsid structural subunits has built-in conformational flexibility; this allows it to maintain slightly different intersubunit contacts and facilitates acquisition of distinct spatial conformations (3, 4). In addition, viruses can use cell (membrane-specific regions, chaperones, etc.) or viral components (scaffolding, accessory, and proteolytic proteins) to facilitate subunit interactions, resulting in the generation of transient conformations (5, 6). Spatial and transient conformations are achieved by the intrinsic structural polymorphism of the viral proteins. Here, we addressed the molecular basis of birnavirus spherical capsid assembly, which is necessary for infectious particle production. We describe the minimal elements that, through electrostatic interactions, allow many copies of the same infectious bursal disease virus (IBDV)5 capsid protein to acquire different conformations.
Birnaviruses are a family of nonenveloped double-stranded (ds) RNA viruses with an icosahedral capsid (7). Most of our understanding of birnaviruses is based on studies of IBDV, an avian pathogen, and of infectious pancreatic necrosis virus, a fish pathogen.
IBDV has a polyploid bipartite dsRNA genome (segments A and B) (8) that is packaged into a single virus particle. Segment A (3.2 kbp) has two partially overlapping open reading frames; the first codes for the nonessential VP5 protein, and the second encodes a polyprotein. Segment B (2.8 kbp) is monocistronic and encodes VP1, the RNA-dependent RNA polymerase (9, 10). The polyprotein (1012 residues) is co-translationally self-cleaved by the viral protease VP4. This yields most of the structural proteins, including the capsid precursor protein pVP2, VP3, and VP4 (11, 12). pVP2 (512 residues) undergoes a variety of defined C-terminal processing events by VP4 at three secondary targets (positions 487, 494, and 501) (13). The resulting intermediates are further cleaved by VP2 itself (14) between residues Ala441 and Phe442, to yield the mature VP2 polypeptide (441 residues) and several C-terminal fragments. The released C-terminal segments remain associated with the capsid (15) and, by promoting the disruption of host cell membranes, appear to be involved in the entry mechanism (16, 17). VP3 is a multifunctional protein that interacts with itself (18, 19), pVP2 (20, 21), VP1 (22–24), and the dsRNA (25, 26).
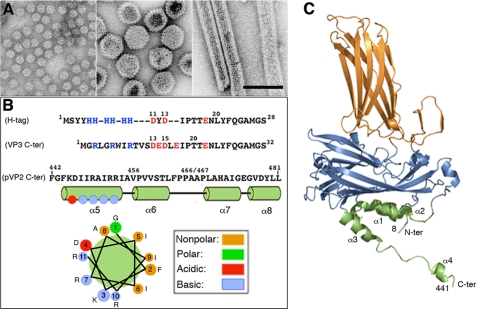
VP2 and a variable amount of pVP2 assemble into 260 trimers, the basic structural units that form a complex T = 13 levo capsid (protein mass ∼38 MDa, diameter ∼700 Å) made up of 12 pentamers and 120 hexamers (27–29). By strict geometric considerations, VP2 trimers adopt five distinct conformations. Although the most abundant assembly in an IBDV-infected cell corresponds to T = 13 particles, other minor aberrant assemblies are found in vivo, such as tubular structures and smaller T = 1 icosahedral capsids (Fig. 1A). Expression of mature VP2 alone results in the spontaneous assembly of T = 1 subviral particles built of 20 VP2 trimers arranged into pentamers, whereas pVP2 or intermediate pVP2 variant expression leads to tubular structures with a hexagonal lattice (20, 27). These IBDV-related structures are formed by the same structural building block, (p)VP2 trimers. The pVP2 71-residue C-terminal domain has an important role in allowing the formation of diverse VP2 conformations (27). This ability resides specifically in the 443GFKDIIRAIR452 amphipathic α-helix, referred to as helix α5. VP2 polypeptides with this α-helix assemble into genuine virus-like particles (VLPs) only when expressed as a chimeric protein with an N-terminal His tag (chimeric proteins His-VP2-456 and His-VP2-466) (20). Previous observations suggested that VP3 must also contribute to the inherent ability of pVP2 to acquire distinct conformations. VP3 mutants with alterations in or deletions of C-terminal residues (30) or entirely lacking the last 13 residues (18) thus fail to support assembly of (p)VP2 into T = 13 capsids, and hexagonal tubular or aberrant assemblies are formed. It was hypothesized that the His tag emulates the role of VP3 during morphogenesis, mimicking the VP3 C-terminal region closely (Fig. 1B). Although the α5-helix is a conformational switch responsible for the inherent structural polymorphism of VP2, its presence alone is insufficient to support this polymorphism. A His tag is nonetheless essential for production of genuine virus-like structures and acts as an α5-helix-triggering factor.
FIGURE 1.
(p)VP2-based chimeric protein system to test VP2 structural polymorphism. A, left, major (p)VP2 regular assemblies isolated from IBDV-infected avian cells or, using heterologous expression systems, from mammalian cells with an inducible rVV and in insect cells with the baculovirus expression system. Icosahedral T = 1 subviral particles (∼23-nm diameter) are built by VP2 trimers associated in an all-pentamer capsid. Center, T = 13 virion particles (65–70-nm diameter) are formed by 260 VP2 trimers making 12 pentamers and 120 hexamers. Right, tubular structures (45–60-nm diameter, depending on the number of helical rows) with a hexagonal lattice are all-hexamer structures. B, VP3 and pVP2 polypeptide segments involved in VP2 structural polymorphism are shown. His tag (H-tag) and VP3 C-terminal (VP3 C-ter) sequences are aligned based on polarity. The His tag used here is composed of a tract of four residues (Met1-Tyr4), six His residues, a linker region (segment Asp11-Thr17), the TEV cleavage site (Glu18-Gly24), and the tract Asn25-Ser28, to which the (p)VP2-based protein is bound. The VP3 C terminus includes the last 16 VP3 residues (242GRLGRWIRTVSDEDLE257; for simplicity, Glu257 is referred to here as Glu17, and so on) and the linker region (except the sequence 11DYD13), the TEV cleavage site (Glu18-Gly24), and the tract Asn25-Ser28. The C-terminal (p)VP2 Phe442-Leu481 sequence (pVP2 C-ter) is shown. The amphipathic α-helix (residues 443–452) is referred as helix α5; α6-, α7-, and α8-helices were also determined by NMR analysis (16). A helical wheel representation of the amphipathic α-helix is shown (bottom). C, VP2 protein x-ray model (Protein Data Bank code 2GSY). Domains P, S, and B are colored orange, blue, and green, respectively. The α-helices (1–4) of domain B are indicated. Note that N and C termini are relatively close.
The VP2 atomic structure was solved by x-ray crystallography of the T = 1 subviral particle capsids (28, 31, 32). The VP2 subunit folds into three domains termed projection (P), shell (S), and base (B). Domains S and P are β-barrels with a jelly roll topology. The B domain is formed by α-helices facing the interior of the shell, corresponding to its N and C termini. The helical C-terminal arm of VP2 establishes domain swapping and mediates interactions between adjacent VP2 trimers, increasing their stability (Fig. 1C).
Here, we show that assembly control of the IBDV T = 13 capsid requires electrostatic interactions between two morphogenetic polypeptide elements, provided independently by the major structural proteins (p)VP2 and VP3. In addition, acidic stretches in the His tag and the VP3 C-terminal residues are preceded by a basic segment, which has an additional role during capsid assembly.
EXPERIMENTAL PROCEDURES
Cells and Viruses
IBDV infections were carried out with the Soroa isolate (24), isolated from an infected chicken in the Soroa region (Cuba) and adapted for growth in chicken embryonic fibroblast primary cultures. It was then adapted for growth in QM7 quail muscle cells. Recombinant vaccinia virus (rVV) VT7/POLY and recombinant baculoviruses (rBV) FB/His-VP2-466 and FB/His-VP2-456 were described previously (20, 27). Expression experiments were carried out in QM7 cells for rVV infections and Trichoplusia ni (H5) insect cells (Invitrogen) for rBV infections. QM7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, H5 cells in TC-100 medium (Invitrogen) with 10% fetal calf serum. rVV and rBV were grown and titrated as described in Refs. 24 and 33.
Construction of Recombinant Baculoviruses
The pFB/His-VP2-466 (20, 27) and pFB/POLY (21) plasmids were used as a template for PCR synthesis to generate the N-terminal deletion mutants rBV/HisΔ2–10-VP2-466 and rBV/HisΔ11–25-VP2-466. PCR was performed with Vent DNA polymerase (Biolabs) and 5′ end primer specific for each mutant (5′-GCGCAGATCTATGGATTACGATATCCCACGACC to generate rBV/HisΔ2–10-VP2-466 from pFB/His-VP2-466, and 5′-CGCAGATCTATGTCNTAYTAYCAYCAYCAYCAYCAYCAYATGGGATCTATGACAAACCTGTCAGATCAA for rBV/HisΔ11-25-VP2-466 from pFB/POLY) and a common 3′ end primer (5′-GCGCAAGCTTAGGCAGGTGGGAACAATGTGG). BglII-HindIII-digested PCR fragments were cloned into pFastBac (Invitrogen) previously digested with BamHI and HindIII.
pFB/CtVP3-VP2-466 and pFB/CtVP3-VP2-456 fusions were constructed by generation of two overlapping PCR fragments. The first used pVOTE.2/VP3 (34) as a template in combination with primers 5′-GCGCCGGTCCGAAACCATGGGTCGGCTGGGCCGCTGGATCAGG and 5′-GTCTCTGATGAGGACCTTGAGATCCCAACGACCGAAAACCTG. The second fragment was generated using primers 5′-CAGGTTTTCGGTCGTTGGGATCTCAAGGTCCTCATCAGAGACGG and 5′-AAGACAATTAGCCCTG, and pFB/HTVP2-466 or pFB/HTVP2-456 (20, 27) as templates, respectively. PCR overlap extension was carried out with primers 5′-GCGCCGGTCCGAAACCATGGGTCGGCTGGGCCGCTGGATCAGG and 5′-AAGACAATTAGCCCTG. The resulting DNA fragment was digested with RsrII and used to replace the original RsrII fragment of the pFB/HTVP2-466 or pFB/HTVP2-456 plasmids.
Using PCR overlap extension, four different mutants for pFB/His-VP2-466 were generated with the following single mutations: D11R, D13R in the His tag, and K445D and R449D in the amphipathic α-helix. PCRs were carried out using the pFB/His-VP2-466 plasmid as template, and the set of mutator primers (supplemental Table 1) in combination with primers 5′-GTATTTTACTGTTTTCGTAACAG and 5′-AAGACAATTAGCCCTG for the His tag mutations and 5′-GGCCGCAAACAAGGGCTGACG and 5′-GCGCAAGCTTAGGCAGGTGGGAACAATGTGG for the α-helix of pVP2 mutations. The resulting DNA fragments were digested with RsrII for the His tag, and BamHI and HindIII for the pVP2 α-helix, and used to replace the original pFB/His-VP2-466 plasmid sequences. Double mutants D11R/K445D, D11R/R449D, D13R/K445D, and D13R/R449D were generated by ligation of the appropriate mutated BamHI/HindIII α-helix fragment into the single mutant His tag plasmid.
Similarly, four mutants were generated for pFB/CtVP3-VP2-466 with the following single mutations: D13R (Asp253) and D15R (Asp255) of the VP3 C-terminal segment and K445D and R449D for VP2 amphipathic α-helix. PCRs were carried out using the pFB/CtVP3-VP2-466 plasmid as template, and the set of mutator primers (supplemental Table 1) in combination with primers described above for the His tag and pVP2 α-helix mutations. The resulting DNA fragments were digested with RsrII for the CtVP3, and BamHI and HindIII for the α-helix of pVP2, and used to replace the original pFB/CtVP3-VP2-466 plasmid sequences. Double mutants D13R/K445D, D13R/R449D, D15R/K445D, and D15R/R449D were generated similarly to the double mutations in pFB/His-VP2-466 described above.
Selection of derived bacmids from the DH10Bac Escherichia coli strain and preparation for lipofectine transfection were done according to the manufacturer's protocols (Invitrogen). The constructs were expressed in H5 insect cells (18).
Generation of the Recombinant Viruses VT7/LacOI
Using PCR overlap extension, three mutant polyprotein genes were constructed with single mutations in the VP3 C-terminal domain: R249D, R246D, and R243D. PCRs were carried out using the polyprotein gene, cloned in the pVOTE.2/POLY plasmid as template, and the set of mutator primers (supplemental Table 2) in combination with primers 5′-GCGCGCATGCAGAGAAGAGCCGGTTGGCA and 5′-GCGCGCTCAGCGGTGGCAGCAGCCA. The resulting DNA fragments were digested with SphI and BlpI and used to replace the original SphI-BlpI fragment of the pVOTE.2/POLY plasmid (33). The plasmids pVOTE/R249D, pVOTE/R246D, and pVOTE/R243D were used to obtain rVV VT7/LacOI/R249D, VT7/LacOI/R246D, and VT7/LacOI/R243D, respectively. BSC-1 cells were infected with rVV VT7LacOI (35) and transfected with the plasmids described above. Selection and amplification of VT7LacOI/R249D, VT7LacOI/R246D, and VT7LacOI/R243D were carried out as described (36).
Purification of IBDV Polyprotein-derived Structures and IBDV Capsids
QM7 and H5 cells (2–5 × 108 cells) were infected with appropriate rVV or rBV (multiplicity of infection 1–5 plaque-forming units/cell). At 72 h postinfection (h.p.i.) for rVV assays and 48 h.p.i. for rBV assays, cells were harvested, lysed in PES buffer (25 mm PIPES, pH 6.2, 150 mm NaCl, and 20 mm CaCl2) plus 1% IGEPAL CA-630 (Sigma) and 1% protease inhibitors (Complete Mini; Roche Applied Science) on ice for 30 min. The lysate was clarified by centrifugation (1,000 × g, 10 min), and the supernatant was processed on a 25% sucrose cushion (170,000 × g, 150 min). The pellet was resuspended in PES buffer, centrifuged in a microfuge (16,000 × g, 1 min), and the supernatant was processed in a linear 20–50% sucrose gradient (200,000 × g, 45 min). The particulate material containing polyprotein-derived structures was collected in 12 fractions and concentrated 20-fold by ultracentrifugation (240,000 × g, 120 min). All purification steps were performed at 4 °C.
IBDV Soroa strain was purified from QM7 cells by a standard protocol (8). Briefly, IBDV particles from the cell medium were harvested 48 h.p.i., precipitated with 3.5% polyethylene glycol 6000 and 0.5 m NaCl, and the resulting pellet was resuspended in PES buffer. Particles were then pelleted through a 25% sucrose cushion (170,000 × g, 150 min) followed by CsCl equilibrium gradient centrifugation after adjusting the initial density of the solution to 1.33 g/ml by CsCl addition (130,000 × g, 14 h, 4 °C). E5 virus particles (the major band of six) were collected, dialyzed against PES buffer, and stored at 4 °C.
SDS-PAGE and Western Blotting
Concentrated sucrose gradient fractions (2–5 μl) were added to Laemmli sample buffer to a 1× final concentration and heated (100 °C, 3 min). Electrophoresis was performed in 11% polyacrylamide gels followed by Western blot analyses using an anti-VP2 and anti-VP3 antibodies (24).
Electron Microscopy
Samples (2–5 μl) of concentrated sucrose gradient fractions were applied to glow-discharged carbon-coated grids (2 min) and negatively stained with 2% aqueous uranyl acetate. Micrographs (Kodak SO-163) were recorded with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of ×40,000.
RESULTS
Analysis of the His Tag as a Triggering Factor: Role of Acidic and Basic Segments
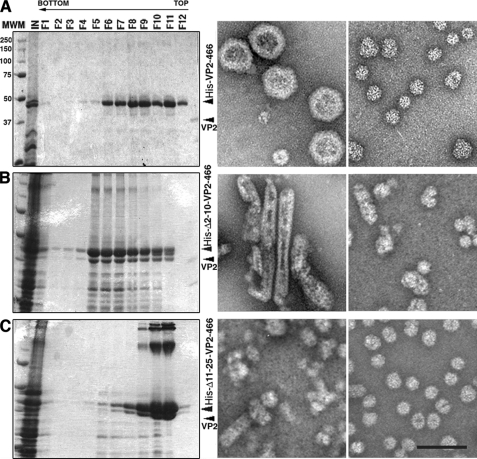
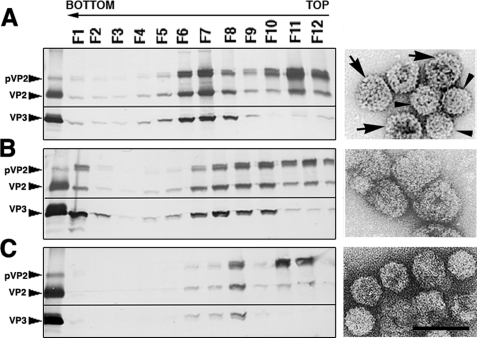
We developed a (p)VP2 chimeric protein system to discern the factors involved in virion capsid assembly (20). Expression of the chimeric His-VP2-466 gene in insect cells leads to spontaneous assembly of T = 13 capsids (Fig. 2A). We hypothesized that the His tag fused to the (p)VP2 N terminus emulates VP3 C terminus function during the morphogenesis of VLP that are structurally indistinguishable from IBDV virions. The sequence of the His tag in these chimeric VP2 constructs has an acidic segment with clear similarity in the disposition of its charged residues to that of the VP3 C-terminal region (Fig. 1B). Our hypothesis of virus capsid assembly assumed that electrostatic interactions are required for adoption of different conformational states because charge complementarity is evident between the acid segment (of the His tag or the VP3 C terminus) and the basic side of the (p)VP2 amphipathic α5-helix. In the context of the His tag and the VP3 C terminus, the acidic segment is preceded by a basic segment (the six His residues and an Arg-rich region for VP3 C terminus). Whether both acidic and basic regions have a role in VLP assembly remains to be elucidated. We therefore generated two deletion mutants of the (p)VP2-based chimeric protein that affect the His tag: the HisΔ2–10-VP2-466 deletion mutant that lacks the 2–10 tract of the His tag (containing the His6 stretch) and the HisΔ11–25-VP2-466 mutant lacking residues of tract 11–25 (containing the acidic residues, among others). These genes were expressed in insect cells, and the extracts were used to purify VLP. Expression of the HisΔ2–10-VP2-466 and HisΔ11–25-VP2-466 genes resulted in the assembly of tubular structures and irregularly sized particles (Fig. 2, B and C), but negative staining showed no VLP similar to those produced by the wild-type His-VP2-466 chimeric gene (Fig. 2A).
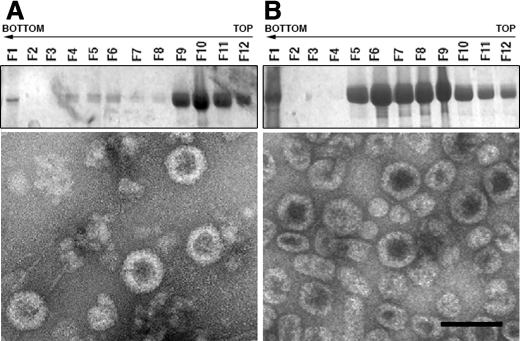
FIGURE 2.
Acidic and basic regions of the molecular triggering factor, the His tag, are essential for T = 13 capsid assembly. A, wild-type His-VP2-466 chimeric protein was expressed in insect cells, and the assemblies were purified on sucrose gradients; 12 fractions were collected, concentrated, and analyzed by SDS-PAGE and Coomassie staining. The direction of sedimentation was right to left, with fraction 12 representing the gradient top. Images on the right correspond to representative electron micrographs (negative staining) of wild-type His-VP2-466 assemblies; T = 13 capsid-like particles are found in the middle fractions (left), and T = 1 capsid-like structures in upper fractions (right). B and C, His tag deletion mutants HisΔ2–10-VP2-466 (B) and HisΔ11–25-VP2-466 (C) were analyzed as described above. HisΔ2–10-VP2-466 assemblies are tubular structures (detected in lower fractions) and isometric irregularly sized particles (detected in upper fractions). HisΔ11–25-VP2-466 rendered small isometric particles. Scale bar, 100 nm.
The His Tag Functionally Emulates the Role of the VP3 C Terminus
We showed previously that expression of the shortest His-tagged pVP2 variant able to assemble into VLP similar to IBDV virions has at least 456 residues. This construct has the amphipathic α5-helix at its C-terminal domain. His-VP2-456 protein represents an abrupt limit; shorter chimeric proteins, such as His-VP2-441, only assemble into T = 1. The His-VP2-466 polypeptide assembles into structures morphologically similar to true T = 13 infectious capsids with higher efficiency than His-VP2-456. We therefore used these constructs to determine whether the His tag in fact mimics the role of the VP3 C-terminal region. We generated two chimeric proteins (with 456 or 466 pVP2 residues) bearing the VP3 C-terminal segment in place of the original His tag; CtVP3-VP2-456 and CtVP3-VP2-466 were inserted into pFastBac and used to generate the corresponding rBV. Expression of CtVP3-VP2-456 and CtVP3-VP2-466 constructs is similar and results in assembly of structures related to the original His-tagged chimeric particles. pVP2 variants fused to an N-terminal VP3 C terminus are able to assemble into structures morphologically similar to authentic T = 13 capsids, which banded at the middle of the gradient (as predicted for this kind of assembly), but there was also a complex mixture of other, related assemblies (Fig. 3A). Most CtVP3-VP2-456 (as for His-VP2-456) was located at the top of the gradient as T = 1 capsids and other isometric small assemblies (20). The tendency to form T = 13 capsids is further improved with the CtVP3-VP2-466 protein (as for His-VP2-466); although the fractions containing T = 13 capsid-like particles were more abundant, they also contained related assemblies of different sizes (Fig. 3B). These results show that exchange of His tag and VP3 C terminus does not affect T = 13 capsid assembly.
FIGURE 3.
His tag emulates the VP3 C terminus. Cells infected with FB/CtVP3-VP2-456 (A) and FB/CtVP3-VP2-466 rBV (B) were harvested at 48 h.p.i. and lysed. C-terminal VP3-tagged pVP2 chimeric assemblies were purified in sucrose gradients; 12 fractions were collected, concentrated, and analyzed by SDS-PAGE and Coomassie staining. The direction of sedimentation was right to left, with fraction 12 representing the gradient top. Images show representative electron microscopy micrographs (negative staining) of chimeric construct assemblies at the middle gradient fractions. Scale bar, 100 nm.
Point Mutations That Alter the Electrostatic Character of the pVP2 Amphipathic α5-Helix and the Triggering Factor Sequences Abolish Virion Capsid Assembly
To study further the role of the pVP2 amphipathic α-helix positive-charged face in VP2 polymorphism, we tested whether T = 13 capsid assembly is abolished by altering the electrostatic character of this face. The amphipathic α-helix has two basic residues, Lys445 and Arg449, which are highly conserved among birnavirus homologues. We engineered two single-point mutants with K445D and R449D substitutions and analyzed their effect in the context of VP3 C terminus- and His-tagged VP2-466. We also introduced single point mutations within the triggering factor sequences of the His tag or the VP3 C terminus. For this, acidic residues Asp11 and Asp13 of the His tag and Asp13 (corresponding to Asp253 of VP3) and Asp15 (corresponding to Asp255) of the VP3 C terminus were replaced with basic residues. Another set of four mutants was generated by substitutions D13R or D15R in the VP3 C terminus- and D11R or D13R in the His-tagged versions.
Side-by-side comparison of the resulting viral-related assemblies generated by expression of the mutant polypeptides with those of the wild-type versions showed that none of the mutants assembled T = 13 capsids (Fig. 4). Whereas amphipathic α-helix mutants assembled as tubular structures, triggering factor mutants tended to assemble as T = 1 capsids. These data further support our hypothesis that the two morphogenetic peptides mediate VP2 polymorphism, when both are present in our model chimeric protein.
FIGURE 4.
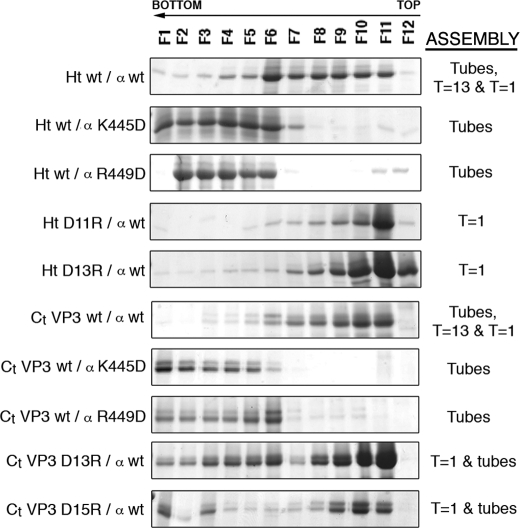
Single-point mutant analysis of the conformational switch and the molecular triggering factor of the pVP2 structural polymorphism. His-tagged (Ht) and VP3 C terminus-tagged (Ct VP3) VP2-466 chimeric proteins with single-point substitution mutations (D11R or D13R in the His tag; D13R (Asp253) or D15R (Asp255) in the VP3 C-terminal segment, and K445D or R449D in the (p)VP2 amphipathic α-helix) were expressed in insect cells, and the assemblies were purified using sucrose gradients; 12 fractions were collected and analyzed by SDS-PAGE and Coomassie staining. Wild-type chimeric protein profiles are shown (Ht wt/α wt, Ct VP3 wt/α wt). The direction of sedimentation was right to left, with fraction 12 representing the top of each gradient. The assemblies are indicated: T = 1, T = 1 subviral particles; T = 13, T = 13 virion-like particles; Tubes, tubular structures with a hexagonal lattice.
Compensatory Molecular Switch/Triggering Factor Double Mutants Restore Virion Capsid Assembly
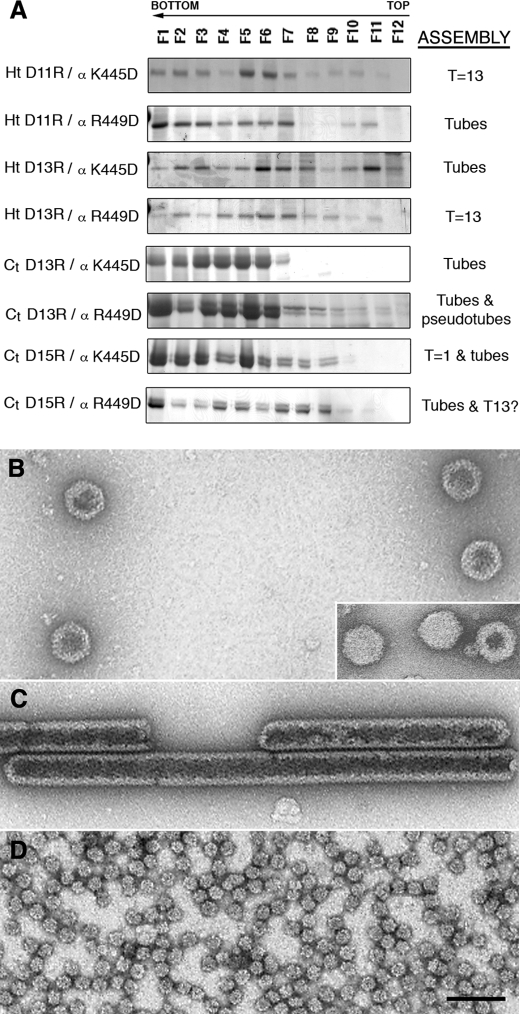
Although our mutational analysis indicated that positive charges on the basic side of the pVP2 α5-helix and the acidic residues of the VP3 C terminus (or the His tag) are critical for acquiring several structural conformations of the capsid protein, it did not confirm that these structural elements interact. We therefore constructed a collection of double-mutant polypeptides from the single-mutant gene versions, in which both morphogenetic elements were mutated simultaneously. If our hypothesis is correct, some of these double mutants should show compensatory effects, leading to restoration of the wild-type phenotype, with recovery of VLP assembly. Some of these new chimeric proteins harboring two individually disadvantageous mutations recovered the wild-type phenotype and assembled as VLP morphologically similar to virion particles (Fig. 5). The His-tagged double-mutant chimeric proteins showed rescue of T = 13 assembly ability more clearly than the VP3 C terminus-tagged proteins. These results indicate that the VP3 C terminus (or the His tag), together with the pVP2 α5-helix, cannot only be modified independently or disengaged, but can also be complemented by generation of double mutants. Our data show that electrostatic interactions are established between VP2 Arg449 and His tag Asp13 and probably between VP2 Arg449 and VP3 Asp255 (or VP3 C-terminal Asp15). Although this interaction is clearly necessary for VP2 structural polymorphism, these results must be treated with caution. Our approach imposes steric restrictions because the VP3 C terminus (or the His tag) is covalently bound to the pVP2 N terminus; that is, when VP3 is a free protein as found in an infected cell, VP3 Asp253 might play the role we have assigned to Asp255.
FIGURE 5.
Rescue of T = 13 virus-like particle assembly by compensatory double mutants of the conformational switch and the molecular triggering factor. A, His- and VP3 C terminus-tagged VP2-466 chimeric proteins with double mutations, in which Asp residues of the His tag or the VP3 C-terminal segment were replaced by Arg (for the His tag, Ht D11R and Ht D13R; for the VP3 C-terminal segment, Ct D13R (Asp253) and Ct D15R (Asp255)), and Lys or Arg residues of the (p)VP2 amphipathic α-helix were replaced by Asp (α K445D or α R449D), expressed in insect cells. The assemblies were purified in sucrose gradients; 12 fractions were collected and analyzed by SDS-PAGE and Coomassie staining. The assemblies are indicated as in Fig. 4. B, electron microscopy of the Ht D13R/α R449D double-mutant assemblies: T = 13 capsid-like particles were found in the middle gradient fractions. Inset, T = 13 virion particles. C and D, assemblies of Ct D13R/α wild-type single-point mutant: tubular structures with a hexagonal lattice in the lower fractions (C) and T = 1 capsids in the upper fractions (D). Scale bar, 100 nm.
Arg-rich Region at the VP3 C Terminus Has a Role in Correct T = 13 Capsid Assembly
We next analyzed whether the basic region preceding the last acidic residues at the C terminus of VP3 affects VP2 structural polymorphism. Expression of the IBDV polyprotein in mammalian cells using an inducible rVV results in high production levels of VLP similar to authentic IBDV particles (24, 37). IBDV polyprotein expression using rBV leads to the assembly of tubular structures and not of VLP. Insect cells might have important cellular factor(s) that interfere with VLP assembly. The rVV vector thus provided an appropriate framework in which to study the role of residues Arg243, Arg246, and Arg249. Three single-point mutants of the IBDV polyprotein gene were inserted into the vaccinia virus genome, giving rise to rVV VT7/LacOI/R249D, VT7/LacOI/R246D, and VT7/LacOI/R243D, in which the Arg residues under study were replaced by Asp. Viral-related assemblies from these mutants were analyzed by ultracentrifugation on sucrose gradients followed by Western blotting using VP2- and VP3-specific antibodies and negative staining. Samples from cells expressing polyprotein R243D assembled into T = 13 (∼65-nm diameter, arrows) and T = 7 capsids (∼53 nm, arrowheads) similar to wild-type polyprotein assemblies (Fig. 6A). In contrast, polyprotein R246D assemblies were unstable and collapsed during purification or after negative staining analysis (Fig. 6B), and polyprotein R249D assembled mainly into T = 7 capsids (Fig. 6C). Together, our findings highlight the importance of the VP3 C-terminal Arg-rich region in the structural polymorphism of the VP2 capsid polypeptide.
FIGURE 6.
VP3 C-terminal basic region is important for T = 13 capsid assembly. Western blot analysis of proteins expressed in cells infected with VT7/LacOI/R243D (A), VT7/LacOI/R246D (B), or VT7/LacOI/R249D rVV (C) is shown. Infected cultures were harvested at 72 h.p.i., and assemblies were purified by two-step centrifugation; 12 fractions were collected, concentrated, and analyzed by SDS-PAGE and Western blotting using anti-VP2 (top) or anti-VP3 (bottom) antibodies. The direction of sedimentation was right to left, with fraction 12 representing the gradient top. Images shown on the right correspond to representative electron micrographs (negative staining) of single-point polyprotein mutant assemblies: Poly/R243D T = 13 (arrows) and T = 7 (arrowheads) (A) capsids, Poly/R246D collapsed T = 13 capsid-like particles (B), and Poly/R249D T = 7 capsids (C). Scale bar, 100 nm.
DISCUSSION
Assembly of large macromolecular complexes is frequently assisted by one or more auxiliary proteins that act as morphogenetic factors, controlling structural polymorphism and triggering structural changes. IBDV capsid assembly provides an excellent model system in which to study the coordination of factors involved in a multistep process that rarely leads to nonviable structures in vivo. Understanding these molecular interactions is necessary for the design of strategies that inhibit viral assembly or misdirect it into noninfectious aberrant structures (38).
The T = 13 IBDV capsid (20, 28, 29) is formed exclusively by VP2 trimers in five distinct conformations. This conformational flexibility is due to the transient presence of the pVP2 C-terminal region, specifically the 443–452 amphipathic α-helix (20), which acts as an inherent molecular switch. VP3, the other major structural protein, also participates in this process, acting as a scaffold protein (21).
Assembly pathways have classically been analyzed in detail from the purified critical proteins which, when mixed together in vitro, assemble into particles that closely resemble procapsids and/or capsids. Using a simple chimeric protein system, we found previously that in the absence of VP3, short VP2 variants containing the amphipathic α-helix assemble into T = 13 capsids only when fused to an N-terminal His tag (20). Here, we show that the His tag effectively emulates the function of the VP3 C terminus because both peptides can be functionally interchanged. We found that basic residues on the charged face of the pVP2 amphipathic α5-helix, whose replacement with aspartic acid abrogates VP2 polymorphism, interact with acidic residues at the VP3 C terminus. These electrostatic interactions were inferred by testing predictable compensatory double mutants in our minimal chimeric protein system. Additional mutation analysis of VP3 at its Arg-rich region, which precedes the acidic C-terminal residues, revealed an additional segment involved in capsid assembly. The results suggest that there are at least two distinct sets of interaction sites for the VP3 scaffold protein; one subset is needed for VP2 structural polymorphism, and the other might have a role in structural stabilization and/or preventing formation of nonviable structures. A similar situation was demonstrated for the scaffolding protein of the Salmonella phage P22 (39, 40).
Many studies over the last few years have shed light on IBDV capsid assembly and maturation. A spherical procapsid is initially assembled, as is also the case of infectious pancreatic necrosis virus (41). According to our hypothesis, the pVP2 coat protein coassembles with the VP3 scaffold polypeptide to form a double-layered structure, with pVP2 on the outer surface and VP3 facing the capsid interior. These contacts could be explained by electrostatic interactions between the VP3 C termini and the amphipathic α5-helices. Initial support for this hypothesis was derived from immunofluorescence and confocal microscopy experiments in insect cells expressing the IBDV polyprotein, which assembled as tubular structures visualized using VP2- and VP3-specific antibodies (33). The stable VP3-pVP2 interaction directs assembly of pVP2 trimers into hexamers and retards pVP2 C-terminal domain processing. In this context, the slow postassembly maturation step sidesteps the natural tendency of mature VP2 to assemble into all-pentameric T = 1 subviral particle. If processing is rapid, the VP3 C-terminal region cannot interact with pVP2 or does so only weakly, and a pentamer is formed from VP2 trimers. We consider that the pentamers could be the nucleating centers for capsid assembly and that additional (host) factors might participate, for example, to supply pentameric VP2 trimers that bind VP3 molecules transiently for the growing procapsid.
This assembly model is consistent with the puzzling VP3 stoichiometry inside the virion, which remains constant at ∼450 copies but is not icosahedrally ordered (8). Five classes of (p)VP2 trimeric capsomers (quasi-equivalent trimers) have been described (29) based on their local environments in the T = 13 capsid (classes a–e). Class a trimers lie on the 5-fold icosahedral axis and make close contact with class b trimers, generating a-b dimers. Class e trimers, located at the strict 3-fold icosahedral axis, interact closely with surrounding class d trimers. Class c trimers make pairwise contact (c-c dimers) similar to a-b dimers. VP3 can assemble as a regular scaffold structure beneath the pVP2 procapsid, with a similar VP3:(p)VP2 ratio except at the pentameric vertices, where a-b dimers do not interact with counterpart VP3 molecules, resembling the incomplete T = 13 shells of reovirus (42).
During the initial steps of procapsid growth, many amino acids on the VP3 outer surface must be in close proximity with the inner basement of the VP2 S domain. The spatial disposition of VP3 might modulate the conformational flexibility of the amphipathic α5-helix, which is found in a “closed state” as a helical bundle in (p)VP2 pentamers or in an “open state” as a star in hexamers (43). The numerous VP3 interactions with pVP2 are thus intimately coordinated with other molecular events that determine the coat protein maturation rate and consequently, pentamer and/or hexamer assembly.
The numerous VP3 scaffold activities associated with its highly hydrophilic C terminus indicate that VP3 is a multitasking protein in capsid morphogenesis. There are many similarities with two well described phage systems, the E. coli phage ϕX174 and the phage P22 (44). VP3 function is much more complex, however, because it also has numerous activities during the viral life cycle. The VP3 C terminus, which also participates in VP1 incorporation into the capsid, serves as a transcriptional activator (22) because it removes the inherent structural blockade of RNA polymerase activity. In addition, the VP3 oligomerization domain maps within the 42 C-terminal residues of the polypeptide (23). Finally, VP3 is also an RNA-binding protein associated with the dsRNA genome (34, 45). Viral maturation through defined steps might allow this VP3 multifunctionality; for example, most VP3 molecules are engaged to (p)VP2, but a small subset of molecules could be involved in VP1 incorporation into the capsid. Once the procapsid structure is fully assembled and closed, VP3 could be released and bind to genomic dsRNA. Studies currently under way suggest that host factors might also participate in the capsid assembly process.
Supplementary Material
Acknowledgment
We thank C. Mark for editorial assistance.
This work was supported by Spanish Ministry of Science and Innovation Grants BFU 2008-02328/BMC and S-0505-Mat-0238 (to J. L. C.), BIO2006-09407 (to J. F. R.), and BIO2008-02361 (to J. R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- IBDV
- infectious bursal disease virus
- dsRNA
- double-stranded RNA
- VP
- viral protein
- VLP
- virus-like particle
- rVV
- recombinant vaccinia virus
- rBV
- recombinant baculovirus
- Ct
- C-terminal
- h.p.i.
- hours postinfection
- PIPES
- piperazine-N,N′-bis(2-ethanesulfonic acid).
REFERENCES
- 1.Zlotnick A. (2005) J. Mol. Recognit. 18, 479–490 [DOI] [PubMed] [Google Scholar]
- 2.Johnson J. E. (2008) J. Struct. Biol. 163, 246–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gertsman I., Gan L., Guttman M., Lee K., Speir J. A., Duda R. L., Hendrix R. W., Komives E. A., Johnson J. E. (2009) Nature 458, 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardone G., Purdy J. G., Cheng N., Craven R. C., Steven A. C. (2009) Nature 457, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. (2005) Curr. Opin. Struct. Biol. 15, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dokland T. (2000) Structure Fold. Des. 8, R157–R162 [DOI] [PubMed] [Google Scholar]
- 7.Delmas B., Kibenge F. S. B., Leong J. C., Mundt E., Vakharia V. N., Wu J. L. (2005) in Virus Taxonomy (Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. eds) pp. 561–569, Elsevier, Amsterdam [Google Scholar]
- 8.Luque D., Rivas G., Alfonso C., Carrascosa J. L., Rodríguez J. F., Castón J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2148–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Einem U. I., Gorbalenya A. E., Schirrmeier H., Behrens S. E., Letzel T., Mundt E. (2004) J. Gen. Virol. 85, 2221–2229 [DOI] [PubMed] [Google Scholar]
- 10.Pan J., Vakharia V. N., Tao Y. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7385–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birghan C., Mundt E., Gorbalenya A. E. (2000) EMBO J. 19, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman A. R., Lee J., Delmas B., Paetzel M. (2006) J. Mol. Biol. 358, 1378–1389 [DOI] [PubMed] [Google Scholar]
- 13.Sánchez A. B., Rodríguez J. F. (1999) Virology 262, 190–199 [DOI] [PubMed] [Google Scholar]
- 14.Irigoyen N., Garriga D., Navarro A., Verdaguer N., Rodríguez J. F., Castón J. R. (2009) J. Biol. Chem. 284, 8064–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Da Costa B., Chevalier C., Henry C., Huet J. C., Petit S., Lepault J., Boot H., Delmas B. (2002) J. Virol. 76, 2393–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galloux M., Libersou S., Morellet N., Bouaziz S., Da Costa B., Ouldali M., Lepault J., Delmas B. (2007) J. Biol. Chem. 282, 20774–20784 [DOI] [PubMed] [Google Scholar]
- 17.Chevalier C., Galloux M., Pous J., Henry C., Denis J., Da Costa B., Navaza J., Lepault J., Delmas B. (2005) J. Virol. 79, 12253–12263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maraver A., Oña A., Abaitua F., González D., Clemente R., Ruiz-Díaz J. A., Castón J. R., Pazos F., Rodríguez J. F. (2003) J. Virol. 77, 6438–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casañas A., Navarro A., Ferrer-Orta C., González D., Rodríguez J. F., Verdaguer N. (2008) Structure 16, 29–37 [DOI] [PubMed] [Google Scholar]
- 20.Saugar I., Luque D., Oña A., Rodríguez J. F., Carrascosa J. L., Trus B. L., Castón J. R. (2005) Structure 13, 1007–1017 [DOI] [PubMed] [Google Scholar]
- 21.Oña A., Luque D., Abaitua F., Maraver A., Castón J. R., Rodríguez J. F. (2004) Virology 322, 135–142 [DOI] [PubMed] [Google Scholar]
- 22.Garriga D., Navarro A., Querol-Audí J., Abaitua F., Rodríguez J. F., Verdaguer N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maraver A., Clemente R., Rodríguez J. F., Lombardo E. (2003) J. Virol. 77, 2459–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardo E., Maraver A., Castón J. R., Rivera J., Fernández-Arias A., Serrano A., Carrascosa J. L., Rodríguez J. F. (1999) J. Virol. 73, 6973–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjalmarsson A., Carlemalm E., Everitt E. (1999) J. Virol. 73, 3484–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luque D., Saugar I., Rejas M. T., Carrascosa J. L., Rodríguez J. F., Castón J. R. (2009) J. Mol. Biol. 386, 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castón J. R., Martínez-Torrecuadrada J. L., Maraver A., Lombardo E., Rodríguez J. F., Casal J. I., Carrascosa J. L. (2001) J. Virol. 75, 10815–10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulibaly F., Chevalier C., Gutsche I., Pous J., Navaza J., Bressanelli S., Delmas B., Rey F. A. (2005) Cell 120, 761–772 [DOI] [PubMed] [Google Scholar]
- 29.Böttcher B., Kiselev N. A., Stel'Mashchuk V. Y., Perevozchikova N. A., Borisov A. V., Crowther R. A. (1997) J. Virol. 71, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chevalier C., Lepault J., Da Costa B., Delmas B. (2004) J. Virol. 78, 3296–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C. C., Ko T. P., Chou C. C., Yoshimura M., Doong S. R., Wang M. Y., Wang A. H. (2006) J. Struct. Biol. 155, 74–86 [DOI] [PubMed] [Google Scholar]
- 32.Garriga D., Querol-Audí J., Abaitua F., Saugar I., Pous J., Verdaguer N., Castón J. R., Rodríguez J. F. (2006) J. Virol. 80, 6895–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Torrecuadrada J. L., Castón J. R., Castro M., Carrascosa J. L., Rodríguez J. F., Casal J. I. (2000) Virology 278, 322–331 [DOI] [PubMed] [Google Scholar]
- 34.Kochan G., Gonzalez D., Rodríguez J. F. (2003) Arch. Virol. 148, 723–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward G. A., Stover C. K., Moss B., Fuerst T. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6773–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earl P. L., Moss B. (1993) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., More D. D., Seidman J. G., Smith J. A., Struhl K. eds) pp. 16.17.1–16.18.10, John Wiley & Sons, New York [Google Scholar]
- 37.Castón J. R., Rodríguez J. F., Casrrascosa J. L. (2008) in Segmented Double-stranded RNA viruses: Structure and Molecular Biology (Patton J. T. ed), Caister Academic Press, Norfolk, UK [Google Scholar]
- 38.Zlotnick A., Lee A., Bourne C. R., Johnson J. M., Domanico P. L., Stray S. J. (2007) Nat. Protoc. 2, 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker M. H., Brouillette C. G., Prevelige P. E., Jr. (2001) Biochemistry 40, 8962–8970 [DOI] [PubMed] [Google Scholar]
- 40.Parent K. N., Doyle S. M., Anderson E., Teschke C. M. (2005) Virology 340, 33–45 [DOI] [PubMed] [Google Scholar]
- 41.Villanueva R. A., Galaz J. L., Valdés J. A., Jashés M. M., Sandino A. M. (2004) J. Virol. 78, 13829–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dryden K. A., Wang G., Yeager M., Nibert M. L., Coombs K. M., Furlong D. B., Fields B. N., Baker T. S. (1993) J. Cell Biol. 122, 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luque D., Saugar I., Rodríguez J. F., Verdaguer N., Garriga D., Martín C. S., -Muriel J. A., Trus B. L., Carrascosa J. L., Castón J. R. (2007) J. Virol. 81, 6869–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fane B. A., Prevelige P. E., Jr. (2003) Adv. Protein Chem. 64, 259–299 [DOI] [PubMed] [Google Scholar]
- 45.Pedersen T., Skjesol A., Jørgensen J. B. (2007) J. Virol. 81, 6652–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.