Abstract
Adiposity is commonly associated with adipose tissue dysfunction and many overnutrition-related metabolic diseases including type 2 diabetes. Much attention has been paid to reducing adiposity as a way to improve adipose tissue function and systemic insulin sensitivity. PFKFB3/iPFK2 is a master regulator of adipocyte nutrient metabolism. Using PFKFB3+/− mice, the present study investigated the role of PFKFB3/iPFK2 in regulating diet-induced adiposity and systemic insulin resistance. On a high-fat diet (HFD), PFKFB3+/− mice gained much less body weight than did wild-type littermates. This was attributed to a smaller increase in adiposity in PFKFB3+/− mice than in wild-type controls. However, HFD-induced systemic insulin resistance was more severe in PFKFB3+/− mice than in wild-type littermates. Compared with wild-type littermates, PFKFB3+/− mice exhibited increased severity of HFD-induced adipose tissue dysfunction, as evidenced by increased adipose tissue lipolysis, inappropriate adipokine expression, and decreased insulin signaling, as well as increased levels of proinflammatory cytokines in both isolated adipose tissue macrophages and adipocytes. In an in vitro system, knockdown of PFKFB3/iPFK2 in 3T3-L1 adipocytes caused a decrease in the rate of glucose incorporation into lipid but an increase in the production of reactive oxygen species. Furthermore, knockdown of PFKFB3/iPFK2 in 3T3-L1 adipocytes inappropriately altered the expression of adipokines, decreased insulin signaling, increased the phosphorylation states of JNK and NFκB p65, and enhanced the production of proinflammatory cytokines. Together, these data suggest that PFKFB3/iPFK2, although contributing to adiposity, protects against diet-induced insulin resistance and adipose tissue inflammatory response.
Keywords: Metabolism, Metabolism/Glucose, Metabolism/Glycolysis, Metabolism/Lipid, Tissue/Organ Systems/Adipocyte, Inflammatory Response, Insulin Resistance
Introduction
As demonstrated by numerous data generated from high fat diet (HFD)3 -fed rodents, overnutrition is associated with adiposity and systemic insulin resistance. For example, feeding an HFD to rats for 10 weeks causes a significant increase in visceral fat mass, which is accompanied by a decrease in systemic insulin sensitivity as evidenced by decreased rate of clamp glucose infusion (1). Similarly, after a feeding of HFD for 20 weeks, both of the two different strains of wild-type mice gain a significant increase in fat mass, which is associated with systemic insulin resistance and glucose intolerance (2). A recent study even indicates that feeding an HFD to mice for only 6 weeks is sufficient to induce adiposity and systemic insulin resistance (3). Furthermore, as documented in mice lacking the myotonic dystrophy protein kinase, a larger augmentation in adiposity is accompanied by a greater increase in the severity of systemic insulin resistance (3). Because of this, adiposity has been generally viewed as an important contributor of systemic insulin resistance (3–7). On the other hand, reducing adiposity has been considered as an effective way to reverse systemic insulin resistance (1, 8). However, adiposity is not necessarily associated with systemic insulin resistance. This is particularly true in genetically modified mice and in mice treated with pharmacological agents (9–13). Following the investigation into the insight of altered systemic insulin sensitivity, it has been suggested that adipose tissue dysfunction is far more important than adiposity in terms of causing systemic insulin resistance (13–16).
Mounting evidence points to a pivotal role for overnutrition-related inflammation in causing adipose tissue dysfunction and thereby systemic insulin resistance. In mice fed an HFD, chronic low grade inflammation in adipose tissue is evident and characterized by an increase in macrophage infiltration and proinflammatory cytokine production (17, 18). This brings about adipose tissue dysfunction, demonstrated by an increase in the production pro-hyperglycemic factors such as free fatty acids and resistin and a decrease in the production of anti-hyperglycemic factors such as adiponectin (4, 15, 19, 20). These changes, along with increased production of proinflammatory cytokines such as tumor necrosis factor α (TNFα) and interleukin 6 (IL-6) from both adipocytes and adipose tissue macrophages, impair insulin signaling in insulin-sensitive tissues including the liver and skeletal muscle, leading to systemic insulin resistance (21–26). In contrast, treatment with thiazolidinediones ameliorates adipose tissue inflammation, which in turn contributes, at least in part, to the reversal of diet-induced adipose tissue dysfunction and systemic insulin resistance (13, 27). For this reason, adipose tissue inflammation is of particular importance to the regulation of systemic insulin sensitivity.
PFKFB3 is the gene that codes for the inducible 6-phosphofructo-2-kinase (iPFK2) that is highly expressed in adipose tissue (28). PFKFB3/iPFK2 generates fructose-2,6-bisphosphate (F26P2), which in turn activates 6-phosphofructo-1-kinase (6PFK1) to enhance glycolysis (29, 30). This effect is involved in adipocyte lipogenesis and triglyceride synthesis (28). However, it is unknown whether the metabolic properties of PFKFB3/iPFK2 are related to the regulation of adipose tissue function, in particular adipose tissue inflammatory response. The present study provides evidence to support a novel role for PFKFB3/iPFK2 in regulating diet-induced systemic insulin resistance and adipose tissue inflammatory response in a manner independent of adiposity.
EXPERIMENTAL PROCEDURES
Animal Experiments
Homozygous disruption of PFKFB3/iPFK2 is embryonic lethal (31). Thus, PFKFB3+/− mice, generated as previously described (31), were used in the present study. Wild-type littermates (C57BL/6J background) were used as the control. All mice were maintained on a 12:12-h light-dark cycle (lights on at 06:00). At the age of 5–6 weeks, mice were fed an HFD (60% fat calories, 20% protein calories, and 20 carbohydrate calories) or low-fat diet (LFD) (10% fat calories, 20% protein calories, and 70 carbohydrate calories) for 12 weeks. Both diets are products of Research Diets, Inc (New Brunswick, NJ) and contain the same of amount of casein, l-cysteine, cellulose, soybean oil, and minerals. However, the HFD contains much more lard and maltodextrin but much less sucrose and no corn starch compared with the LFD. During the 12-week feeding period, body weight, and food intake of the mice were recorded every 4 days. After the feeding regimen, mice were fasted for 4 h before sacrifice for collection of blood and tissue samples (32–34). After anesthesia with pentobarbital (50 mg/kg body weight) via intraperitoneal injection, the abdomen was quickly opened. Epididymal, mesenteric, and perinephric fat depots were dissected and weighed as visceral fat content (33). After weighing, adipose tissue samples were either fixed and embedded for histological and immunohistochemical analyses or frozen in liquid nitrogen and then stored at −80 °C for further analyses. Some mice were fasted similarly and used for insulin and glucose tolerance tests and insulin signaling analyses. All study protocols were reviewed and approved by the Institutional Animal Care and Use Committees of Texas A&M University and the University of Minnesota.
Determination of PFKFB3 mRNA, iPFK2 Amount, and F26P2 Level
PFKFB3 mRNA and iPFK2 amount in the adipose tissue were determined using real-time RT-PCR and Western blot, respectively, as described below. Similarly, the amount of iPFK2 in the brown adipose and liver tissues was determined using Western blot. The levels of F26P2 were measured using the 6PFK1 activation method (32).
Measurement of Adipose Tissue Lipolysis
Assays were conducted in the same ways as described by Berger (35) and Haemmerle (36). Briefly, freshly isolated adipose tissue samples were washed several times with phosphate-buffered saline and incubated in a final volume of 1 ml of high glucose Dulbecco's modified Eagle's medium (DMEM) containing 2% fatty acid-free bovine serum albumin with or without 10 μm isoproterenol at 37 °C for 3 h. Aliquots of the medium were collected hourly to quantify glycerol content using metabolic kits (BioVision, Mountain View, CA). Lipolysis was estimated as the efflux of glycerol.
Insulin and Glucose Tolerance Tests
Mice were fasted for 4 h and received an intraperitoneal injection of insulin (1 unit/kg) or d-glucose (2 g/kg). For insulin tolerance tests, blood samples (5 μl) were collected from the tail vein before and at 15, 30, 45, and 60 min after the bolus insulin injection. Similarly, for glucose tolerance tests, blood samples were collected from the tail vein before and at 30, 60, 90, and 120 min after the glucose bolus injection (34).
Measurement of Plasma Metabolic Parameters
The levels of plasma glucose, triglycerides, and free fatty acids were measured using metabolic assay kits (Sigma and BioVision, Mountain View, CA). The levels of plasma insulin and leptin were measured using ELISA kits (Crystal Chem Inc., Downers Grove, IL).
Histological and Immunohistochemical Analyses of Adipose Tissue
The paraffin-embedded adipose tissue blocks were cut into sections of 5-μm thickness and stained with hematoxylin and eosin. In addition, the sections were stained for the expression of F4/80 with rabbit anti-F4/80 (1:100) (AbD Serotec, Raleigh, NC) as previously described (17, 37). The fraction of F4/80-expressing cells for each sample is calculated as the sum of the number of nuclei of F4/80-expressing cells divided by the total number of nuclei in sections of a sample. Six fields per slide were included, and a total of 4–6 mice per group were used.
Isolation of Stromal Vascular Cells (Macrophages) and Adipocytes from Adipose Tissue
Stromal vascular cells (SVCs) and adipocytes were isolated using the collagenase digestion method as previously described (13). After digestion and centrifugation, the pelleted cells were collected as SVCs, and the floating cells were harvested as adipocytes.
Cell Culture and Treatment
3T3-L1 cells were maintained in high glucose DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin. To differentiate 3T3-L1 cells, the 2-day post-confluent cells were incubated in growth medium supplemented with 10 μg/ml insulin, 1 μm dexamethasone, and 0.5 mm 3-isobutyl-1-methylxanthine for 48 h, followed by incubation for an additional 6–8 days in growth medium supplemented with 10 μg/ml insulin. To knock down PFKFB3/iPFK2, predifferentiated 3T3-L1 cells were transfected with the plasmid containing shRNA against mouse PFKFB3 (iPFK2-KD) (OriGene, Rockville, MD) with LipofectamineTM 2000 Transfection Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Similarly, predifferentiated 3T3-L1 cells were transfected with shRNA vector (iPFK2-Ctrl) and served as the control. After transfection for 24 h, the cells were induced for differentiation for 6–8 days. Thereafter, iPFK2-KD adipocytes and iPFK2-Ctrl adipocytes, as well as un-transfected adipocytes were used for the following assays. Some transfected predifferentiated 3T3-L1 cells were selected with puromycin (5 μg/ml) to establish stable iPFK2-KD and iPFK2-Ctrl cell lines for further analyses.
To verify PFKFB3/iPFK2 knockdown, cell lysates were prepared and used to determine iPFK2 amount using Western blot. To determine the rate of glucose incorporation into lipid, each well (6-well plate) of the cells was incubated with DMEM supplemented with 1 μCi of [U-14C]-glucose for 24 h as previously described (38). After sequential extraction with 30% KOH, 95% ethanol, 9 m H2SO4, and petroleum ether, the amount of 14C lipids was quantified using a Beckman liquid scintillation counter. To quantify adipocyte lipid content, the cells were stained with Oil Red O. The lipid-associated dye was extracted with isopropanol for 15 min. The optical density (OD) of the extraction solution was measured using spectrophotometer at 510 nm (9). To determine the status of adipocyte oxidative stress, the cells were treated with palmitate (250 μm) or vehicle for 24 h and used to measure the production of reactive oxygen species (ROS) using the nitroblue tetrazolium (NBT) assay as previously described (39). Additionally, after differentiation, stable iPFK2-KD and iPFK2-Ctrl adipocytes were treated with or without palmitate (250 μm) for 24 h. In the last hour of treatment, 100 μm etomoxir (an inhibitor of carnitine palmitoyltransferase-1, CPT1) or 10 mm N-acetyl-l-cysteine (an antioxidant) was added, and the production of ROS was determined. To determine adipocyte expression of adipokines and proinflammatory cytokines, the total RNA of the cells was prepared and used for real-time RT-PCR. To determine changes in inflammatory signaling, the cells were treated with or without lipopolysaccharides (LPS, 100 ng/ml) for 1 h prior to harvest. Cell lysates were prepared and used to measure the levels and phosphorylation states of JNK and NFκB p65 using Western blots. To determine changes in insulin signaling, the cells were treated with or without insulin (100 nm) for 30 min prior to harvest. Cell lysates were prepared and used to measure the levels and phosphorylation state of Akt using Western blots.
RNA Isolation, Reverse Transcription, Real-time PCR, and Microarray
The total RNA was isolated from frozen tissue samples and cultured/isolated cells. RNA isolation and real-time RT-PCR were conducted as previously described (34). The mRNA levels were analyzed for PFKFB3, F4/80, TNFα, IL-6, resistin, adiponectin, peroxisome proliferator-activated receptor α (PPARα), PPARγ co-activator 1 (PGC1), and CPT1 in adipose tissue samples and/or cell samples. Additionally, the RNA of adipose tissue samples from HFD-fed PFKFB3+/− mice and wild-type littermates were used for mouse Agilent 44K microarray using methods as previously described (40, 41).
Western Blots
Lysates were prepared from frozen tissue samples and cultured cells. Western blots were conducted as previously described (33, 34). The levels of iPFK2, Akt1/2, phospho-Akt (Ser473), JNK, phospho-JNK, NFκB p65, and phospho-p65 were analyzed.
Statistical Methods
Numeric data are presented as means ± S.E. Statistical significance was assessed by unpaired, two-tailed analysis of variance or Student's t test. Differences were considered significant at the two-tailed p < 0.05.
RESULTS
Disruption of PFKFB3/iPFK2 Decreases iPFK2 Expression and Activity in Adipose Tissue
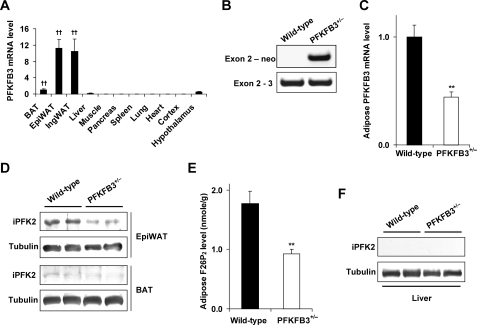
The expression profile of PFKFB3/iPFK2 was determined in various tissues in wild-type mice. Among the key tissues that are involved in the regulation of systemic insulin sensitivity and metabolic homeostasis, PFKFB3/iPFK2 is expressed at high levels in the adipose tissue and expressed at very low levels in the liver and muscle (Fig. 1A). These data suggest that PFKFB3/iPFK2-associated metabolic changes are due primarily to alteration of PFKFB3/iPFK2 in the adipose tissue. In this regard, PFKFB3+/− mice were used to examine changes in PFKFB3 mRNA and iPFK2 amount and activity in the adipose tissue. Disruption of PFKFB3/iPFK2 was confirmed using PCR (Fig. 1B). This resulted in decreased PFKFB3/iPFK2 expression at both the mRNA and protein levels (Fig. 1, C and D). Because PFKFB3/iPFK2 determines the production of F26P2, the levels of adipose tissue F26P2 were quantified as a direct indicator of iPFK2 activity. In PFKFB3+/− mice, the levels of adipose tissue F26P2 were significantly lower than those in wild-type littermates (Fig. 1E). Since the PFKFB3 mRNA was also expressed in both the liver and brown adipose tissue, the amount of iPFK2 in these two tissues was determined. Compared with that in epididymal adipose tissue, the amount of iPFK2 was less abundant in the brown adipose tissue (Fig. 1D). Additionally, the amount of iPFK2 was undetectable in the liver (Fig. 1F).
FIGURE 1.
Disruption of PFKFB3/iPFK2 decreases iPFK2 expression and activity in adipose tissue. A, wild-type C57BL/6J mice were used for analyses. PFKFB3 is abundantly expressed in epididymal (Epi) and inguinal (Ing) white adipose tissue. BAT, brown adipose tissue. Data are means ± S.E., n = 4. ††, p < 0.01 versus liver. B, PCR analyses of mouse genomic DNA using an exon 2-specific primer with a neomycin-specific primer (+/−, heterozygous) or an exon 3-specific primer (+/+, wild-type). For C and E, data are means ± S.E., n = 4 - 6. **, p < 0.01 PFKFB3+/− versus wild-type. C, levels of PFKFB3 mRNA in epididymal adipose tissue were quantified using real-time RT-PCR. D, amount of iPFK2 in both epididymal adipose tissue and brown adipose tissue was measured using Western blot. E, levels of F26P2 in epididymal adipose tissue were determined using the 6PFK1 activation method. F, amount of iPFK2 in the liver was determined using Western blot.
Disruption of PFKFB3/iPFK2 Blunts HFD-induced Adiposity
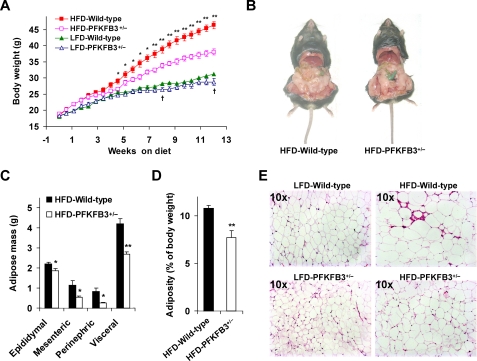
Feeding an HFD to mice induces adiposity (2, 16). To determine the effect of PFKFB3/iPFK2 disruption on diet-induced adiposity, PFKFB3+/− mice and wild-type littermates were started on an HFD. As the control, PFKFB3+/− mice and wild-type littermates were fed a LFD. Following the diet feeding for a period of 12 weeks, PFKFB3+/− mice weighed only slightly lesser than wild-type littermates on a LFD. However, on an HFD, PFKFB3+/− mice exhibited a much smaller gain in body weight than did wild-type littermates (Fig. 2A). This was attributed, at least in part, to a smaller increase in visceral fat content (Fig. 2, B–D). Consistently, the adipocytes in PFKFB3+/− mice were markedly less enlarged than in wild-type littermates in response to HFD feeding as determined by histology (Fig. 2E). To determine the possible contribution of food intake to the difference in body weight and adiposity, food intake of the mice was also monitored. However, there was no difference in food intake (data no shown).
FIGURE 2.
Disruption of PFKFB3/iPFK2 blunts HFD-induced adiposity. At the age of 5–6 weeks, PFKFB3+/− mice and wild-type littermates were fed an HFD or LFD for 12 weeks. For A, C, and D, data are means ± S.E., n = 6. *, p < 0.05 and **, p < 0.01 HFD-PFKFB3+/− versus HFD-wild-type; †, p < 0.05 LFD-PFKFB3+/− versus LFD-wild-type. A, changes in body weight. B–D, changes in visceral fat content. The sum of epididymal, mesenteric, and perinephric fat mass was estimated as visceral fat content. E, adipose tissue histology. The sections of epididymal fat pad were stained with hematoxylin and eosin.
Disruption of PFKFB3/iPFK2 Exacerbates HFD-induced Adipose Tissue Dysfunction and Systemic Insulin Resistance
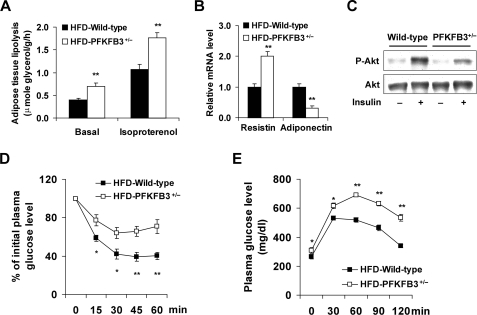
Feeding an HFD to mice leads to adipose tissue dysfunction, which contributes to the development of systemic insulin resistance (42, 43). To determine the effect of PFKFB3/iPFK2 disruption on HFD-induced adipose tissue dysfunction, changes in adipose tissue lipolysis, adipokine expression, and adipose tissue insulin signaling were analyzed. Compared with those in HFD-fed wild-type littermates, the rates of adipose tissue lipolysis were significantly higher in HFD-fed PFKFB3+/− mice under both basal and isoproterenol-stimulated conditions (Fig. 3A). When the expression of adipokines was measured, the mRNA levels of resistin were increased, and the mRNA levels of adiponectin were decreased in the adipose tissue in HFD-fed PFKFB3+/− mice compared with their respective levels in HFD-fed wild-type littermates (Fig. 3B). When insulin signaling was measured, HFD-fed PFKFB3+/− mice exhibited a marked decrease in insulin-stimulated Akt phosphorylation (Fig. 3C), indicating exacerbation of adipose tissue insulin resistance. In combination, these data suggest that disruption of PFKFB3/iPFK2 increases the severity of HFD-induced adipose tissue dysfunction.
FIGURE 3.
Disruption of PFKFB3/iPFK2 exacerbates HFD-induced adipose tissue dysfunction and systemic insulin resistance. At the age of 5–6 weeks, PFKFB3+/− mice and wild-type littermates were fed an HFD for 12 weeks. For A, B, D, and E, data are means ± S.E., n = 6. *, p < 0.05 and **, p < 0.01 HFD-PFKFB3+/− versus HFD-wild-type. A, rates of adipose tissue lipolysis were measured under both basal and isoproterenol-stimulated conditions. B, changes in the mRNA levels of adipose tissue resistin and adiponectin. C, adipose tissue insulin signaling. After anesthesia by an intraperitoneal (i.p.) injection of pentobarbital (50 mg/kg body weight), mice were injected with insulin (1 units/kg) or PBS into the inferior vena cava (i.v.) and epididymal adipose tissue samples were collected 5-min later. For D and E, mice were fasted for 4 h and received an intraperitoneal injection of insulin (1 units/kg) (D) or d-glucose (2 g/kg) (E).
To determine changes in systemic insulin sensitivity, insulin and glucose tolerance tests were conducted. On an HFD, insulin resistance and glucose intolerance were more severe in PFKFB3+/− mice than in wild-type littermates (Fig. 3, D and E), which were correlated with adipose tissue dysfunction well. Additionally, the levels of plasma glucose were significantly higher in PFKFB3+/− mice than in wild-type littermates on both a LFD and HFD (Table 1). Consistent with changes in adiposity, the levels of plasma leptin were lower in PFKFB3+/− mice than in wild-type littermates on an HFD (Table 1). This may also contribute to exacerbation of insulin resistance in PFKFB3+/− mice.
TABLE 1.
Changes in the levels of plasma metabolites and insulin
At the age of 5–6 weeks, PFKFB3+/− mice and wild-type littermates were fed an HFD or LFD for 12 weeks. After the feeding regimen, mice were fasted for 4 h before collection of plasma samples. Data are means ± S.E., n = 6.
| Wild-type | PFKFB3+/− | |
|---|---|---|
| Glucose (mg/dl) | ||
| LFD | 156 ± 16 | 202 ± 11a |
| HFD | 238 ± 6b | 308 ± 8bc |
| Insulin (ng/ml) | ||
| LFD | 2.1 ± 0.3 | 3.2 ± 1.3 |
| HFD | 5.3 ± 1.6d | 8.1 ± 0.8ab |
| Leptin (ng/ml) | ||
| LFD | 4.2 ± 1.3 | 2.0 ± 0.7 |
| HFD | 65.0 ± 6.6b | 43.8 ± 3.7ab |
| Free fatty acids (mm) | ||
| LFD | 0.1 ± 0 | 0.2 ± 0a |
| HFD | 0.2 ± 0.1d | 0.3 ± 0d |
| Triglycerides (mg/dl) | ||
| LFD | 34 ± 1 | 40 ± 3a |
| HFD | 48 ± 4d | 51 ± 2d |
a p < 0.05 PFKFB3+/− vs. wild-type on the same diet.
b p < 0.01 HFD vs. LFD for the same genotype.
c p < 0.01 PFKFB3+/− vs. wild-type on the same diet.
d p < 0.05 HFD vs. LFD for the same genotype.
Disruption of PFKFB3/iPFK2 Exacerbates HFD-induced Adipose Tissue Inflammatory Response and Increases Adipose Expression of Genes Involved in Fatty Acid Oxidation
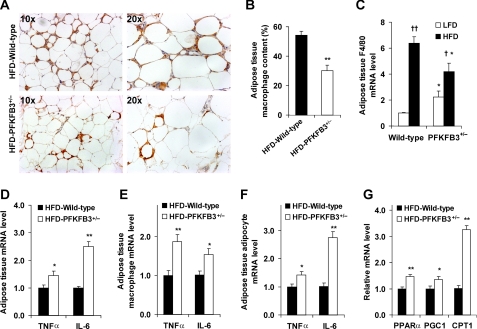
Feeding an HFD to mice induces adipose tissue inflammation, which is characterized by macrophage infiltration and increased proinflammatory cytokine expression (17, 18). These indicators were determined to analyze the effect of PFKFB3/iPFK2 disruption. Compared with LFD-fed mice, HFD-fed PFKFB3+/− mice, and wild-type littermates both revealed macrophage infiltration into the adipose tissue (data of LFD-fed mice not shown). However, PFKFB3+/− mice exhibited a smaller increase in HFD-induced macrophage infiltration in the adipose tissue compared with wild-type littermates (Fig. 4, A and B). Additionally, HFD feeding did not increase adipose tissue mRNA levels of F4/80 in PFKFB3+/− mice as did in wild-type mice (Fig. 4C), which is consistent with changes in macrophage infiltration in the adipose tissue. In contrast, the mRNA levels of TNFα and IL-6 in adipose tissue were higher in HFD-fed PFKFB3+/− mice than in HFD-fed wild-type littermates (Fig. 4D). Furthermore, similar trends were also observed in the mRNA levels of TNFα and IL-6 in macrophages and adipocytes isolated from the adipose tissue (Fig. 4, E and F). These data suggest that disruption of PFKFB3/iPFK2 increases HFD-induced adipose tissue inflammatory response although it causes less macrophage infiltration. Consistent with increased adipose tissue inflammatory response, HFD-fed PFKFB3+/− mice showed a greater increase in the severity of adipose tissue dysfunction (see above). Given that HFD-fed PFKFB3+/− mice exhibited a lesser increase in adiposity than did HFD-fed wild-type littermates, disruption of PFKFB3/iPFK2 appears to increase HFD-induced adipose tissue inflammatory response in a manner independent of adiposity.
FIGURE 4.
Disruption of PFKFB3/iPFK2 exacerbates HFD-induced adipose tissue inflammatory response and increases adipose expression of genes involved in fatty acid oxidation. At the age of 5–6 weeks, PFKFB3+/− mice and wild-type littermates were fed an HFD for 12 weeks. A, macrophage infiltration in adipose tissue. The sections of epididymal fat pad were immunostained for F4/80. For B–G, data are means ± S.E., n = 4 - 6. *, p < 0.05 and **, p < 0.01 HFD-PFKFB3+/− versus HFD-wild-type (in B and D–G) or PFKFB3+/− versus wild-type on the same diet (in C); †, p < 0.05 and ††, p < 0.01 HFD versus LFD for the same genotype (in C). B, fraction of adipose tissue macrophages. C–G, mRNA levels of inflammatory markers and genes involved in fatty acid oxidation were quantified using real-time RT-PCR. C, mRNA levels of F4/80 in adipose tissue. D–F, mRNA levels of TNFα and IL-6 in adipose tissue (D), as well as in macrophages (E) and adipocytes (F) isolated from adipose tissue. G, mRNA levels of genes involved in fatty acid oxidation in adipose tissue.
To explore the role played by PFKFB3/iPFK2 in linking nutrient metabolism and the inflammatory response, the adipose tissue of HFD-fed mice was used for microarray analyses. Compared with wild-type littermates, PFKFB3+/− mice exhibited changes in the expression of 495 genes from a total of 23694 genes (p < 0.01). Of interest, PFKFB3+/− mice showed an increase in the expression of PPARα and PGC1, two master regulators that stimulate fatty acid oxidation (44–47), as well as an increase in the expression of oxidative-stress responsive 1 (Table 2). Using real-time RT-PCR, the increase in the mRNA levels of PPARα and PGC1 was confirmed (Fig. 4G). Furthermore, the mRNA levels of CPT1, a key enzyme that controls the rate-limiting step of fatty acid oxidation (48, 49), was markedly increased in PFKFB3+/− mice (Fig. 4G). These data, in combination with those obtained from iPFK2-KD adipocytes, suggest that PFKFB3/iPFK2 controls the status of adipocyte oxidative stress by regulating the balance between glycolysis and fatty acid oxidation.
TABLE 2.
Selected adipose genes that are altered by disruption of PFKFB3/iPFK2
Adipose mRNA transcriptome was analyzed in HFD-fed PFKFB3+/− mice and wild-type littermates using a microarray approach. Changes in the mRNA levels of adipose genes were expressed as the PFKFB3+/−/wild-type ratios, n = 4.
| Gene symbol | Gene name | Fold of change |
|---|---|---|
| Gcgr | Glucagon receptor | 2.56a |
| Retn | Resistin | 2.15a |
| Ppara | Peroxisome proliferator-activated receptor α | 1.68a |
| Ptprb | Protein-tyrosine phosphatase, receptor type, B | 1.63a |
| Ppargc1a | Peroxisome proliferative-activated receptor, gamma, coactivator 1 α | 1.56a |
| Rbp4 | Retinol-binding protein 4 | 1.54a |
| Ptprm | Protein-tyrosine phosphatase, receptor type, M | 1.52a |
| Oxidative-stress responsive 1 | 1.24a | |
| Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | 0.67a |
| Stat3 | Signal transducer and activator of transcription 3 | 0.66a |
| Il1rn | Interleukin 1 receptor antagonist | 0.65a |
| Srxn1 | Sulfiredoxin 1 homolog (S. cerevisiae) | 0.64a |
| Tnfrsf11a | Tumor necrosis factor receptor superfamily, member 11a | 0.60a |
| Acsl4 | Acyl-CoA synthetase long-chain family member 4 | 0.60a |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 0.44a |
a p < 0.05 PFKFB3+/− versus wild-type.
Knockdown of PFKFB3/iPFK2 Suppresses Adipocyte Lipid Accumulation, Impairs Adipocyte Function, and Increases the Adipocyte Inflammatory Response
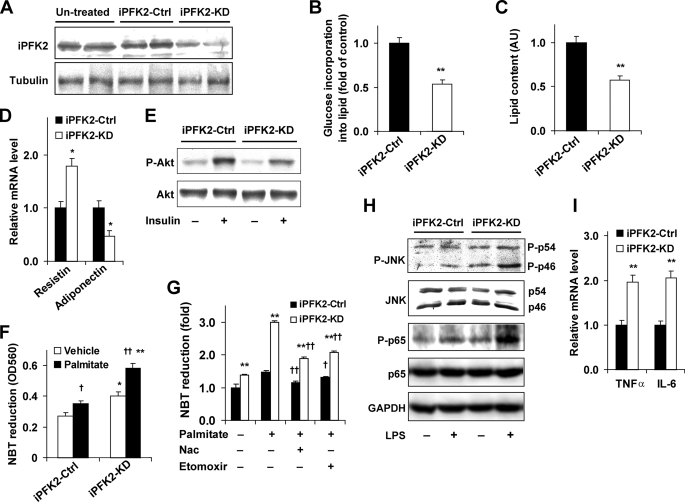
The direct role of PFKFB3/iPFK2 in regulating adipocyte metabolic and inflammatory responses was explored in 3T3-L1 adipocytes that were treated with shRNA against PFKFB3. As expected, knockdown of PFKFB3/iPFK2 was evidenced by a decrease in iPFK2 amount (Fig. 5A). This resulted in a decrease in the incorporation of glucose into lipid (Fig. 5B) and adipocyte lipid accumulation compared with iPFK2-Ctrl adipocytes (Fig. 5C), suggesting that knockdown of PFKFB3/iPFK2 decreases glycolysis and glycolysis-derived lipogenesis and triglyceride synthesis. Additionally, the metabolic changes in iPFK2-KD adipocytes were accompanied by an increase in the mRNA levels of resistin and a decrease in the mRNA levels of adiponectin, as well as a decrease in insulin-stimulated phosphorylation of Akt (Fig. 5, D and E), indicating impaired adipocyte function.
FIGURE 5.
Knockdown of PFKFB3/iPFK2 suppresses adipocyte lipid accumulation, impairs adipocyte function, and increases the adipocyte inflammatory response. Predifferentiated 3T3-L1 cells were transfected with the plasmid containing shRNA against PFKFB3/iPFK2 (iPFK2-KD) or shRNA vector (iPFK2-Ctrl) for 24 h and induced for differentiation for 6–8 days. Untransfected cells were differentiated similarly and used as the untreated control. Some transfected 3T3-L1 cells were selected by puromycin (5 μg/ml) to establish stable iPFK2-KD or iPFK2-Ctrl cell lines. A, Western blot for iPFK2. For B–D, F, G, and I, data are means ± S.E. n = 4. *, p < 0.05 and **, p < 0.01 iPFK2-KD versus iPFK2-Ctrl in B–D, and I, and in F and G for the same treatment (palmitate or vehicle in F and palmitate or vehicle with or without etomoxir or Nac in G); †, p < 0.05 and ††, p < 0.01 palmitate versus vehicle in F and palmitate in the presence of etomoxir or Nac versus palmitate alone in G under the same condition (iPFK2-KD or iPFK2-Ctrl). B, changes in the rates of glucose incorporation into lipid. C, changes in adipocyte lipid accumulation (arbitrary unit). D, changes in the mRNA levels of adipokines were quantified using real-time RT-PCR. E, adipocyte insulin signaling was analyzed using Western blot. Before harvest, the cells were incubated with or without insulin (100 nm) for 30 min. F and G, the production of ROS was measured using the NBT assay. F, after differentiation, iPFK2-KD and iPFK2-Ctrl adipocytes were treated with or without palmitate (250 μm) for 24 h. G, after differentiation, the stable cell lines were treated with or without palmitate (250 μm) for 24 h. In the last hour of treatment, 100 μm etomoxir (an inhibitor of carnitine palmitoyltransferase-1, CPT1) or 10 mm N-acetyl-l-cysteine (an antioxidant) was added. H, adipocytes were incubated with or without LPS (100 ng/ml) for 1 h prior to harvest. The levels and phosphorylation states of JNK and NFκB p65 were determined using Western blots. I, mRNA levels of proinflammatory cytokines were quantified using real-time RT-PCR.
Because the in vivo data suggested a potential role for PFKFB3/iPFK2 in linking fuel metabolism and oxidative stress, the production of ROS in cultured 3T3-L1 cells was determined to further illustrate the underlying mechanisms. Compared with iPFK2-Ctrl adipocytes, iPFK2-KD adipocytes produced much more ROS under both basal and palmitate-stimulated conditions (Fig. 5F), indicating an increase in the status of oxidative stress. To determine the contribution of elevated fatty acid oxidation to the increased production of ROS, the effect of etomoxir on ROS production was examined in the stable iPFK2-KD and iPFK2-Ctrl adipocytes. Treatment with etomoxir lowered down the production of ROS in palmitate-stimulated iPFK2-KD adipocytes to the level comparable with those brought about by treatment with N-acetyl-l-cysteine, a compound that inhibits most general pathways for ROS production (Fig. 5G). These data suggest that elevated fatty acid oxidation is the major cause of the increased production of ROS when PFKFB3/iPFK2 is disrupted. Considering the role of increased oxidative stress in impairing insulin signaling and triggering inflammatory signaling through JNK and/or NFκB pathways (50, 51), changes in the levels and phosphorylation states of JNK and NFκB p65 were determined. Compared with iPFK2-Ctrl adipocytes, iPFK2-KD adipocytes showed an increase in the phosphorylation states of JNK and NFκB p65 under both basal and LPS-stimulated conditions (Fig. 5H). Additionally, the mRNA levels of TNFα and IL-6 were higher in iPFK2-KD adipocytes than in iPFK2-Ctrl adipocytes (Fig. 5I). In combination, these data indicated that the inflammatory response in iPFK2-KD adipocytes was increased compared with iPFK2-Ctrl adipocytes. To determine whether the observed changes in the adipocyte inflammatory response were associated with possible alterations in adipocyte differentiation, the expression of CCAAT/enhancer-binding protein α, PPARγ, and aP2, all of which are determinants of adipocyte differentiation (52), was measured and showed no difference (data not shown). Furthermore, because changes in the metabolic and inflammatory parameters of adipocytes were nearly identical to those observed in PFKFB3+/− mice, disruption of PFKFB3/iPFK2 in adipocytes was likely responsible, to a large extent, for adipose tissue dysfunction in HFD-fed PFKFB3+/− mice.
DISCUSSION
Feeding an HFD to mice induces adiposity, which is associated with adipose tissue dysfunction, a key contributor of systemic insulin resistance (42, 43). This is the case in wild-type littermates. However, in PFKFB3+/− mice, although bringing about a much smaller increase in adiposity, feeding an HFD caused a much greater increase in the severity of HFD-induced adipose tissue dysfunction and systemic insulin resistance than in wild-type littermates. These changes were attributed, at least in part, to the increased adipose tissue inflammatory response, which was evidenced by higher levels of proinflammatory cytokines in both isolated adipose tissue macrophages and adipocytes in PFKFB3+/− mice. Consistently, in cultured adipocytes, knockdown of PFKFB3/iPFK2 caused a decrease in lipid accumulation and an increase in the status of oxidative stress, which were accompanied by enhanced inflammatory signaling, increased mRNA levels of TNFα and IL-6, and decreased insulin signaling. Together, these data argue in favor of a novel and unique role for PFKFB3/iPFK2 in regulating HFD-induced adipose tissue dysfunction and systemic insulin resistance in a manner independent of adiposity.
The unique role of PFKFB3/iPFK2 in dissociating HFD-induced adiposity and adipose tissue dysfunction is attributed, at a large extent, to the metabolic properties of PFKFB3/iPFK2. Notably, PFKFB3/iPFK2 stimulates adipocyte glycolysis (28). An increase in PFKFB3/iPFK2-stimulated glycolysis not only provides lactate and pyruvate (which are converted into acetyl-CoA and used for lipogenesis to provide free fatty acids), but also increases the production of dihydroxyacetone phosphate, which is converted into glycerol-3-phosphate as a required substrate for adipocyte triglyceride synthesis. Upon disruption of PFKFB3/iPFK2, both glycolysis and glycolysis-derived lipogenesis and triglyceride synthesis are impaired, which is supported by the data that PFKFB3/iPFK2-knockdown adipocytes exhibited a decrease in the incorporation of glucose into lipid, and thereby adipocyte lipid accumulation. This contributes to a smaller gain in adiposity in PFKFB3+/− mice after HFD feeding. Importantly, the impairment in using glucose as a fuel due to disruption of PFKFB3/iPFK2 likely causes a compensatory increase in fatty acid oxidation, which is supported by increased expression of CPT1, as well as PPARα and PGC1 in adipose tissue of PFKFB3+/− mice. The compensatory increase in fatty acid oxidation not only contributes to a smaller gain in adiposity in PFKFB3+/− mice after HFD feeding, but more importantly, appears to trigger oxidative stress. In support of this concept, PFKFB3/iPFK2-knockdown adipocytes exhibited an increase in the production of ROS under both basal and palmitate-stimulated conditions. Furthermore, inhibition of fatty acid oxidation by etomoxir caused a marked decrease in palmitate-stimulated production of ROS in PFKFB3/iPFK2-knockdown adipocytes. In agreement with the role oxidative stress in disturbing adipokine expression and initiating the inflammatory response in adipocytes (15), the increased production of ROS in PFKFB3/iPFK2-knockdown adipocytes was associated, on the one hand, with inappropriately altered expression of resistin and adiponectin, and on the other hand, with increased phosphorylation states of JNK and NFκB p65 and increased mRNA levels of TNFα and IL-6 in PFKFB3/iPFK2-knockdown adipocytes. Apparently, PFKFB3/iPFK2 links nutrient metabolism and adipocyte function.
The role of adipose tissue dysfunction in causing systemic insulin resistance has been well documented (17, 18). Indeed, this role is illustrated by at least two plausible mechanisms (4, 15, 19, 20). In the first mechanism, adipose tissue dysfunction causes an increase in the production of free fatty acids, resistin, and retinol-binding protein 4 (RBP4) and a decrease in the production of adiponectin. These factors are carried to the insulin-sensitive tissues including the liver and skeletal muscle through circulation to impair insulin signaling and ultimately bring about systemic insulin resistance (24–26, 53). In the second mechanism, adipose tissue-derived proinflammatory cytokines are similarly carried to insulin-sensitive tissues through circulation to directly impair insulin signaling in the tissues (22, 54, 55). In the present study, PFKFB3+/− mice exhibited an increase in the severity of HFD-induced systemic insulin resistance and glucose intolerance. This was attributed, at least in part, to increased adipose tissue dysfunction, as evidenced by increased adipose tissue lipolysis, inappropriate adipokine expression, and decreased insulin signaling, as well as increased expression of proinflammatory cytokines including TNFα and IL-6 in the adipose tissue. These data support a pivotal role for PFKFB3/iPFK2 in protecting against HFD-induced adipose tissue dysfunction, and thereby systemic insulin resistance. Moreover, because PFKFB3/iPFK2 disruption-associated changes in adipose tissue dysfunction were nearly identical to those in PFKFB3/iPFK2-knockdown adipocytes, PFKFB3/iPFK2 in adipocytes is thereby responsible largely for the regulation of adipose tissue function. It should be pointed out that PFKFB3/iPFK2 is also expressed in tissues other than adipose tissue (31). Thus, a possible role for PFKFB3/iPFK2 in non-adipose tissue in regulating systemic insulin sensitivity cannot be ruled out. However, considering the profile of PFKFB3/iPFK2 expression, the role of PFKFB3/iPFK2 in non-adipose tissue would not be as important as that in adipose tissue. Additionally, in terms of regulating systemic glucose homeostasis, dysregulated liver glucose metabolism is thought to contribute to hyperglycemia and glucose intolerance (32–34). Given that the amount of iPFK2 was undetectable in the liver, dysregulated liver glucose metabolism, if existed in PFKFB3+/− mice, was likely due to a secondary effect of PFKFB3 disruption in extrahepatic tissues, in particular adipose tissue.
It is also a novel finding that disruption of PFKFB3/iPFK2 exacerbated HFD-induced adipose tissue inflammatory response, which was also independent of adiposity. Additionally, the increased adipose tissue inflammatory response was independent of macrophage accumulation in adipose tissue. In a generally accepted concept, HFD-induced adipose tissue inflammatory is characterized by increased production of proinflammatory cytokines, which is positively correlated with macrophage infiltration (17, 18). However, several lines of new evidence suggest that the inflammatory status of macrophages is more important than the number of macrophages in terms of controlling the production of proinflammatory cytokines in the adipose tissue. For example, mice that lack PPARγ in macrophages exhibit fewer macrophages in adipose tissue but have higher mRNA levels of IL-6 than wild-type control mice (56). In contrast, treatment with rosiglitazone, a PPARγ agonist, increases the abundance of macrophages in adipose tissue, but decreases the production of proinflammatory cytokines such as IL-18 (13). Considering this, disruption of PFKFB3/iPFK2 appears to increase the inflammatory activity of the infiltrated macrophages. This notion is indeed supported by the data that the mRNA levels of TNFα and IL-6 were higher in macrophages isolated from PFKFB3+/− mice than in wild-type littermates. Therefore, disruption of PFKFB3/iPFK2 exacerbates HFD-induced adipose tissue inflammatory response in a manner depending on increasing macrophage inflammatory status rather than macrophage infiltration. However, further study is required to elucidate the underlying mechanisms by which disruption of PFKFB3/iPFK2 blunts HFD-induced macrophage infiltration into adipose tissue.
In summary, the present study demonstrates a novel and unique role for PFKFB3/iPFK2 in regulating adiposity and adipose tissue function, and thereby systemic insulin sensitivity. This role is manifested by the fact that disruption of PFKFB3/iPFK2 ameliorates HFD-induced adiposity, but exacerbates HFD-induced adipose tissue dysfunction, in particular, adipose tissue inflammatory response, which contributes to an increase in the severity of systemic insulin resistance. Because of this, a potential risk of inducing systemic insulin resistance and enhancing adipose tissue inflammatory response should be taken into account when inhibition of PFKFB3/iPFK2 is considered as therapeutic approach.
Acknowledgment
We thank Dr. Alice Villalobos for assistance with image analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants HL78679 and HL080569 (to Y. H.). This work was also supported by AHA Grant 0430151N (to Y. H.) and the William W. Allen Foundation (to C. W. and J. L.).
- HFD
- high fat diet
- iPFK2
- inducible 6-phosphofructo-2-kinase
- F26P2
- fructose-2,6-bisphosphate
- 6PFK1
- 6-phosphofructo-1-kinase
- PPARα
- peroxisome proliferator-activated receptor α
- PGC1
- peroxisome proliferative-activated receptor γ, coactivator 1
- CPT1
- carnitine palmitoyltransferase-1
- JNK
- c-Jun N-terminal kinase
- NFκB
- nuclear factor κB
- TNFα
- tumor necrosis factor α
- IL
- interleukin
- DMEM
- Dulbecco's modified Eagle's medium
- ROS
- reactive oxygen species
- LFD
- low fat diet.
REFERENCES
- 1.Kusunoki M., Hara T., Tsutsumi K., Nakamura T., Miyata T., Sakakibara F., Sakamoto S., Ogawa H., Nakaya Y., Storlien L. H. (2000) Diabetologia 43, 875–880 [DOI] [PubMed] [Google Scholar]
- 2.Gallou-Kabani C., Vigé A., Gross M. S., Rabès J. P., Boileau C., Larue-Achagiotis C., Tomé D., Jais J. P., Junien C. (2007) Obesity 15, 1996–2005 [DOI] [PubMed] [Google Scholar]
- 3.Llagostera E., Carmona M. C., Vicente M., Escorihuela R. M., Kaliman P. (2009) FEBS Letters 583, 2121–2125 [DOI] [PubMed] [Google Scholar]
- 4.Kahn B. B., Flier J. S. (2000) J. Clin. Invest. 106, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki Y., Glass L., Triplitt C., Wajcberg E., Mandarino L. J., DeFronzo R. A. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E1135–E1143 [DOI] [PubMed] [Google Scholar]
- 6.Ghibaudi L., Cook J., Farley C., van Heek M., Hwa J. J. (2002) Obesity 10, 956–963 [DOI] [PubMed] [Google Scholar]
- 7.Krekoukia M., Nassis G. P., Psarra G., Skenderi K., Chrousos G. P., Sidossis L. S. (2007) Metabolism 56, 206–213 [DOI] [PubMed] [Google Scholar]
- 8.Air E. L., Strowski M. Z., Benoit S. C., Conarello S. L., Salituro G. M., Guan X. M., Liu K., Woods S. C., Zhang B. B. (2002) Nat. Med. 8, 179–183 [DOI] [PubMed] [Google Scholar]
- 9.Zhang J., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Song Q., Chen Y. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10703–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Combs T. P., Pajvani U. B., Berg A. H., Lin Y., Jelicks L. A., Laplante M., Nawrocki A. R., Rajala M. W., Parlow A. F., Cheeseboro L., Ding Y. Y., Russell R. G., Lindemann D., Hartley A., Baker G. R., Obici S., Deshaies Y., Ludgate M., Rossetti L., Scherer P. E. (2004) Endocrinology 145, 367–383 [DOI] [PubMed] [Google Scholar]
- 11.Muurling M., Mensink R. P., Pijl H., Romijn J. A., Havekes L. M., Voshol P. J. (2003) Metabolism 52, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 12.Gavrilova O., Haluzik M., Matsusue K., Cutson J. J., Johnson L., Dietz K. R., Nicol C. J., Vinson C., Gonzalez F. J., Reitman M. L. (2003) J. Biol. Chem. 278, 34268–34276 [DOI] [PubMed] [Google Scholar]
- 13.Stienstra R., Duval C., Keshtkar S., van der Laak J., Kersten S., Müller M. (2008) J. Biol. Chem. 283, 22620–22627 [DOI] [PubMed] [Google Scholar]
- 14.Evans R. M., Barish G. D., Wang Y. X. (2004) Nat. Med. 10, 355–361 [DOI] [PubMed] [Google Scholar]
- 15.Qatanani M., Lazar M. A. (2007) Genes Dev. 21, 1443–1455 [DOI] [PubMed] [Google Scholar]
- 16.Bullen J. W., Jr., Bluher S., Kelesidis T., Mantzoros C. S. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1079–E1086 [DOI] [PubMed] [Google Scholar]
- 17.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr. (2003) J. Clin. Invest. 112, 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lumeng C. N., DeYoung S. M., Bodzin J. L., Saltiel A. R. (2007) Diabetes 56, 16–23 [DOI] [PubMed] [Google Scholar]
- 19.Rosen E. D., Spiegelman B. M. (2006) Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badman M. K., Flier J. S. (2007) Gastroenterology 132, 2103–2115 [DOI] [PubMed] [Google Scholar]
- 21.Hotamisligil G. S., Peraldi P., Budavari A., Ellis R., White M. F., Spiegelman B. M. (1996) Science 271, 665–668 [DOI] [PubMed] [Google Scholar]
- 22.Cheung A. T., Ree D., Kolls J. K., Fuselier J., Coy D. H., Bryer-Ash M. (1998) Endocrinology 139, 4928–4935 [DOI] [PubMed] [Google Scholar]
- 23.Cheung A. T., Wang J., Ree D., Kolls J. K., Bryer-Ash M. (2000) Diabetes 49, 810–819 [DOI] [PubMed] [Google Scholar]
- 24.Boden G., Cheung P., Stein T. P., Kresge K., Mozzoli M. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E12–E19 [DOI] [PubMed] [Google Scholar]
- 25.Berg A. H., Combs T. P., Du X., Brownlee M., Scherer P. E. (2001) Nat. Med. 7, 947–953 [DOI] [PubMed] [Google Scholar]
- 26.Kabir M., Catalano K. J., Ananthnarayan S., Kim S. P., Van Citters G. W., Dea M. K., Bergman R. N. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E454–E461 [DOI] [PubMed] [Google Scholar]
- 27.Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atsumi T., Nishio T., Niwa H., Takeuchi J., Bando H., Shimizu C., Yoshioka N., Bucala R., Koike T. (2005) Diabetes 54, 3349–3357 [DOI] [PubMed] [Google Scholar]
- 29.Okar D. A., Wu C., Lange A. J. (2004) in Advances in Enzyme Regulation Vol. 44, pp. 123–154, Elsevier, New York: [DOI] [PubMed] [Google Scholar]
- 30.Rider M. H., Bertrand L., Vertommen D., Michels P. A., Rousseau G. G., Hue L. (2004) Biochem. J. 381, 561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesney J., Telang S., Yalcin A., Clem A., Wallis N., Bucala R. (2005) Biochem. Biophys. Res. Commun. 331, 139–146 [DOI] [PubMed] [Google Scholar]
- 32.Wu C., Okar D. A., Newgard C. B., Lange A. J. (2001) J. Clin. Invest. 107, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Kang J. E., Peng L., Li H., Khan S. A., Hillard C. J., Okar D. A., Lange A. J. (2005) Cell Metabolism 2, 131–140 [DOI] [PubMed] [Google Scholar]
- 34.Wu C., Khan S. A., Peng L. J., Li H., Camella S. G., Lange A. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E536–E543 [DOI] [PubMed] [Google Scholar]
- 35.Berger J. J., Barnard R. J. (1999) J. Appl. Physiol. 87, 227–232 [DOI] [PubMed] [Google Scholar]
- 36.Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. (2002) J. Biol. Chem. 277, 4806–4815 [DOI] [PubMed] [Google Scholar]
- 37.Wang H., Zhang W., Zhu C., Bucher C., Blazar B. R., Zhang C., Chen J. F., Linden J., Wu C., Huo Y. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stansbie D., Brownsey R. W., Crettaz M., Denton R. M. (1976) Biochem. J. 160, 413–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subauste A. R., Burant C. F. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E159–E164 [DOI] [PubMed] [Google Scholar]
- 40.Li X., Chiang H. I., Zhu J., Dowd S. E., Zhou H. (2008) BMC Genomics 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang H. I., Swaggerty C. L., Kogut M. H., Dowd S. E., Li X., Pevzner I. Y., Zhou H. (2008) BMC Genomics 9, 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haugen F., Drevon C. A. (2007) Proc. Nutr. Soc. 66, 171–182 [DOI] [PubMed] [Google Scholar]
- 43.Morin C. L., Eckel R. H., Marcel T., Pagliassotti M. J. (1997) Endocrinology 138, 4665–4671 [DOI] [PubMed] [Google Scholar]
- 44.Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. (1998) J. Biol. Chem. 273, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 45.Li P., Zhu Z., Lu Y., Granneman J. G. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E617–E626 [DOI] [PubMed] [Google Scholar]
- 46.Puigserver P., Spiegelman B. M. (2003) Endocr. Rev. 24, 78–90 [DOI] [PubMed] [Google Scholar]
- 47.Benton C. R., Wright D. C., Bonen A. (2008) Appl. Physiol. Nutr. Metab. 33, 843–862 [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen B. B., Holmbäck U. C., Volpi E., Morio-Liondore B., Paddon-Jones D., Wolfe R. R. (2002) J. Clin. Invest. 110, 1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mercader J., Ribot J., Murano I., Felipe F., Cinti S., Bonet M. L., Palou A. (2006) Endocrinology 147, 5325–5332 [DOI] [PubMed] [Google Scholar]
- 50.Nakamura S., Takamura T., Matsuzawa-Nagata N., Takayama H., Misu H., Noda H., Nabemoto S., Kurita S., Ota T., Ando H., Miyamoto K., Kaneko S. (2009) J. Biol. Chem. 284, 14809–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans J. L., Goldfine I. D., Maddux B. A., Grodsky G. M. (2002) Endocr. Rev. 23, 599–622 [DOI] [PubMed] [Google Scholar]
- 52.Farmer S. R. (2006) Cell Metabolism 4, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao-Borengasser A., Varma V., Bodles A. M., Rasouli N., Phanavanh B., Lee M. J., Starks T., Kern L. M., Spencer H. J., 3rd, Rashidi A. A., McGehee R. E., Jr., Fried S. K., Kern P. A. (2007) J. Clin. Endocrinol. Metab. 92, 2590–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002) Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- 55.Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M. J., Ye J. (2002) J. Biol. Chem. 277, 48115–48121 [DOI] [PubMed] [Google Scholar]
- 56.Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]