Abstract
The transforming growth factor-β (TGF-β) maintains epithelial homeostasis and suppresses early tumor formation, but paradoxically at later stages of tumor progression, TGF-β promotes malignancy. TGF-β activates phosphorylation of Smad2 and -3 effectors. Smad2 and -3 are known to have different functions, but differential regulation of their phosphorylation has not been described. Here we show that upon hypoxia, the TGF-β-induced phosphorylation of Smad3 was inhibited, although Smad2 remained phosphorylated. The inhibition of Smad3 phosphorylation was not due to TGF-β receptor inactivation. We show that Smad3 was dephosphorylated by PP2A (protein phosphatase 2A) specifically under hypoxic conditions. The hypoxic Smad3 dephosphorylation required intact expression of the essential scaffold component PR65 of PP2A. PP2A physically interacted with Smad3 that occurred only in hypoxia. Accordingly, Smad3-associated PP2A activity was found under hypoxic conditions. Hypoxia attenuated the nuclear accumulation of TGF-β-induced Smad3 but did not affect Smad2. Moreover, the influence of TGF-β on a set of Smad3-activated genes was attenuated by hypoxia, and this was reversed by chemical PP2A inhibition. Our data demonstrate the existence of a Smad3-specific phosphatase and identify a novel role for PP2A. Moreover, our data implicate a novel mechanism by which hypoxia regulates growth factor responses.
Keywords: Cancer, Oxygen/Hypoxia, Phosphorylation/Phosphatases/Serine-Threonine, Protein/Protein-Protein Interactions, Signal Transduction/Phosphoprotein Phosphatases/PP1/PP2A, Transcription/Smad, HIF-1, Metastasis
Introduction
TGF-β2 is a multifunctional growth factor that is required for normal embryonic development, is involved in immunological responses, and maintains epithelial homeostasis as well as restricts the growth of normal cells and limits tumor formation (1, 2). However, at later stages of cancer development, cells become resistant to the growth inhibitory properties of TGF-β and begin to exploit it as a growth-promoting cytokine (1, 3, 4). TGF-β operates by activating TGF-β receptor complexes on the cell surface. TGF-β brings type 1 receptor (TGF-βRI, i.e. ALK5) to close proximity of the type 2 receptor that activates the type 1 receptor, subsequently leading to the activation of R-Smads (Smad2 and -3) by phosphorylation of serine residues at the C-terminal ends (5, 6). R-Smads bind to a common Smad4 mediator, are accumulated in the nucleus, and activate transcriptional responses (5, 7, 8).
The TGF-β signaling through Smad2/3 is antagonized by several mechanisms, such as the activation of inhibitory Smads (I-Smads, Smad6 and -7) and dephosphorylation of R-Smads and TGF-β receptor. I-Smads operate by preventing R-Smad binding to the activated receptor, by competing with R-Smad binding to Smad4, and by inducing internalization and proteosomal degradation of TGF-βRI resulting in the attenuation of TGF-β signaling (7–9). Smad7 has also been shown to mediate PP1C (protein phosphatase 1C) binding to the activated TGF-βRI and subsequent inactivation of the receptor by dephosphorylation (10). Independently from I-Smads, PP2A can inhibit the TGF-β superfamily receptors and BMP R-Smad (11, 12). Moreover, Smad2 and -3 activity may be attenuated by a negative feedback mechanism independently from the TGF-βRI activity through dephosphorylation of the C-terminal serines by the PPM1A (PP2C) phosphatase (13). The PPM1A phosphatase is a general inhibitor of TGF-β and BMP signaling. It may inhibit the activity of several Smad proteins either by dephosphorylation (Smad1–3) (13, 14) or by regulating their degradation (Smad1, -5, and -8) (15). Noticeably, in each case the attenuation of TGF-βRI signaling results in the attenuation of both Smad2 and -3 phosphorylation. PPM1A belongs to a different class of phosphatases than PP1C and PP2A. PPM1A is a monomer, and PP1C and PP2A multimers. PP2A (protein phosphatase 2A) belongs to the same conserved gene family as PP1C, but it has distinct substrate specificity and expression pattern and is differentially inhibited by phosphatase inhibitors such as okadaic acid (16, 17). Their requirement on divalent cations differs (17). The substrate specificity of PP2A multimer complex depends on regulatory β-subunits. PP2A may form ∼70 different forms and thereby may differentially target a number of substrates. Accordingly, PP2A has been reported to exert different activity toward the TGF-β superfamily receptors (11).
The lack of sufficient tissue oxygenation, i.e. hypoxia, occurs during development and is a common feature of ischemic diseases and tumors. The best characterized mechanisms by which hypoxia signals are mediated are through the hypoxia-inducible factor (HIF). HIF is stabilized upon hypoxia and activates the transcription of a wide range of genes required to counteract the reduced oxygen availability (18–20). Solid tumors contain hypoxic regions because of aberrant or limited amounts of vasculature. Hypoxia in tumors occurs at later stages of tumor growth, at the time when the tumors reach the size of ∼1 mm. Similarly to TGF-β, hypoxia acts as a progression factor in carcinomas, and the occurrence of hypoxia coincides with the conversion of the TGF-β response (1). Cooperation between hypoxia and TGF-β signaling has been reported to occur at least by two mechanisms. Hypoxia activates the expression of TGF-β1 (21). Moreover, TGF-β has been reported to cooperate with hypoxia, and hypoxic signaling is affected by Smad3, which binds the α-subunit of the HIF complex and thereby enhances the hypoxic gene expression (22–24). However, whether hypoxia may modulate TGF-β signaling remains under investigation. Here, we show that hypoxia leads to imbalance in TGF-β response as hypoxia selectively blocks the TGF-β-induced phosphorylation of Smad3 without influencing Smad2 phosphorylation. This is due to dephosphorylation of Smad3 by protein phosphatase 2A selectively under hypoxia.
MATERIALS AND METHODS
Cell Culture and Transfections
Cells were routinely cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum, penicillin, streptomycin, and l-glutamine. Transfections were performed with Effectene (Qiagen) or FuGENE HD (Roche Applied Science) according to the manufacturer's protocol. Hypoxic treatments were performed in Invivo2 400 incubator (Ruskinn Technologies, UK) in 1% O2, 5% CO2, 90% moisture. Oxygen was replaced with 99.5% pure N2 (AGA, Finland). For growth factor treatment, cells were either serum-starved or grown in minimal serum (CMS: fetal bovine serum passed through 2% charcoal/dextran) (Hyclone) for 24 h prior to addition of TGF-β1 or BMP2 (Promocell) at 5 ng/ml. Okadaic acid (Sigma) or Fostriecin (Sigma) was added 1 h prior to TGF-β1 or in experiments using constitutively active TGF-βRI 90 min prior to sample collection. For siRNA experiments, double-stranded siRNA oligonucleotides (siPR65 (number 1), 5′-CCACCAAGCACAUGCUACC(dTdT)-3′; siPR65 (number 2), 5′-GGCUGAACAUCAUCUCUAA(dTdT)-3′; PPM1A, 5′-GCGUGAUUUCAAACCAUAA(dTdT)-3′, and nontarget 5′-CCUACAUCCCGAUCGAUGAUG (dTdT)-3′) were used at 200 nm final concentration on 12-well cell culture plates. Transfections were performed with Oligofectamine (Invitrogen) following the manufacturer's protocol. TGF-βR1 inhibitor SB431542 (Sigma) was used at 5 μm. For wounding assay, near-confluent HaCaT cell layer was serum-starved for 6 h and wounded with a pipette tip.
Detection of Proteins, Antibodies, and Affinity Purification
For protein analysis, cells were lysed and samples run on SDS-PAGE in a mini-gel chamber or Criterion precast gels (Bio-Rad) and transferred onto polyvinylidene difluoride membrane (Millipore). Western blot analyses were performed in 5% bovine serum albumin in Tris-buffered saline with 0.1% Tween 20 antibody using the following antibodies: rabbit polyclonal t-Smad2 (Zymed Laboratories Inc. and Cell Signaling) at 1:2000; t-Smad3 (Zymed Laboratories Inc. and Cell Signaling) at 1:2000; PR65 (Santa Cruz Biotechnology) at 1:2000; P-Smad3 (Cell Signaling Technology) at 1:2500 for Western blot; at 1:600 for immunohistochemistry; another P-Smad3 antibody (a kind gift from Dr. Moustakas) (48) at 1:2500; P-Smad2 (Cell Signaling Technology) (48) at 1:2500; rat anti-HA (Roche Applied Science) at 1:2500; mouse anti-Strep-tactin (IBA) at 1:2500; PPM1A (Abcam) at 1:2000; and mouse monoclonal AC-74 actin antibody (Sigma) at 1:5000 dilution. All primary antibody treatments were done overnight at 4 °C followed by secondary antibody treatment for 2 h at room temperature. Anti-mouse horseradish peroxidase, anti-rat horseradish peroxidase, and anti-rabbit horseradish peroxidase antibodies (DAKO) were used at 1:10,000 dilutions. Proteins were detected with enhanced chemiluminescence reagent (Amersham Biosciences). As a secondary antibody, either Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch) or Alexa Fluor488-labeled donkey anti-rabbit (Invitrogen) was used. Hoechst 33342 (Invitrogen) was used for staining the nuclei. Visualization was performed with LSM 510 confocal microscope or Lumar stereo microscope (Zeiss). For quantifying the P-Smad2/3 expression at the wound edges, an open source image analysis software (ImageJ, rsb.info.nih.gov) was used.
The Strep-tactin III affinity purification methods have been described in detail elsewhere (32). Strep-tactin III-tagged bait protein transfections were performed in HeLa cells on 10-cm dishes followed by 16 h of hypoxia and 30 min of TGF-β1 exposure. Cells were lysed in 1 ml by drawing through an 18-gauge needle. 100 μl for interaction studies and 50 μl for phosphatase assay of cell lysate was used for input, and the rest was transferred into 200 μl of Strep-tactin purification column. After washes, the samples were eluted in six 100-μl fractions. Fractions were analyzed separately, or all fractions were combined and concentrated to 100 μl with Microcon YM-30 (Millipore) filtration columns.
For mRNA expression analysis, samples were lysed according to the RNeasy mini kit protocol (Qiagen). RNA purity and concentrations were measured with a NANOdrop UV-visible spectrophotometer. For Q-RT-PCR (TaqMan), cDNA was prepared from 0.5 μg of total RNA using 100 units of Moloney murine mammary tumor virus reverse transcriptase (H−) (Promega) and NTP mixture (Fermentas) at 0.5 mm final concentration and analyzed according to the manufacturer's protocol in optical 96-well plates (Applied Biosystems).
Luciferase Assay
For reporter gene assay, HaCaT cells stably transfected with Smad3-specific luciferase reporter gene (CAGA12-Luc) and a constantly active TK-Renilla gene were used. The assay was performed with Dual-Luciferase® reporter assay system (E1910, Promega) according to the manufacturer's protocol and measured with VICTORTM 2 plate reader (PerkinElmer Life Sciences). The cells were lysed passively into 20 μl of lysis buffer, and luminescence from each well was measured for 8 s. The luciferase activity was normalized to Renilla activity.
PP2A Phosphatase Assays
HeLa cells were transfected with the indicated Strep-tactin III fusion proteins on 10-cm dishes. Cells were grown in minimal serum, and hypoxic treatment was started 8 h after serum starvation. After 16 h of hypoxia, TGF-β1 was applied to the cells for 30 min. Cells were lysed, passed through purification columns, and washed twice. The second wash was performed with PP2A washing buffer supplemented with 125 μg/ml bovine serum albumin. Half of the samples were used for direct PP2A activity measurement, and half of the samples were exposed to okadaic acid 30 min before the phosphatase activity assay. Assay was performed with RediPlateTM 96 EnzChek® serine-threonine phosphatase assay kit (R33700, Invitrogen) according to manufacturer's protocol and measured with Envision plate reader (PerkinElmer Life Sciences).
In vitro dephosphorylation of Smad3 was performed by transfecting HeLa cells separately with Strep-tactin III-Smad3 or PR65-Strep-tactin III fusion proteins on 10-cm dishes. PR65- Strep-tactin III-transfected cells were treated either in normoxia or 16 h in 1% O2 prior to purification. Strep-tactin III-Smad3-transfected cells were kept in normoxic conditions and treated with TGF-β 45 min prior to purification. Samples were purified with affinity chromatography and washed with PP2A reaction buffer without bovine serum albumin (adapted from RediPlateTM 96 EnzChek® serine-threonine phosphatase assay kit (R33700, Invitrogen)) and concentrated to a 110-μl volume. Purified PP2A samples were split in half, and other half was treated with 20 nm okadaic acid for 15 min prior to dephosphorylation reaction. Strep-tactin III-Smad3 was combined with equal amounts of PP2A samples or pure reaction buffer. Reactions were incubated in 37 °C for 1 h. Phosphatase reaction was stopped by adding SDS sample buffer, and results were analyzed by Western blotting.
RESULTS
Hypoxia Inhibits Smad3, but Not Smad2, Phosphorylation
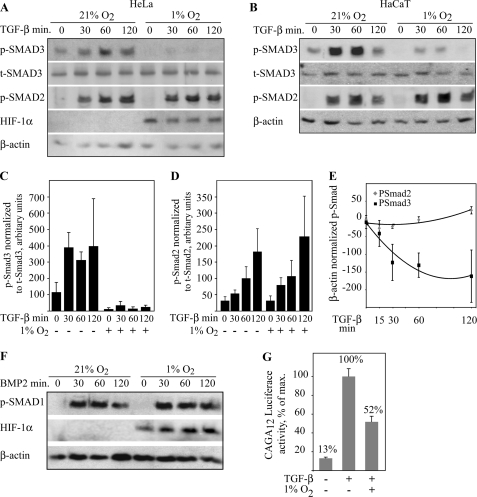
To investigate the effect of hypoxia on the activity of the TGF-β pathway, phosphorylation of the C-terminal serines of Smad2 and -3 were analyzed by phospho-specific antibodies in HeLa cells. The cells were pre-exposed to either normoxia (21% O2) or 16-h hypoxia (1% O2) followed by TGF-β stimulus (5 ng/ml) for 30–120 min at corresponding oxygen tension. In the normoxic HeLa cells, Smad2 and -3 phosphorylation was activated by TGF-β as expected, peaking approximately at 60 min. In the cells exposed to hypoxia, the TGF-β-induced Smad3 phosphorylation was abolished or markedly decreased at all time points up to 2 h (Fig. 1A). Similar results were obtained with two independent phospho-Smad3 antibodies (not shown). Surprisingly, however, hypoxia had no effect on the TGF-β-induced Smad2 phosphorylation. Unlike the phosphorylated Smad3, the total Smad3 levels remained unchanged regardless of the hypoxic exposure. Similarly to HeLa cells, the TGF-β-elicited phosphorylation of Smad3 in immortalized HaCaT cells was inhibited by hypoxia, but hypoxia had no effect on Smad2 phosphorylation (Fig. 1B). Quantification of the data in HeLa cells demonstrated strong reduction in the phosphorylated Smad3 level by hypoxia as compared with total Smad3 level (Fig. 1C). In contrast, Smad2 phosphorylation was not reduced by hypoxia (Fig. 1D). Comparison of the phosphorylated Smad3 and Smad2 levels illustrated hypoxic reduction only on Smad3 that persisted up to 2 h (Fig. 1E). Similarly to hypoxia, a chemical hypoxia mimetic CoCl2 abolished the TGF-β-induced phosphorylation of Smad3 (data not shown).
FIGURE 1.
Smad3 phosphorylation is inhibited in hypoxia but other R-Smads are not affected. HeLa (A) and HaCaT cells (B) were exposed to either ambient oxygen (21% O2) or hypoxia (1% O2) for 16 h followed by TGF-β treatment and Western blot analysis using phospho-specific Smad (p-Smad) and total Smad3 (t-Smad3) antibodies. TGF-β treatment was performed in the indicated oxygen tension. Both cell lines show TGF-β-activated Smad2 and -3 phosphorylation (0 versus 30–60 min). In hypoxia, Smad3 phosphorylation is reduced, but Smad2 phosphorylation remains unaffected. Hypoxia does not affect the t-Smad3 levels. C, quantification of p-Smad3 normalized to t-Smad3 level in HeLa cells upon normoxia (−) and hypoxia (+) at the indicated times of TGF-β stimulus. D, quantification of p-Smad2 normalized to t-Smad2 level in HeLa cells upon normoxia and hypoxia. E, comparison of hypoxic Smad2 and -3 phosphorylation at indicated time points of TGF-β stimulation in HaCaT cells. For each data point, the normoxic value was subtracted from the corresponding hypoxic value. The means ± S.E. from three independent experiments are shown. F, BMP-2 induced Smad1 phosphorylation is not affected by hypoxia. HaCaT cells were exposed to hypoxia followed by BMP-2 stimulus and Western blot analysis for phospho-Smad1 expression. G, HaCaT cells stably transfected with Smad3-specific luciferase reporter gene (CAGA12-Luc) were exposed to either ambient oxygen or hypoxia for 16 h followed by 3-h TGF-β treatment. Luciferase activity is shown as fold change from the maximal activity. At least four independent experiments ± S.E. were performed.
By virtue of the high homology between Smad1 and -3 at the C-terminal ends, the phospho-Smad3 antibodies recognize phosphorylated Smad1 as well. We therefore exposed cells to BMP2 (bone morphogenetic protein 2), which is a well characterized inducer of Smad1 phosphorylation (25). BMP2 induced robust phosphorylation of the protein recognized by the phospho-Smad1/3 antibody, but the induction was not altered by hypoxia (Fig. 1F). The data indicated that hypoxia specifically blocks the Smad3 phosphorylation induced by TGF-β, but it leaves other R-Smad phosphorylation unaffected. We further studied the effect of hypoxia on Smad3-activated transcription by using a HaCaT cell line expressing Smad3-activated luciferase reporter gene (CAGA12-Luc) (26, 27). Smad3 can directly bind CAGA12 and hence the CAGA12-Luc reporter is activated by Smad3 more robustly than by Smad2. The cells were exposed to normoxia or hypoxia followed by TGF-β stimulus. TGF-β activated the CAGA12-Luc activity severalfold as expected. This, however, was strongly reduced by hypoxia demonstrating a biological end point for the hypoxic effect on Smad3 phosphorylation (Fig. 1G).
Hypoxic Inhibition of Smad3 Phosphorylation Is TGF-β Receptor-independent
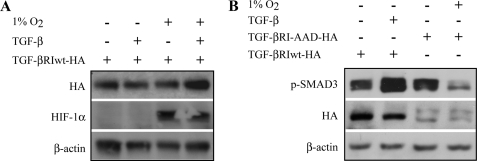
Many of the mechanisms reported to antagonize TGF-β signaling occur by blocking the TGF-βRI activation. The observed inhibition of Smad3 phosphorylation in hypoxia was unlikely to occur through receptor inactivation as this should result in the inactivation of both Smad2 and -3. However, we wanted to exclude a receptor level regulation in the hypoxic inhibition of Smad3. First, HaCaT cells were transfected with a vector encoding wild type TGF-βRI hemagglutinin fusion protein (TGF-βRIwt-HA) and exposed to hypoxia or normoxia with or without TGF-β1 treatment. Western blot analysis showed that the TGF-βRI protein level remained constant regardless of the treatment used, indicating that hypoxia does not induce receptor degradation (Fig. 2A). Next, HaCaT cells were transfected with an HA-tagged constitutively active mutant form of TGF-βRI (TGF-βRI-AAD-HA) (28) and exposed to hypoxia or normoxia. As expected, TGF-βRI-AAD effectively activated Smad3 phosphorylation in the absence of TGF-β ligand. Yet hypoxia strongly reduced the Smad3 phosphorylation induced by the constitutively active receptor (Fig. 2B). Moreover, the use of a chemical inhibitor of TGF-βRI demonstrated that hypoxia accelerates the disappearance of TGF-β-induced Smad3 phosphorylation after receptor blockade (supplemental Fig. 1). The data demonstrated that the hypoxic inhibition in Smad3 phosphorylation is not due to inactivation of the TGF-β receptor.
FIGURE 2.
Hypoxic inhibition of Smad3 phosphorylation is TGF-β receptor-independent. A, HaCaT cells were transfected with wild type TGF-βRI HA fusion protein (TGF-βRIwt-HA) and exposed to either ambient oxygen or hypoxia for 16 h followed by 30 min of TGF-β treatment. Western blot analysis shows that TGF-βRI is not degraded upon hypoxia. B, HaCaT cells were transfected with wild type TGF-βRI or with a constitutively active form of TGF-βRI (TGF-βRI-AAD-HA) HA fusion proteins. The phosphorylation of Smad3 was inhibited by hypoxia in the presence of the constitutively active TGF-βRI-AAD.
Hypoxic Dephosphorylation of Smad3 by PP2A
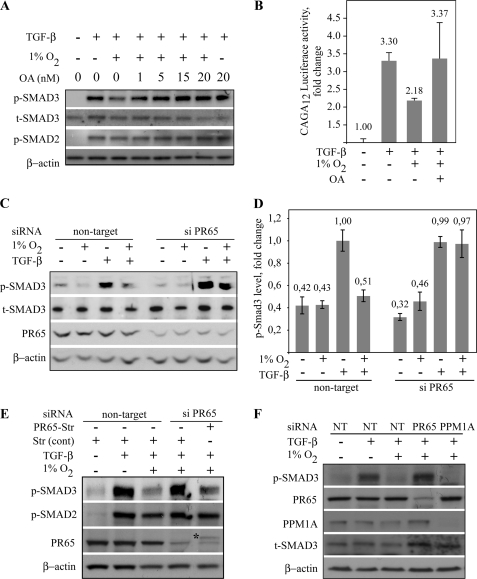
The results above suggested that hypoxia induced dephosphorylation of Smad3. We next tested OA, a noncompetitive inhibitor of protein phosphatase family, including the ubiquitously expressed PP2A (29). HaCaT cells were exposed to normoxia or hypoxia followed by application of increasing concentrations of OA and treated with TGF-β for 30 min to induce Smad phosphorylation. As shown previously, hypoxia strongly diminished the phosphorylation of Smad3 induced by TGF-β. However, a pretreatment of cells with OA reversed the hypoxic attenuation on the Smad3 phosphorylation indicating that hypoxia exerts its effects on Smad3 by dephosphorylation. Partial reversal of Smad3 phosphorylation was seen already at 1 nm OA, and the reversal was complete between 15 and 20 nm, although the highest used OA concentration induced only a minor increase in the normoxic Smad3 phosphorylation (Fig. 3A). In line with this, OA reversed the hypoxia-induced block on the TGF-β-induced CAGA12-Luc activity (Fig. 3B). These suggested that the dephosphorylation of Smad3 under hypoxic conditions was due to PP2A, because PP2A is more sensitive to OA than other protein phosphatase family members, such as PP1C or PPM1A, which is insensitive to OA (30).
FIGURE 3.
PP2A dephosphorylates Smad3 under hypoxia. A, HaCaT cells were exposed to 16 h of hypoxia followed by increasing concentrations of OA for 90 min (3rd to 5th lanes) and TGF-β for 30 min. 1 nm OA partially rescued the hypoxia-induced Smad3 phosphorylation, and the rescue was complete with 10–20 nm OA. B, HaCaT cells stably transfected with Smad3-specific luciferase reporter gene (CAGA12-Luc) exposed to the indicated conditions. Luciferase activity is shown as fold change to nontreated normoxic exposure. Four independent experiments ± S.E. are shown. C, HaCaT cells transfected with nontarget or PR65-specific double-stranded oligonucleotide siRNAs, followed by the indicated stimuli and analysis of phosphorylated and total Smad3 levels. The dephosphorylation of Smad3 by hypoxia (nontarget, 4th lane) was reversed by PR65 siRNA (siPR65, 4th lane). D, quantification of the level of Smad3 phosphorylation upon indicated stimuli and siRNA treatment in HaCaT cells. Means ± S.E. of four independent experiments are shown. E, HaCaT cells transfected with nontarget or PR65-specific siRNAs together with control or PR65 expressing plasmids followed by hypoxic and TGF-β treatment as indicated. The inhibition of hypoxic Smad3 dephosphorylation by PR65 siRNA (4th lane) was restored by exogenous PR65 expression (5th lane). Smad2 phosphorylation was not affected by any treatment. Asterisk points to the exogenous PR65. F, HaCaT cells transfected with nontarget (NT), PR65, or PPM1A-specific siRNAs followed by hypoxic and TGF-β treatment. PPM1A expression was abolished by siRNA, but this had no effect on the hypoxic Smad3 dephosphorylation (5th lane).
PP2A is a multiprotein complex consisting of a regulatory, a catalytic, and an essential scaffold (PR65) subunit (16). To study the involvement of PP2A specifically in the inhibition of hypoxic Smad3 phosphorylation, we used two independent PR65 siRNAs. First, cells were transfected with either nontarget or PR65-specific siRNA, which has previously been validated elsewhere and shown to be specific for PR65 (31). We further validated the siRNA function upon hypoxic and TGF-β exposure. The PR65 siRNA reduced the PR65 protein expression to 20–40% of the basal level in HaCaT cells regardless of the treatment (supplemental Fig. 2A). Following siRNA transfection, the cells were subjected to either normoxia or 16 h of hypoxia followed by a TGF-β stimulus for 1 h. In the cells with PR65 knockdown, the hypoxic suppression of TGF-β-elicited Smad3 phosphorylation was abolished or markedly reduced (Fig. 3C). Densitometric quantification of four independent experiments demonstrated a nearly complete loss of Smad3 dephosphorylation when exposed to siPR65 in hypoxia (Fig. 3D). Importantly, siPR65 did not have an effect on the normoxic TGF-β-induced Smad3 phosphorylation. Another independent PR65-targeted siRNA (siPR65 number 2) also demonstrated reduction of hypoxic Smad3 dephosphorylation (supplemental Fig. 2B). To further validate the involvement of PP2A, we performed a rescue experiment by simultaneously transfecting PR65-expressing plasmid and PR65 siRNA. As shown previously, PR65 siRNA blocked the hypoxic inhibition in Smad3 phosphorylation. This was reversed by exogenous PR65 expression, i.e. PR65 specifically restored the dephosphorylation of Smad3 in hypoxia (Fig. 3E). Again, no effect was seen on Smad2 phosphorylation. PPM1A has been suggested to dephosphorylate both Smad2 and -3 at the C-terminal serines under normoxia (13). We asked whether PPM1A was involved in the hypoxic regulation of Smad3. Exposure of HaCaT cells to PPM1A siRNA abolished the PPM1A expression, but this had no effect on the hypoxic Smad3 phosphorylation (Fig. 3F). This implied that PPM1A is not involved in the hypoxia-specific Smad3 dephosphorylation.
Smad3 Interacts with PP2A in Hypoxia
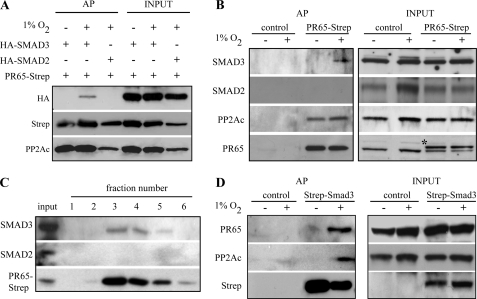
We next sought for a physical interaction between PP2A and Smad3 in hypoxia. Cells were co-transfected with a PR65 Strep-tactin fusion protein and HA-tagged Smad2 or -3. Samples were subjected to Strep-tactin affinity purification (32), and co-purification of Smad2 and -3 was detected by Western blot analysis. Exogenously expressed Smad3 was co-purified with PR65. Noticeably, this occurred only in hypoxic conditions but not in normoxic conditions. The overexpressed Smad2 did not co-purify with PR65. The catalytic subunit of PP2A (PP2Ac) was used as a positive control for PR65 interaction (Fig. 4A). Next, the interaction of endogenous Smad2 and -3 with exogenous PR65-Strep-tactin fusion protein was analyzed from cells exposed to either normoxia or 16 h of hypoxia prior to the TGF-β treatment. The endogenous Smad3 co-purified with PR65, similarly to PP2Ac. Again, the interaction between PR65 and endogenous Smad3 was seen only under hypoxic conditions. Endogenous Smad2, in contrast, did not co-purify with PR65-Strep-tactin under any condition (Fig. 4B). The results were further confirmed by affinity purification of endogenous Smad2 and -3 from cells exposed to a hypoxia mimetic CoCl2. The endogenous Smad3 co-purified with PR65-Strep-tactin and was detected in equimolar amounts in the elution fractions (Fig. 4C), whereas Smad2 did not co-purify with PR65. In a reciprocal experiment, we used Strep-tactin-Smad3 fusion protein as a bait and analyzed the endogenous components of PP2A after exposure of the cells to either normoxia or hypoxia. Smad3 fusion protein captured the endogenous PR65 and, noticeably, also the endogenous PP2A catalytic subunit PP2Ac (Fig. 4D). In agreement with the previous findings, the interaction between Smad3 and PP2A was specifically detected in cells exposed to hypoxia but not in normoxic cells.
FIGURE 4.
PP2A interacts with Smad3 under hypoxia. A, HeLa cells were co-transfected with PR65-Strep-tactin fusion protein and HA-tagged Smad2 or -3. Cells were exposed to normoxia or 16 h of hypoxia followed by 30 min of TGF-β treatment. Samples were affinity-purified with Strep-tactin columns and analyzed by Western blotting. Exogenous Smad3 was co-purified with PR65-Strep-tactin, and this was seen only in hypoxia (2nd lane). B, HeLa cells transfected with a control or PR65-Strep-tactin vector and exposed to normoxia or 16 h of hypoxia followed by TGF-β treatment. After bait capture and elution, the indicated proteins were detected by Western blotting demonstrating that endogenous Smad3 interacts with PR65 in hypoxic conditions. The catalytic subunit of PP2A (PP2Ac) was used as a positive control for PR65 interaction. The upper species in the PR65 blot represents the exogenous PR65-Strep-tactin (shown by an asterisk). C, detection of endogenous Smad2/3 interaction with PR65 upon chemical hypoxia. HeLa cells transfected with a PR65-Strep-tactin fusion protein were exposed to CoCl2 and TGF-β. PR65-Strep-tactin was captured on Strep-tactin columns. After washes, six elution fractions were collected and analyzed by Western blotting demonstrating that endogenous Smad3, but not Smad2, interacts with PR65. D, HeLa cells were transfected with Strep-tactin -Smad3 fusion protein or Strep-tactin control vector and exposed to normoxia or 16 h of hypoxia followed by TGF-β stimulus. Samples were affinity-purified with Strep-tactin columns and analyzed by Western blot. Endogenous PP2A subunits PR65 and PP2Ac were co-purified in hypoxia with Strep-tactin-Smad3.
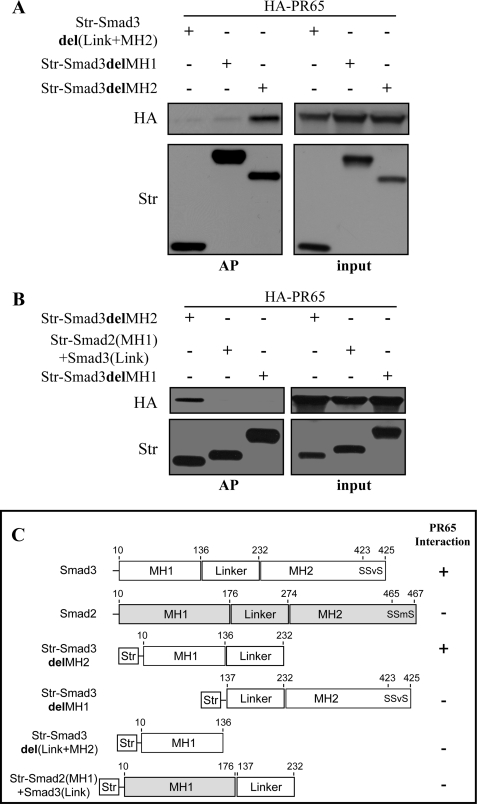
To map the Smad3 domain required for PP2A interaction and to confirm the ability of PP2A to discriminate between Smad2 and -3, we created Strep-tactin-tagged deletion mutants of Smad3. First, constructs lacking Smad3 MH1, MH2, or MH2 domain together with the linker domain were used (Fig. 5C). The Smad deletion constructs were co-transfected with full-length HA-PR65 into HeLa cells. 24 hours post-transfection, the cells were exposed to hypoxia for 16 h followed by lysis, affinity purification, and Western blot analysis. Smad3 interaction with PP2A required the N-terminal MH1 domain as well as the linker region of Smad3 but not the MH2 domain (Fig. 5A). To study the specificity of the interaction of PP2A with Smads, we used a chimeric mutant consisting of an MH1 domain from Smad2 fused to the linker region of Smad3. The interaction with PR65 was abolished when MH1 domain of Smad3 was switched to the MH1 domain of Smad2 (Fig. 5B). This further demonstrated that the Smad-PR65 interaction is specific for Smad3.
FIGURE 5.
Smad3 MH1 domain with the linker region confer PP2A interaction. A, HeLa cells were co-transfected with the indicated Strep-tactin-tagged (Str) Smad3 deletion mutants and HA-tagged PR65. Cells were exposed to 1% O2 for 1 h followed by affinity purification (AP) and Western blot analysis. HA-PR65 was co-purified only with Smad3delMH2. No interaction was detected with Smad3delMH1 or Smad3del(Link+MH2) mutants. B, substituting Smad3 MH1 domain with Smad2 MH1 domain abolishes the interaction with PR65. Affinity purification shows that PR65 co-purifies only with Smad3delMH2 and not with Smad2(MH1)+Smad3(Link) or Smad3delMH1. C, schematic illustration of the used constructs Smad3 in white and Smad2 in gray. Summary of PR65 interaction with each Smad construct is marked by + or − sign.
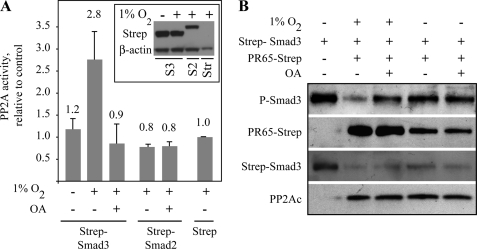
To further confirm that the hypoxic Smad3 dephosphorylation is achieved by PP2A, we analyzed the Smad3-associated PP2A activity in normoxic and hypoxic conditions. HeLa cells were transfected with Strep-tactin-tagged Smad2, Smad3, or with a control Strep-tactin vector. After affinity purification, the capture of PP2A activity was studied using a PP2A-specific phosphatase assay with divalent cation chelation (Fig. 6A). Phosphatase activity was captured only by Smad3. Importantly, the Smad3-associated phosphatase activity was detected specifically in the cells exposed to hypoxia. Smad2, in contrast, did not capture PP2A phosphatase activity even under hypoxia. Furthermore, okadaic acid completely inhibited the Smad3-associated hypoxic phosphatase activity. Finally, we performed an in vitro dephosphorylation assay of Smad3 by PP2A (Fig. 6B). PR65-Strep-tactin and Strep-tactin-Smad3 were separately transfected into HeLa cells. PR65 transfectants were exposed to normoxia or hypoxia without TGF-β. Strep-tactin-Smad3 transfectant was kept in normoxia and treated with TGF-β to induce phosphorylation prior to purification. After Strep-tactin affinity purification, Smad3 was exposed in vitro to PP2A derived from different conditions with or without the addition of OA to the reaction. Hypoxia-treated PP2A efficiently dephosphorylated the purified Smad3 This was completely inhibited by OA. In contrast, normoxia-exposed PP2A did not dephosphorylate Smad3 nor did OA have an effect on the phosphorylation of Smad3 in these conditions. The data further validated our finding of a PP2A-elicited Smad3 dephosphorylation that occurred specifically under hypoxia and implied that PP2A is a bona fide phosphatase responsible for the dephosphorylation of Smad3 under hypoxia.
FIGURE 6.
Hypoxic PP2A activity associates with Smad3. A, HeLa cells were transfected either with a Strep-tactin control vector or Strep-tactin-Smad3 or -2 fusion proteins and exposed to hypoxia and TGF-β. Samples were Strep-tactin-purified and analyzed by PP2A phosphatase assay. Phosphatase activity above background level was captured only by Smad3 in hypoxia. The captured activity was completely inhibited by OA. No phosphatase activity was co-purified with Smad2. The results from at least three biological repetitions (mean ± S.E.) are presented as fold change from hypoxic control vector. B, hypoxia-purified PP2A dephosphorylates Smad3 in vitro. HeLa cells were separately transfected with Strep-tactin-Smad3 or PR65-Strep-tactin fusion proteins. Smad3-expressing cells were kept in normoxia and treated with TGF-β to induce Smad3 C-terminal phosphorylation (1st lane), and PR65-expressing cells were exposed to normoxia or 16 h of hypoxia prior to Strep-tactin column purification. Purified Smad3 was left untreated (1st lane) or treated with purified PP2A with or without okadaic acid as indicated (2nd to 5th lanes).
PP2A Is Required for Hypoxic Loss of Smad3 Activity
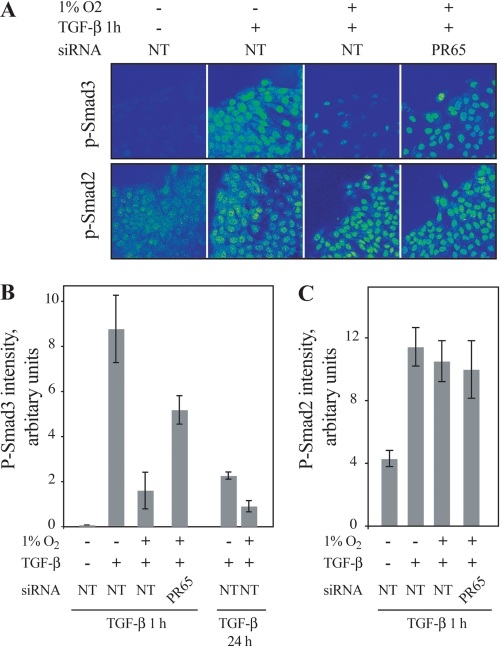
We next investigated the nuclear accumulation of phosphorylated Smad3 that occurs upon TGF-β induction and the role of PP2A in it. Wounded monolayers of HaCaT cells transfected with control siRNA were used to activate maximal Smad2/3 nuclear accumulation. The cells were pre-exposed to normoxia or hypoxia followed by 1 h of TGF-β stimulus and stained for P-Smad2 and -3. Strongly reduced nuclear expression of phosphorylated Smad3 at the wound edge was detected in the cells pre-exposed to hypoxia as compared with TGF-β stimulus alone. In contrast to control siRNA, a pretreatment of the cells with PR65 siRNA rescued the effect of hypoxia on the phosphorylation of Smad3 at the wound edge further supporting the involvement of PP2A in the hypoxia-elicited phenotypic change (Fig. 7A). Quantification of P-Smad3 expression using image analysis from different conditions demonstrated strong hypoxia-induced attenuation of P-Smad3 expression in the nuclei (Fig. 7B). As shown previously, no effect of hypoxia nor PP2A inhibition on the TGF-β-induced Smad2 phosphorylation was detected (Fig. 7, A and C).
FIGURE 7.
Hypoxia inhibits nuclear accumulation of P-Smad3 through PP2A. A, HaCaT cells were pre-exposed to siRNA (NT, nontarget or siPR65) prior to wounding and exposure to the indicated stimuli. Cells were stained for phospho-Smad3 (upper panels) and for phospho-Smad2 (lower panels). B, quantification of phosphorylated Smad3 level at wound edges upon the indicated stimuli and pre-exposure to siRNA. Cells were stained and analyzed for phospho-Smad3 expression at the wound edge by image analysis software (ImageJ). A threshold level was set from normoxic samples without TGF-β stimulus. The means ± S.E. from three optical fields (each containing ∼40 cells) are shown. C, quantification of phosphorylated Smad2 level at wound edges upon the indicated stimuli and pre-exposure to siRNA.
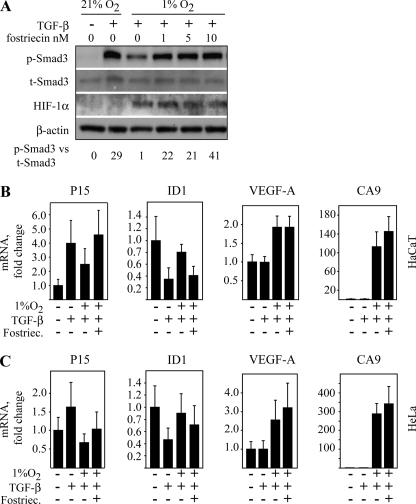
To ask whether hypoxia has influence on the TGF-β-induced gene expression, we performed Q-RT-PCR analysis in HaCaT and HeLa cells of known TGF-β-activated genes. We further asked how the inhibition of PP2A activity might affect the hypoxia-modulated gene expression. First, we tested the effect of a highly selective PP2A inhibitor fostriecin, which blocks the catalytic activity of PP2A with higher specificity than OA (33). Similarly to okadaic acid, fostriecin reversed the hypoxic attenuation on Smad3 phosphorylation with the reversal being complete at 10 nm (Fig. 8A), and it had no effect on the normoxic Smad3 phosphorylation even at high (100 nm) concentrations (supplemental Fig. S3A).
FIGURE 8.
Effect of hypoxia on the gene expression induced by TGF-β and Smad3. A, HaCaT cells were exposed to 16 h of hypoxia followed by increasing concentrations of fostriecin for 90 min and TGF-β for 30 min. Partial rescue of hypoxic Smad3 phosphorylation was seen at 1 nm fostriecin, and the rescue was complete with 10 nm fostriecin. p-Smad3 levels normalized to t-Smad3 are shown at the bottom of the panel. B, Q-RT-PCR analysis of two TGF-β- and Smad3-responsive genes (P15 and ID1) and two hypoxia-activated genes (VEGF and CA9) in HaCaT cells upon the indicated exposure of a combination of TGF-β, hypoxia, and fostriecin (Fostriec.) (10 nm). C, Q-RT-PCR analysis of two TGF-β- and Smad3-responsive genes (P15 and ID1) and two hypoxia-activated genes (VEGF and CA9) in HeLa cells upon the indicated exposure of a combination of TGF-β, hypoxia, and fostriecin (10 nm). The cells were exposed to TGF-β for 1 h in the case of p15, CA9, and VEGF-A and in the case of ID1 for 4 h.
Based on the literature, we selected two cell cycle-regulating genes reported to be under the influence of Smad3, p15 (34) and ID1 (34, 35). For controls, we chose two well characterized hypoxia-activated genes, the CA9 (carbonic anhydrase 9) (36) and VEGF (37). In HaCaT cells, TGF-β activated the expression of p15 and down-regulated ID1 expression without having any effect on VEGF or CA9, which, however, were strongly induced by hypoxia (Fig. 8B). Noticeably, hypoxia reversed the effect of TGF-β on both p15 and ID1 expression. Moreover, fostriecin completely blocked the effect of hypoxia on both TGF-β responsive genes but did not affect the expression of the hypoxic genes (Fig. 8B). Similarly to HaCaT cells, the effect of TGF-β on p15 and ID1 was reversed by hypoxia in HeLa cells, which in turn was attenuated by PP2A inhibition (Fig. 8C). The data together with the reporter gene assays indicated that hypoxia can influence the TGF-β and Smad3-responsive gene expression and that at least partially this requires PP2A activity.
DISCUSSION
Here we have shown that hypoxia attenuates the TGF-β-induced Smad3 phosphorylation but leaves the Smad2 phosphorylation intact. Mechanistically, this is due to dephosphorylation of Smad3 by PP2A. To our knowledge, this is the first demonstration of a Smad phosphatase that is selective to one R-Smad, leading to a shutdown of only one arm of the TGF-β signaling.
The TGF-β activated canonical R-Smad pathway may be inhibited by various mechanisms, including the inhibition of R-Smad receptor interaction as well as receptor degradation by I-Smads. In normoxic conditions, this type of inhibition and subsequent attenuation in R-Smad phosphorylation is well established. Moreover, PP1C can dephosphorylate TGF-βRI (10). Dephosphorylation events have been shown to occur on several other components in the TGF-β superfamily signaling cascade. BMP and TGF-β signaling are regulated by SCP1–3 phosphatases, which target the C-terminal phosphorylation of Smad1 and the linker phosphorylation of Smad2 and -3 (38, 39). BMP receptor associates with one form of PP2A leading to dephosphorylation of BMP R-Smad, Smad1, in normoxia, and different PP2A forms can dephosphorylate different receptors of the TGF-β superfamily and Smad1 (11, 12). Bengtson et al. (12) show that PP2A dephosphorylates Smad1 linker region and only weakly the C terminus in normoxia. They also reported that the linker region dephosphorylation leads to enhanced nuclear localization of Smad1 in normoxia. Noticeably, in the study by Batut et al. (11), the entire TGF-β signaling is regarded to be attenuated as measured by Smad2 C-terminal phosphorylation. These studies clearly indicate that PP2A regulates TGF-β and BMP signaling both at the receptor and R-Smad levels under normoxic conditions. However, our results show that the block in Smad3 C-terminal phosphorylation under hypoxia is achieved independently from the receptor activity and importantly that it is specific for Smad3. The data demonstrate that TGF-βRI protein level is not affected by hypoxia and that forced receptor expression does not overcome the hypoxic attenuation in Smad3 phosphorylation. Importantly, Smad3 phosphorylation continued to be inhibited by hypoxia in the presence of a constitutively active phosphatase-resistant mutantof TGF-βRI. In line with this, after Smad3 phosphorylation was blocked by a chemical TGF-βRI inhibitor, hypoxia accelerated the dephosphorylation process.
Besides the block in essential receptor interactions or the enhancement of receptor degradation, Smad2 and -3 may also be inactivated by dephosphorylation. However, regardless of the mechanism of inhibition, both Smad2 and -3 phosphorylations are affected (13). Smad2 and -3 were recently reported to be dephosphorylated under sustained TGF-β treatment at longer time points (>2 h) by PPM1A (13), indicating together with our data that the protein phosphatases play a major role in regulating TGF-β responses independently of receptor level regulation. Although we do not want to exclude basal level dephosphorylation of Smad3 by PPM1A or any other phosphatase, it is clear that they do not have a major effect on the hypoxic Smad3 dephosphorylation after TGF-β stimulus at shorter time points (<2 h). PPM1A is insensitive to okadaic acid but has an absolute requirement for Mg2+ or Mn2+ ions. In contrast, PP2A is not dependent on divalent ions but is strongly inhibited by okadaic acid. Arguing against the involvement of PPM1A, our data show that the hypoxic Smad3 dephosphorylation is okadaic acid-inhibitable at low concentration. Moreover, in hypoxia Smad3 co-purifies phosphatase activity in the presence of EDTA, a strong Mg2+/Mn2+ chelator. Most importantly, depletion of PPM1A by siRNA could not restore the hypoxic Smad3 dephosphorylation. This was in striking contrast to the depletion of PP2A by siRNA, which did inhibit the Smad3 dephosphorylation. The inhibition occurred in hypoxia but did not have a marked effect on Smad3 phosphorylation in normoxia providing further support for the involvement of PP2A in hypoxia-specific Smad3 dephosphorylation. Moreover, the data demonstrated a physical interaction of PP2A and Smad3 and that the interaction occurs only under hypoxic conditions but not in normoxia. Furthermore, the data demonstrated that the hypoxia-activated PP2A did not interact with the Smad2 MH1 domain. In keeping with this, PP2A activity co-purified with Smad3 but not with Smad2, and PP2A dephosphorylated Smad3 in vitro. These were specifically seen under hypoxic conditions.
Several lines of evidence demonstrate that Smad2 and -3 have separate functions. Knock-out studies show that Smad2 and -3 are not functionally interchangeable (40, 41). Moreover, Smad2 and -3 have been reported to bear distinct functions in transcriptional responses (35) and post-translational protein modification (42). Our data on the hypoxic regulation of R-Smad activity imply a mechanism as to how differential regulation of Smad2 and -3 may occur. Furthermore, the data may shed light to previous findings, such as the phenotype of Smad3 knock-out mice with acceleration of wound healing (40) as well as the enhancement of tumor progression in Smad3 null mice (43).
To our knowledge, PP2A is the first phosphatase demonstrating specificity between Smad2 and -3. PP2A has been implicated in a number of cellular homeostasis maintaining processes, and in the setting of carcinogenesis it is mainly implicated as a tumor suppressor (16, 44). At least 12 (>30, including the splice variants) different regulatory and two scaffold subunits for PP2A exist. Together, these may specify some 70 different the PP2A targets and allow precisely regulated phosphatase activity. In keeping with this, changes in the PP2A regulatory subunits have been reported to affect the ability of PP2A to induce cell transformation (45), and different forms can inhibit different TGF-β superfamily receptors (11). It is plausible that hypoxia induces changes to the PP2A composition by switching regulatory subunits and thereby PP2A target specificity. Furthermore, because hypoxia is known to induce the expression of nearly a hundred different proteins and also to activate post-transcriptional modifications, including hydroxylation, it is possible that the hypoxia-induced proteins and/or modifications may be involved in the recruitment of PP2A to Smad3. Interestingly, hypoxia has been reported to induce the expression of TGF-β (21, 46), and HIF-1α has been shown to cooperate with Smad3 and to modify the Smad-activated transcriptional responses (24). The detailed composition of the Smad3-associating PP2A complex as well as whether HIF may also directly bind to the complex remain to be investigated.
The in vivo consequences of hypoxic Smad3 dephosphorylation by PP2A will require extensive investigation. Based on our data, we speculate that hypoxia has a major effect on the TGF-β-regulated epithelial function through Smad3-selective dephosphorylation. Supporting this, hypoxia led to decreased expression of nuclear P-Smad3 that was restored by PP2A knockdown, although the nuclear accumulation of P-Smad2 was not affected by hypoxia. Consequently, at least a subset of TGF-β-affected genes that regulate epithelial integrity and proliferation such as p15 and ID1 were modulated by hypoxia that in turn was reversed by PP2A inhibition. Strikingly, however, under hypoxia the phosphorylated Smad2 may function and carry normal responses in hypoxia (47). Noticeably also, while in hypoxia PP2A dephosphorylates the C-terminal serines of Smad3 and the phosphorylation at the linker region may still retain functionality and convey some of Smad3 functions.
Supplementary Material
Acknowledgments
Raisa Vuorinen and Taina Kalevo-Mattila are acknowledged for expert technical assistance. Drs. Joan Massague and Carl-Henrik Heldin kindly provided the TGF-βRI constructs.
The work was supported by Academy of Finland Grants 200779, 210282, and 8212695, the Sigrid Juselius Foundation, the Emil Aaltonen Foundation, the Finnish Cancer Research Foundation, and Turku University Hospital Grants EVO 13336 and 13031.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- TGF-β
- transforming growth factor-β
- TGF-βRI
- TGF type 1 receptor
- BMP
- bone morphogenetic protein
- siRNA
- small interfering RNA
- OA
- okadaic acid
- VEGF
- vascular endothelial growth factor
- HA
- hemagglutinin
- t-Smad
- total Smad
- p-Smad
- phospho-specific Smad
- HIF
- hypoxia-inducible factor
- Q-RT-PCR
- quantitative reverse transcription-PCR.
REFERENCES
- 1.Siegel P. M., Massagué J. (2003) Nat. Rev. Cancer 3, 807–821 [DOI] [PubMed] [Google Scholar]
- 2.Derynck R., Akhurst R. J., Balmain A. (2001) Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 3.Cui W., Fowlis D. J., Bryson S., Duffie E., Ireland H., Balmain A., Akhurst R. J. (1996) Cell 86, 531–542 [DOI] [PubMed] [Google Scholar]
- 4.Bierie B., Moses H. L. (2006) Nat. Rev. Cancer 6, 506–520 [DOI] [PubMed] [Google Scholar]
- 5.Moustakas A., Souchelnytskyi S., Heldin C. H. (2001) J. Cell Sci. 114, 4359–4369 [DOI] [PubMed] [Google Scholar]
- 6.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 7.ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 8.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 9.Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 10.Shi W., Sun C., He B., Xiong W., Shi X., Yao D., Cao X. (2004) J. Cell Biol. 164, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batut J., Schmierer B., Cao J., Raftery L. A., Hill C. S., Howell M. (2008) Development 135, 2927–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengtsson L., Schwappacher R., Roth M., Boergermann J. H., Hassel S., Knaus P. (2009) J. Cell Sci. 122, 1248–1257 [DOI] [PubMed] [Google Scholar]
- 13.Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y. G., Meng A., Feng X. H. (2006) Cell 125, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan X., Liang Y. Y., Feng X. H., Lin X. (2006) J. Biol. Chem. 281, 36526–36532 [DOI] [PubMed] [Google Scholar]
- 15.Kokabu S., Nojima J., Kanomata K., Ohte S., Yoda T., Fukuda T., Katagiri T. (2010) J. Bone Miner. Res., in press [DOI] [PubMed] [Google Scholar]
- 16.Janssens V., Goris J. (2001) Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen P. T. (1997) Trends Biochem. Sci. 22, 245–251 [DOI] [PubMed] [Google Scholar]
- 18.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 19.Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) EMBO J. 20, 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av, Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 21.Li J. J., Lu J., Kaur C., Sivakumar V., Wu C. Y., Ling E. A. (2008) Neuroscience 156, 662–672 [DOI] [PubMed] [Google Scholar]
- 22.Sánchez-Elsner T., Botella L. M., Velasco B., Langa C., Bernabéu C. (2002) J. Biol. Chem. 277, 43799–43808 [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Elsner T., Botella L. M., Velasco B., Corbí A., Attisano L., Bernabéu C. (2001) J. Biol. Chem. 276, 38527–38535 [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Akman H. O., Smith E. L., Zhao J., Murphy-Ullrich J. E., Batuman O. A. (2003) Blood 101, 2253–2260 [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar M., Liu F., Hata A., Doody J., Massagué J. (1997) Genes Dev. 11, 984–995 [DOI] [PubMed] [Google Scholar]
- 26.Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C. S. (2007) Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wieser R., Wrana J. L., Massagué J. (1995) EMBO J. 14, 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bialojan C., Takai A. (1988) Biochem. J. 256, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández J. J., Candenas M. L., Souto M. L., Trujillo M. M., Norte M. (2002) Curr. Med. Chem. 9, 229–262 [DOI] [PubMed] [Google Scholar]
- 31.Arnold H. K., Sears R. C. (2006) Mol. Cell. Biol. 26, 2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junttila M. R., Saarinen S., Schmidt T., Kast J., Westermarck J. (2005) Proteomics 5, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 33.Walsh A. H., Cheng A., Honkanen R. E. (1997) FEBS Lett. 416, 230–234 [DOI] [PubMed] [Google Scholar]
- 34.Kretschmer A., Moepert K., Dames S., Sternberger M., Kaufmann J., Klippel A. (2003) Oncogene 22, 6748–6763 [DOI] [PubMed] [Google Scholar]
- 35.Piek E., Ju W. J., Heyer J., Escalante-Alcalde D., Stewart C. L., Weinstein M., Deng C., Kucherlapati R., Bottinger E. P., Roberts A. B. (2001) J. Biol. Chem. 276, 19945–19953 [DOI] [PubMed] [Google Scholar]
- 36.Wykoff C. C., Beasley N. J., Watson P. H., Turner K. J., Pastorek J., Sibtain A., Wilson G. D., Turley H., Talks K. L., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Cancer Res. 60, 7075–7083 [PubMed] [Google Scholar]
- 37.Liu Y., Cox S. R., Morita T., Kourembanas S. (1995) Circ. Res. 77, 638–643 [DOI] [PubMed] [Google Scholar]
- 38.Knockaert M., Sapkota G., Alarcón C., Massagué J., Brivanlou A. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11940–11945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrighton K. H., Willis D., Long J., Liu F., Lin X., Feng X. H. (2006) J. Biol. Chem. 281, 38365–38375 [DOI] [PubMed] [Google Scholar]
- 40.Ashcroft G. S., Yang X., Glick A. B., Weinstein M., Letterio J. L., Mizel D. E., Anzano M., Greenwell-Wild T., Wahl S. M., Deng C., Roberts A. B. (1999) Nat. Cell Biol. 1, 260–266 [DOI] [PubMed] [Google Scholar]
- 41.Heyer J., Escalante-Alcalde D., Lia M., Boettinger E., Edelmann W., Stewart C. L., Kucherlapati R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12595–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stasyk T., Dubrovska A., Lomnytska M., Yakymovych I., Wernstedt C., Heldin C. H., Hellman U., Souchelnytskyi S. (2005) Mol. Biol. Cell 16, 4765–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y., Richardson J. A., Parada L. F., Graff J. M. (1998) Cell 94, 703–714 [DOI] [PubMed] [Google Scholar]
- 44.Mumby M. (2007) Cell 130, 21–24 [DOI] [PubMed] [Google Scholar]
- 45.Chen W., Possemato R., Campbell K. T., Plattner C. A., Pallas D. C., Hahn W. C. (2004) Cancer Cell 5, 127–136 [DOI] [PubMed] [Google Scholar]
- 46.Scheid A., Wenger R. H., Schäffer L., Camenisch I., Distler O., Ferenc A., Cristina H., Ryan H. E., Johnson R. S., Wagner K. F., Stauffer U. G., Bauer C., Gassmann M., Meuli M. (2002) FASEB J. 16, 411–413 [DOI] [PubMed] [Google Scholar]
- 47.Oft M., Akhurst R. J., Balmain A. (2002) Nat. Cell Biol. 4, 487–494 [DOI] [PubMed] [Google Scholar]
- 48.Leivonen S. K., Chantry A., Hakkinen L., Han J., Kahari V. M. (2002) J. Biol. Chem. 277, 46338–46346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.