Abstract
Toll-like receptor 4 (TLR4) is unique among the Toll-like receptors in its ability to utilize TLR/IL1R-domain-containing adaptor protein (TIRAP), which recruits TLR4-MyD88 to phosphatidylinositol 4,5-bisphosphate (PIP2)-rich sites on the plasma membrane, to activate NF-κB and MAPK pathways. Here, we show that AIP1 disrupts formation of the TLR4- TIRAP-MyD88 complex without directly binding to any of the complex components. AIP1 via its pleckstrin homology and C2 domains binds to phosphatidylinositol 4-phosphate, a lipid precursor of PIP2. Knock-out of AIP1 in cells increases and overexpression of AIP1 reduces cellular PIP2 levels. We further show that AIP1 is a novel GTPase-activating protein (GAP) for Arf6, a small GTPase regulating cellular PIP2 production and formation of the TLR4-TIRAP-MyD88 complex. Thus, deletion of the GAP domain on AIP1 results in a loss of its ability to mediate the inhibition of Arf6- and TLR4-induced signaling events. We conclude that AIP1 functions as a novel Arf6-GAP to negatively regulate PIP2-dependent TLR4-TIRAP-MyD88 signaling.
Keywords: Cytokines/Tumor Necrosis Factor, Receptors/Toll-like, Signal Transduction/G-proteins, AIP1, TIRAP, TLR4
Introduction
Toll-like receptors (TLRs)4 play certain critical roles in the innate immune response in mammals through the recognition of conserved molecular patterns associated with different pathogens (1–3). TLR4 is one of the best defined TLRs and the central signaling receptor for lipopolysaccharides (LPS) in mammals (3, 4). TLR4 recruits the TLR/IL1R (TIR) domain-containing adaptor proteins to activate downstream signals upon ligand stimulation. These adaptors in mammals are MyD88, TIRAP (also called Mal), TRIF, TRAM, and SARM (5–14).
TLR4 is unique among the TLRs because it can utilize all of the TIR domain-containing adaptors and mediate activation of both the MyD88-dependent NF-κB and MAPK cascades as well as the MyD88-independent IRF-3 signaling pathway (1, 12, 14). It had been unclear how these two pathways were coordinated until TIRAP and TRAM were found as two sorting adaptors (15, 16). Although the delivery of TLR4 by TRAM from the plasma membrane to endosomes initiates TLR4-mediated IRF-3 activation (16), targeting of TLR4 and MyD88 to phosphatidylinositol 4,5-bisphosphate (PIP2) sites on the plasma membrane by TIRAP launches the TLR4-mediated MyD88-dependent signaling pathway to activate NF-κB and MAPK. TIRAP has an N-terminal PIP2 binding domain, which anchors to the plasma membrane, and a C-terminal TIR domain, which recruits MyD88 and TLR4 to the PIP2 sites to activate downstream signaling. Mutations in the PIP2 binding domain of TIRAP, or depletion of cellular PIP2, can abrogate both the recruitment of TIRAP to the plasma membrane and its capacity to induce cytokine production in response to LPS (15).
The cellular level of PIP2 is dynamically regulated. In mammalian cells, phosphatidylinositol 4-phosphate (PI4P) 5-kinase (PIP5K) phosphorylates PI4P to generate PIP2 (17). In response to growth factors or other stimuli, PIP2 can be hydrolyzed by phosphoinositide-specific phospholipase C to diacylglycerol and inositol 1,4,5-trisphosphate, or it can be further phosphorylated by PI3K to generate PIP3. However, regulation of PIP5K activity, a key step in controlling cellular basal PIP2, is critical for PIP2-dependent TLR4 signaling. The best characterized activator of PIP5K is Arf6 (ADP-ribosylation factor 6), a small GTPase. As the sole member of the class III ADP-ribosylation factor family, Arf6 resides on the plasma membrane and endosomes and regulates the traffic between these compartments (18, 19). Specific guanine nucleotide exchange factors activate Arf6 by promoting its dissociation from GDP and binding of GTP, whereas the GTPase-activating proteins (GAPs) inactivate Arf6 by promoting GTP hydrolysis. The evidence supporting the role of Arf6 in TLR4 signaling includes the following: Arf6 is activated by β2-integrin; Arf6 is an activator of PIP5K; and most importantly, blockade of Arf6 activity blunted TLR4-MyD88-dependent signaling.
Here, we show AIP1 plays an inhibitory role in the LPS-induced NF-κB and MAPK signaling pathways. The results suggest that AIP1, initially identified as a member of Ras-GAP family protein in our laboratory, is a novel Arf6 GAP that can promote Arf6-GTP hydrolysis both in vitro and in vivo. Deletion of AIP1 from mouse endothelial cells can enhance cellular PIP2 levels, in turn enhancing LPS-induced NF-κB and MAPK signaling.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
FLAG-tagged AIP1/2AB plasmid was described previously (20). The GST-tagged AIP1- and GST-tagged mutations of AIP1 were generated by PCR using AIP1/2AB as template and constructed into pDEST15 vector. TLR4/CD4, HA/TLR4, FLAG-MyD88, and HA-TIRAP were gifts from Dr. Ruslan Medzhitov; the FLAG-TIRAP and mutations for TIRAP were generated by PCR using HA-TIRAP as template and constructed into pCDNA3–2AB vector. Myc-IL1-R1 was generated by PCR using IL1-R1 cDNA as template. GST-GGA3VHS-GAT plasmid was a gift from Dr. Juan S. Bonifacino. HA-Arf6, HA-Arf6T27N, and HA-Arf6Q67L were gifts from Dr. James Casanova. GST-Arf6 was generated by PCR using HA-Arf6 as template and constructed into pDEST15 vector. EGFP-PIP5Kα, EGFP-PIP5Kβ, and EGFP-PIP5Kγ were gifts from Dr. Dianqing Wu. Antibody against AIP1 was described previously (20). Antibodies against IκBα, p38, and β-actin were from Sigma. Antibodies against p-IκBα and p-p38 were from Cell Signaling.
Cell Culture and Transfections
Mouse lung microvessel EC cells were cultured in MLEC medium containing 40% low glucose Dulbecco's modified Eagle's medium, 40% Ham's F-12, 20% fetal bovine serum, 2% EC growth factors. Bovine aortic endothelial cells, 293T cells, and Cos7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. 293T and Cos7 cells were transfected by Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). MLEC cells were transfected by Amaxa HMVEC-L Nucleofector kit according to manufacturer's protocol. For reconstitution of AIP1 back to AIP1-KO, cells (1 × 106) were reconstituted with the indicated AIP1 constructs by electroporation. 48 after transfection, cells were treated with LPS for the indicated times.
Immunoblotting and Immunoprecipitation
293T cells after transfection for 24 h were washed twice with cold PBS and harvested in lysis buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Triton X-100, 1 mm sodium orthovanadate, 1 mm sodium pyrophosphate, 10 mg/ml aprotinin, 10 mg/ml leupeptin, 2 mm phenylmethylsulfonyl fluoride, 1 mm EDTA) for 20 min on ice. Immunoprecipitation and immunoblotting were performed as described previously (20).
Electrophoretic Mobility Shift Assays (EMSAs)
The double-stranded oligonucleotide containing a κB consensus site from the immunoglobulin κ gene (Promega) was used for EMSA. Preparation of nuclear extracts and EMSA were performed as described previously (21).
Indirect Immunofluorescence Microscopy
MLEC cells, bovine aortic endothelial cells, or Cos7 cells after transfection for 24 h were washed twice by ice-cold PBS, then fixed in 4% paraformaldehyde for 20 min at 25 °C, and permeabilized with 0.1% Triton X-100 for 1.5 min. Samples were treated with block buffer (1% normal horse serum in PBS) for 30 min, and the appropriate primary antibodies were diluted into PBS and incubated for 1 h. Alexa Fluor 594-phalloidin (Molecular Probes) was used to stain F-actin. Secondary antibodies were incubated after primary antibodies binding and washed three times in PBS. Images were captured under a Leica SP5 confocal microscope.
Reporter Gene Assay
293T cells (1 × 106) were transfected with appropriate reporter (0.l μg) and expression (0.5 μg) plasmids. Reporter gene assays were performed 20 h after transfection by the luciferase assay system (Promega) according to the manufacturer's instructions.
Protein Purification
GST fusion proteins used in GGA3 pulldown assay and lipid binding assays were purified from Escherichia coli BL21DE3 competent cells (Stratagene). Protein expression was induced for 3 h at 37 °C with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation. Cell pellets were resuspended by sonication in 1× PBST buffer in the presence of 1% Triton X-100 and full protease inhibitors. The extracts were clarified by centrifugation at 12,000 rpm for 1 h and then mixed with glutathione-Sepharose 4B (GE Healthcare). Bound proteins were washed three times with the same cell breaking buffer in the presence of 500 mm NaCl. The beads that bind proteins for pulldown assay were stored at 1× PBS with 30% glycerol at −20 °C. Proteins for lipid binding assays were eluted with 20 mm glutathione in 1× PBST. Protein purity and concentration were determined by Coomassie Blue-stained polyacrylamide gel.
Lipid Binding Assay
PIP strips and membrane lipid strips (Echelon Biosciences) were immersed in blocking buffer (1× PBS, 1% fat-free milk, 0.1% Tween 20) for 1 h. Strips were probed for 2 h at 25 °C with the indicated GST fusion protein (50 ng/ml) in the presence of anti-GST antibody (Sigma). Blots were then washed in blocking buffer three times for 10 min each and probed with a horseradish peroxidase-conjugated anti-rabbit IgG (GE Healthcare) for 30 min in blocking buffer. Bound protein was detected using ECL (GE Healthcare).
Lipid Extraction and Phosphatidylinositol 4,5-Bisphosphate Measurement by Enzyme-linked Immunosorbent Assay
Cells (1 × 105) were washed once with ice-cold PBS and lysed in 375 μl of methanol/chloroform/HCl (40:20:1) mixture on ice, followed by 100 μl of chloroform and 225 μl of water. After vortexing for 1 min, samples were centrifuged at 3000 rpm for 2 min at 4 °C, and the lower organic phase was collected and dried by vacuum. Lipid extracts were dissolved directly in ethanol at room temperature, loaded onto a microplate, and dried under vacuum. The membrane was blotted with mouse anti-phosphatidylinositol 4,5-bisphosphate monoclonal antibody (Assay Designs, Ann Arbor, MI) for 1 h and subsequently blotted with goat anti-mouse IgG-horseradish peroxidase (GE Healthcare) for 25 min. The microplate was washed three times with PBS after each incubation. Finally, chemiluminescence substrate was added to the plate (Kirkegaard & Perry Laboratories), and luminescence intensity was determined by a luminometer.
GGA3 Pulldown Assay
Cells for GGA3 pulldown assay (1 × 106) were lysed in lysis buffer, and supernatant was incubated with GST-GGA3 beads or control GST beads at 4 °C for 2 h. Beads were then washed three times in lysis buffer. Bound proteins were eluted in SDS loading buffer and detected by immunoblotting with Arf6 antibody.
GAP Activity Assay
Proteins for GAP activity assay were expressed in BL21DE3 competent cells at the same conditions and were purified at the following conditions. Cell pellets were resuspended by sonication in high salt cell breaking buffer (50 mm Tris-HCl, 10% sucrose, 10 mm EDTA, 600 mm KCl, 0.01% Igepal, 1 mm dithiothreitol, full protein inhibitors, and 1 mm phenylmethylsulfonyl fluoride). Extracts from the cell pellet were mixed with glutathione-Sepharose 4B at 4 °C for 2 h and washed with T buffer (25 mm Tris-HCl, 10% glycerol, 1 mm EDTA) containing 1 m KCl. Proteins were eluted with 20 mm glutathione in T buffer containing 150 mm KCl followed by washing and concentrating with Millipore centrifugal filter devices. Protein purity and concentration were determined by Coomassie Blue-stained polyacrylamide gel. The GTP hydrolysis level of purified Arf6 (1 μm) in the presence of AIP1-GAP or control GST proteins was determined by GTPase assay kit (Innova Biosciences) at A635 m following the manufacturer's protocol.
RESULTS AND DISCUSSION
AIP1 Inhibits LPS-induced NF-κB and MAPK Activation
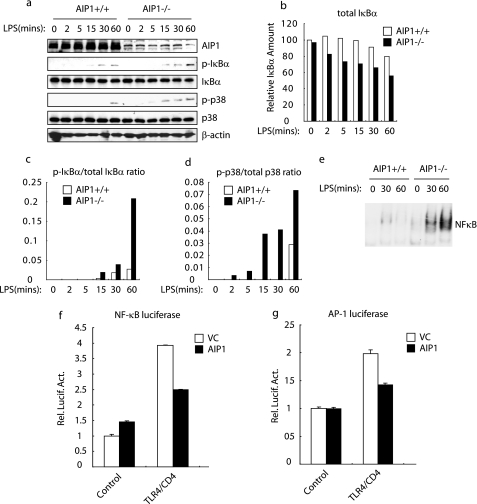
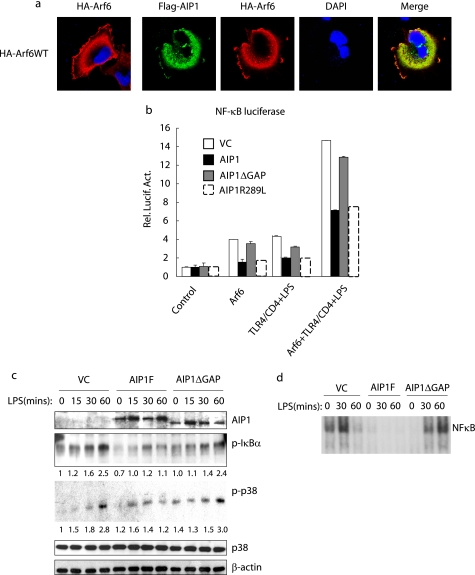
Our previous studies showed AIP1 (ASK1-interacting protein-1) is a positive regulator in tumor necrosis factor-induced c-Jun N-terminal kinase (JNK)/p38 MAPK signaling but is an inhibitor of tumor necrosis factor-α-induced NF-κB activation in vascular endothelial cells (EC). Because tumor necrosis factor-α, IL-1, and LPS are potent proinflammatory stimuli and activate NF-κB signaling pathways in EC, we attempted to determine whether AIP1 plays a role in the IL-1- and LPS-induced signaling in EC. To this end, WT and AIP1-KO mouse EC were treated with IL-1β or LPS for various times, and the activation of MAPK and NF-κB was determined by Western blot with antibodies against phospho-p38 or phospho-IκBα. IL-1β induced rapid phosphorylations of p38 and IκBα. IκBα was strongly degraded, followed by a re-synthesis (supplemental Fig. S1a). LPS induced a delayed but sustained response in p38 activation and NF-κB with a weak degradation of IκBα, which was only detected in the presence of the protein synthesis inhibitor cycloheximide (supplemental Fig. S1b). Similar to the observations in tumor necrosis factor signaling, IL-1β-induced p38 MAPK was reduced, and IL-1β-induced NF-κB signaling was enhanced in AIP1-KO EC (supplemental Fig. S1a). However, LPS-induced activation of both the MAPK and NF-κB signaling was augmented in AIP1-KO EC (Fig. 1a, with quantification in b–d). Accordingly, LPS-induced NF-κB activation as measured by EMSA was also enhanced in AIP1-KO EC (Fig. 1e). Interestingly, Pam3CSK4 (TLR2 ligand) and LPS (TLR4 ligand) showed a similar pattern of p38 MAPK and NF-κB activation (supplemental Fig. S1c). Consistent with these results, overexpressed AIP1 in 293T cells inhibits TLR4-mediated MAPK and NF-κB activation in reporter gene assays (Fig. 1, f and g).
FIGURE 1.
AIP1 inhibits LPS-induced NF-κB and MAPK activation. Effects of AIP1 in LPS-induced NF-κB and MAPK activation in MLEC. WT and AIP1-KO MLEC (1 × 106) were treated with LPS (1 ng/ml) for the indicated times. Phospho- and total IκBβ and p38 were determined by immunoblotting with the respective antibodies (a). The quantification of total IκBβ, and the ratios of p-IκBβ/IκB and p-p38/p38 are presented in b–d. Nuclear extracts from WT and AIP1-KO EC (0, 30, and 60 min) were also prepared for EMSA using a NF-κB probe (e). f and g, effects of AIP1 in TLR4-mediated NF-κB and AP-1 activation. 293T cells (1 × 106) were transfected with the indicated reporter gene (0.l μg) and expression plasmids (0.5 μg). Data are the means ± S.D. from three independent experiments. Rel. Lucif. Act., relative luciferase activity.
AIP1 Disrupts the Complex Formation of TLR4-MyD88- TIRAP
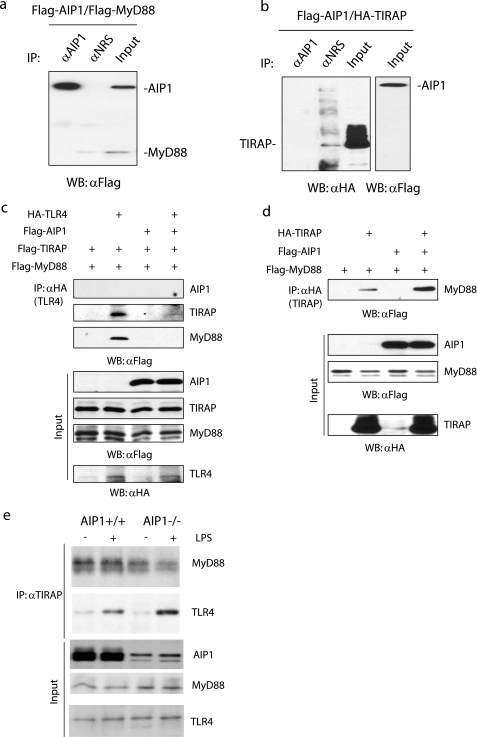
TLR4 and TLR2, but not IL-1R, mediate NF-κB and MAPK activation by recruiting two pivotal adaptor proteins, MyD88 and TIRAP. TIRAP binds to PIP2 sites on the plasma membrane, and then facilitates the association between TLR4 and MyD88, which in turn functions as a platform to recruit other downstream molecules. To determine how AIP1 inhibits TLR4 signaling, we first examined whether AIP1 could block TLR4 signaling complex formation via directly binding to MyD88, TIRAP, or TLR4. Although we could not detect AIP1 associations with any of them, TLR4-TIRAP-MyD88 complex formation was diminished in the presence of AIP1 (Fig. 2, a–c). Interestingly, we found that AIP1 dramatically enhanced the association of TIRAP with MyD88 (Fig. 2d). We next determined the effect of AIP1 on endogenous TIRAP-MyD88 and TLR4-TIRAP-MyD88 complexes in the absence or presence of the TLR4 ligand LPS. WT and AIP1-KO mouse EC were untreated or treated with LPS. TIRAP-MyD88 was readily detected in resting WT EC and was slightly decreased by LPS treatment. Consistent with the overexpression system where AIP1 enhances TIRAP-MyD88 interaction, TIRAP-MyD88 association was decreased in AIP1-KO EC (Fig. 2e). In contrast, TIRAP-TLR4 association was not detected in the resting state but was strongly induced in response to LPS. Moreover, this interaction was augmented in AIP1-KO cells (Fig. 2e). Taken together, these results indicate that TIRAP-MyD88 can form independently of the TLR4-TIRAP-MyD88 complexes, and AIP1 has differential effects on these complexes. Most importantly, these data suggest that AIP1-mediated disruption of the TLR4-TIRAP-MyD88 complex is not dependent on its direct associations with the TLR4 signaling components.
FIGURE 2.
AIP1 disrupts the complex formation of TLR4-MyD88-TIRAP. a, AIP1 does not associate with MyD88. FLAG-AIP1 and FLAG-MyD88 expression plasmids were co-transfected into 293T cells. Association of AIP1 with MyD88 was determined by immunoprecipitation (IP) with anti-AIP1 followed by immunoblotting with anti-FLAG. A normal rabbit serum was used as a control. WB, Western blot. b, AIP1 does not associate with TIRAP. FLAG-AIP1 and HA-TIRAP were co-transfected into 293T cells. Association of AIP1 with TIRAP was determined by immunoprecipitation with anti-AIP1, followed by immunoblotting with anti-HA. c and d, AIP1 reduces TLR4-TIRAP-MyD88 complexes but enhances associations of TIRAP-MyD88. 293T cells were transfected with FLAG-TIRAP, HA-TLR4, and FLAG-MyD88 (c) or HA-TIRAP and FLAG-MyD88 (d) in the absence or presence of FLAG-AIP1. Association of TLR4-TIRAP-MyD88 and TIRAP-MyD88 was determined by immunoprecipitation with anti-HA, followed by immunoblotting with anti-FLAG. e, effect of AIP1 on endogenous TIRAP-MyD88 and TLR4-TIRAP-MyD88 complexes. WT and AIP1-KO mouse EC were untreated or treated with LPS (1 ng/ml for 15 min). Associations of endogenous TIRAP-MyD88 and TIRAP-TLR4 were determined by immunoprecipitation with anti-TIRAP, followed by immunoblotting with anti-MyD88 and TLR4, respectively.
Furthermore, we observed that endogenous AIP1 exhibited plasma membrane localization in WT mouse EC cells (supplemental Fig. S2a). Overexpression of AIP1 in bovine aortic endothelial cells and Cos7 cells showed a similar pattern of membrane localization as well as cytoplasmic staining. This staining pattern is similar to the reported localization of TIRAP in macrophages (15) and in Cos7 cells (supplemental Fig. S2b). Not surprisingly, both AIP1 and TIRAP appeared on the plasma membrane, where they displayed a co-localization pattern (supplemental Fig. S2c).
AIP1 and TIRAP Bind to Distinct Lipids Present on the Plasma Membrane
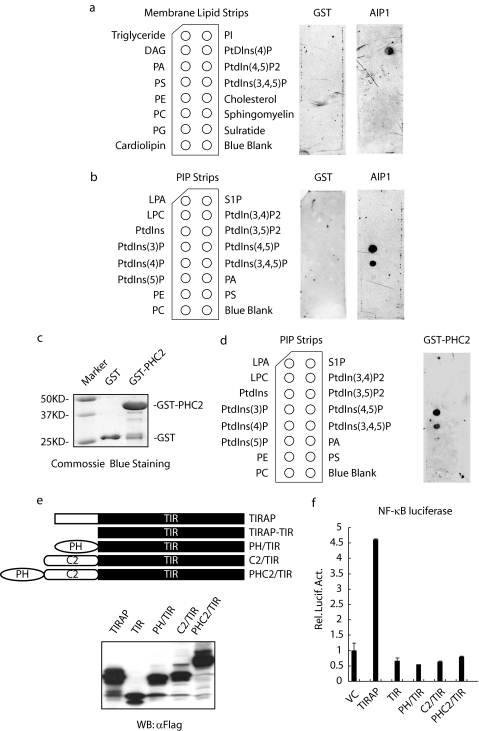
The membrane location of TIRAP is dependent on the lipid composition of PIP2. AIP1 contains two predicted lipid binding domains at its N terminus, the pleckstrin homology (PH) domain and the C2 domain. One possibility is that AIP1 disrupts TLR4-TIRAP-MyD88 by competing with TIRAP for PIP2 binding on the plasma membrane. To test this, we examined the lipid binding profiles of AIP1 in the membrane lipid assay, using recombinant GST-AIP1. The full-length AIP1 specifically associated with PIP4, but not with PIP2 or PIP3 (Fig. 3a). We further analyzed the lipid binding profiles of AIP1 in a PIP strip assay. AIP1 specifically bound to PI4P as well as PI3P, a lipid present in other organelles, including endocytic vesicles (Fig. 3b).
FIGURE 3.
AIP1 and TIRAP bind to distinct lipids present on the plasma membrane. a and b, membrane lipid strip (a) and PIP strip assays (b) were performed using recombinant GST-AIP1 protein. GST was used as a control. The left panel indicates the identity of each lipid spot. DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; LPA, lysophosphatidic acid; LPC, lysophosphocholine. c and d, PH/C2 domains of AIP1 are sufficient for lipid binding. GST-PHC2 was purified (c) and was used for the PIP strip assay (d) as described. S1P, sphingosine 1-phosphate; VC, vector control. e and f, replacement of the PIP2 binding domain in TIRAP by the lipid binding domains (PH, C2, or PHC2 domain) did not restore the TIRAP activity in terms of NF-κB activation. A schematic diagram for truncated AIP1-TIRAP and its expression in 293T cells shown in e. The κB-dependent reporter gene activation by truncated AIP1-TIRAP in 293T cells is presented in f. Rel. Lucif. Act., relative luciferase activity.
To determine whether the PH domain or the C2 domain is required for lipid binding, we created truncated GST-AIP1 with a deletion of the PH domain (ΔPH), the C2 domain (ΔC2), or both (AIP1-C). An in vitro lipid binding assay showed both ΔPH and ΔC2 retained similar lipid binding profiles to the full-length AIP1. AIP1-C failed to bind to PI4P, although it still bound to PI3P, suggesting that the PH/C2 domains are required for PI4P binding (supplemental Fig. S3). To determine whether the PH/C2 domains are sufficient for PI4P binding, GST-PHC2 containing only the PH/C2 domains was used for the same lipid binding assay. GST-PHC2 bound to both PI3P and PI4P. Taken together, these results demonstrate that AIP1 does not bind to PIP2, but instead, AIP1 via the PH and C2 domains, binds to PI4P, a precursor PIP2 (Fig. 3, c and d).
PIP2 binding is required for TIRAP function. The PIP2-binding motif of TIRAP can be replaced by a different PIP2-binding motif, but not by other non-PIP2 membrane-targeting motifs, to restore TIRAP activity in mediating the TLR4 signaling (8, 15). We reasoned that the AIP1 membrane targeting domains would not rescue TIRAP function upon replacing the TIRAP lipid-binding motif. To test this, a series of chimeras in which the lipid binding domain (PH, C2, or both) from AIP1 was fused to the TIRAP-TIR domain, and NF-κB activation was determined in a κB-reporter gene assay. None of the chimeras could restore the activity of TIRAP-TIR in terms of NF-κB activation (Fig. 3, e and f). Taken together, these data confirm that the lipid binding domain of AIP1 is functionally distinct from that of TIRAP. These data suggest that AIP1 is recruited to the plasma membrane via PI4P, a lipid precursor of PIP2, excluding the possibility that AIP1 inhibits the TLR4 signaling pathway by competing with TIRAP for PIP2 binding.
AIP1 Down-regulates the Cellular PIP2 Level by Acting as a Novel Arf6 GTPase-activating Protein
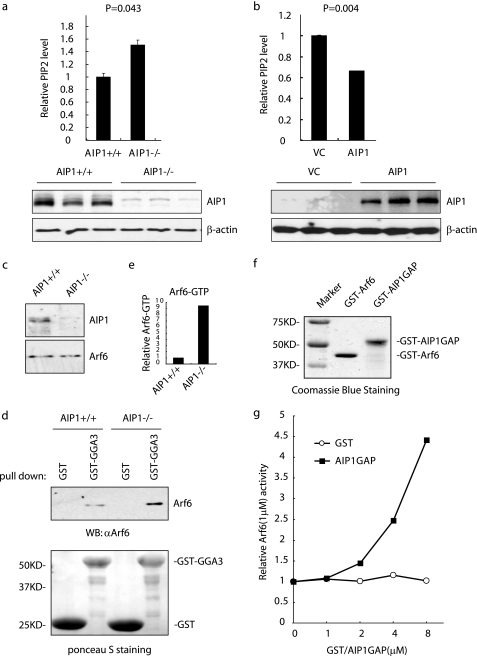
We next examined whether AIP1 alters PIP2 levels in EC to regulate TLR4-TIRAP-MyD88 signaling. The cellular levels of PIP2 were measured in WT and AIP1-KO mouse EC by enzyme-linked immunosorbent assay, and the PIP2 level was significantly increased in AIP1-KO EC (Fig. 4a). Conversely, overexpression of AIP1 in EC strongly reduced the PIP2 level (Fig. 4b). These results indicate that AIP1 disrupts the LPS-TLR4 signaling pathway by down-regulating PIP2 production.
FIGURE 4.
AIP1 down-regulates cellular PIP2 level by acting as a novel Arf6 GTPase-activating protein. a and b, deletion of AIP1 increases and overexpression of AIP1 reduces the PIP levels in EC. Triplicates from each group were performed. AIP1 and β-actin expression from triplicates were detected by Western blot with respective antibodies. a, WT and AIP1-KO MLEC are compared. b, AIP1-KO MLEC were transfected with control (VC) plasmid or AIP1 plasmid by electroporation, and cellular PIP2 levels were measured by enzyme-linked immunosorbent assay. The absorbance values of WT group (a) and AIP1-KO VC group (b) were normalized to 1. Data are the means ± S.D. from three independent experiments. c–e, Arf6 activity is increased in AIP1-KO cells. The total Arf6 in WT and AIP1-KO MLEC was determined by immunoblotting with anti-Arf6 (c). Arf6-GTP form from WT and AIP1-KO MLEC were determined by a GST-GGA3VHS-GAT pulldown assay. Bound Arf6 was determined by immunoblotting with anti-Arf6. GST was used as a control. GST and GST-GGA3VHS-GAT on the membrane were visualized by Ponceau S staining (d). WB, Western blot. Densitometry quantification of bound Arf6 in WT and AIP1-KO MLEC is shown in e with normalization of WT as 1.0. f and g, GAP domain of AIP1 functions as Arf6 GAP. The in vitro GAP activity of Arf6 was determined using 1 mm Arf6 protein in the absence or presence of GST-AIP1 at the concentrations indicated. GST was used as a control. GTP hydrolysis was determined by spectrometry at 635 nm. Relative Arf6 (1 μm) activities were quantified in f, and the basal value without AIP1 was normalized to 1.
Arf6 has been shown to be a critical regulator of PIP2 synthesis by activating PIP5K (22). Therefore, blocking the activity of Arf6 interferes with TLR4-MyD88-dependent signaling (15). We did not detect associations of AIP1 with any of the three isoforms (α, β, and γ) of PIP5K (supplemental Fig. S4). Because AIP1 is a Ras GTPase-activating protein (Ras-GAP), we tested whether AIP1 functions as an Arf6-GAP, reducing cellular PIP2 levels. Arf6 activity was measured by a GST-GGA3 pulldown assay. GGA3VHS-GAT is 1–313-amino acid truncation mutant of ADP-ribosylation factor-binding protein GGA3, which only interacts with an active form of Arf6 (Arf6-GTP) but not the inactive form Arf6-GDP (23). We first verified this assay by using 293T cell lysates overexpressing HA-tagged Arf6WT, Arf6Q67L (a constitutively active form of Arf6), or Arf6T27N (a dominant negative form (of Arf6)). Indeed, GST-GGA3VHS-GAT pulled down more Arf6Q67L than Arf6WT. However, Arf6T27N was not pulled down by GST-GGA3VHS-GAT (supplemental Fig. S5a). We then assessed the endogenous Arf6-GTP level in WT and AIP1-KO mouse EC. The total levels of Arf6 in WT and AIP1-KO mouse EC were similar. However, the level of Arf6-GTP in AIP1-KO mouse EC was much higher than WT mouse EC (Fig. 4c). In contrast, activity of Arf1, another member of the Arf family known to regulate PIP5K, was not affected by AIP1 deletion (supplemental Fig. S5b). These data suggest that AIP1 specifically inhibits Arf6 activity in EC.
To determine whether AIP1 directly functions as an Arf6-GAP, we performed an in vitro GAP activity assay. Like other small GTPases (24), Arf6 has a very low intrinsic GTPase activity. However, the GTP hydrolysis activity of Arf6 was dramatically increased in the presence of AIP1-GAP (Fig. 4d).
Arf6-GAP Activity of AIP1 Is Critical in Regulation of TLR4 Signaling Pathway
To determine the functional significance of Arf6 regulation by AIP1, we first examined the cellular localization of AIP1 and Arf6. Arf6 is localized on the cellular membrane and recycles between the plasma membrane and endocytic compartments during its GTPase cycle. GTP-bound Arf6 resides on the plasma membrane, and the hydrolysis of bound GTP enhanced by Arf6 GAPs is necessary for Arf6 translocation to endocytic organelles. Return of Arf6 to the cell surface occurs upon its activation by Arf6 guanine nucleotide exchange factors (25). Indeed, Arf6-WT was detected in both plasma membrane and endocytic vesicles (Fig. 5a), although Arf6T27N was predominantly detected in endocytic vesicles. The co-localization of AIP1 with Arf6-WT was more pronounced than Arf6T27N (supplemental Fig. S6a), suggesting that AIP1 preferentially targets the active form of Arf6.
FIGURE 5.
Arf6-GAP activity of AIP1 is critical in the regulation of the TLR4 signaling pathway. a, AIP1 is co-localized with Arf6 on both the plasma membrane and endosomes. AIP1 and Arf6 expression plasmids were co-transfected into Cos7 cells. Localization of AIP1 and Arf6 was determined by indirect immunofluorescence with anti-FLAG (AIP1) and anti-HA (Arf6) followed by Alexa Fluor-488 donkey anti-rabbit IgG and Alexa Fluor-599 donkey anti-mouse IgG. DAPI, 4′,6-diamidino-2-phenylindole. b, AIP1 inhibits Arf6-mediated TLR4 signaling. The κB reporter gene was transfected with Arf6 alone or Arf6 plus TLR4 in the absence or presence of AIP1-WT, AIP1 mutant with deletion of the GAP1 domain (AIP1-ΔGAP), or a single mutation at Arg-289 (AIP1-R289). The Arf6/TLR4 group was treated with LPS (1 ng/ml) for 8 h prior to the reporter gene assay analysis. VC, vector control. Rel. Lucif. Act., relative luciferase activity. c, GAP domain is critical for AIP1-mediated inhibition on LPS-induced signaling events. AIP1-KO MLEC were reconstituted with the indicated AIP1 constructs followed by treatment with LPS for the indicated times. LPS-induced activation of NF-κB and MAPK was determined. The relative levels of p-IκBα and ratios of p-p38/p38 are shown below the blots, with the untreated control group as 1.0. d, nuclear extracts from vector control, AIP1-WT, and AIP1-ΔGAP (0, 30, and 60 min) were prepared for EMSA using an NF-κB probe.
We then determined whether AIP1 inhibits Arf6-mediated TLR4 signaling. Overexpression of WT or Arf6Q67L, but not Arf6T27N, induced both basal and TLR4-mediated activation of κB-reporter gene (Fig. 5b and supplemental Fig. S6b). Co-expression of AIP1, but not a mutant AIP1 with a deletion of the GAP domain (AIP1-ΔGAP), significantly suppressed Arf6-WT-mediated NF-κB activation (Fig. 5b). AIP1, initially identified as Ras-GAP (12, 14, 26), shows similarities as well as differences from Arf-GAPs. Specifically, Arg-289 of AIP1, a residue critical for Ras-GAP activity, is absent in Arf-GAPs (supplemental Fig. S6c). To exclude the possibility that AIP1 is acting on Ras to regulate Arf6 activation, we determined the effect of AIP1-R289L, a mutant defective in Ras-GAP activity, on Arf6-WT-mediated NF-κB activation. AIP1-R289L had the same effect on Arf6-induced NF-κB activity (Fig. 5b). These data suggest that AIP1 directly inhibits Arf6.
Finally, we determined if AIP1 GAP activity is critical in inhibiting LPS signaling in mouse EC. To this end, AIP-KO mouse EC was reconstituted with AIP1-F or AIP1-ΔGAP. Cells were treated with LPS and LPS-activated MAPK, and NF-κB was determined by phospho-specific antibodies. Re-expression of AIP1-WT, but not AIP1-ΔGAP in AIP-KO mouse EC, significantly blocked LPS-induced signaling compared with AIP-KO mouse EC reconstituted with a control vector (Fig. 5c). Similar results of AIP1 on NF-κB activation were obtained by EMSA (Fig. 5d). These data confirm that AIP1 suppresses LPS-induced signaling in a GAP activity-dependent manner.
In conclusion, we have identified AIP1 as a novel Arf6-GAP that inhibits Arf6-mediated PIP2 production, in turn leading to an inhibition of the PIP2-dependent and TIPAP-mediated formation of the TLR4-TIRAP-MyD88 complex, and ultimately the activation of the downstream NF-κB and MAPK signaling pathways (supplemental Fig. S7). Consistent with this model, a genetic deficiency of AIP1 in mice (AIP1-KO) causes hypersensitivity to LPS challenge.5 With AIP1-KO mice readily available in our laboratory, this study warrants further investigation of the roles of AIP1 in the LPS-TLR4 response and innate immunity.
Supplementary Material
Acknowledgments
We thank Drs. Rusaln Medzhitov, Dan Wu, and Jordan S. Pober for discussion and critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL-65978-5 and P01HL070295-6. This work was also supported by National Nature Science Foundation of China Grant 30828032 (to W. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
T. Wan, T. Liu, H. Zhang, and W. Min, unpublished observations.
- TLR
- Toll-like receptor
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- MAPK
- mitogen-activated protein kinase
- PH
- pleckstrin homology
- PI4P
- phosphatidylinositol 4-phosphate
- GAP
- GTPase-activating protein
- LPS
- lipopolysaccharide
- TIR
- TLR/IL1R
- TIRAP
- TLR/IL1R-domain-containing adaptor protein
- PIP5K
- phosphatidylinositol 4-phosphate 5-kinase
- WT
- wild type
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- EMSA
- electrophoresis mobility shift assay
- EC
- endothelial cells
- IL-1
- interleukin 1
- HA
- hemagglutinin.
REFERENCES
- 1.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 2.Janeway C. A., Jr., Medzhitov R. (2002) Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 3.Takeda K., Akira S. (2005) Int. Immunol. 17, 1–14 [DOI] [PubMed] [Google Scholar]
- 4.Arbour N. C., Lorenz E., Schutte B. C., Zabner J., Kline J. N., Jones M., Frees K., Watt J. L., Schwartz D. A. (2000) Nat. Genet. 25, 187–191 [DOI] [PubMed] [Google Scholar]
- 5.Carty M., Goodbody R., Schröder M., Stack J., Moynagh P. N., Bowie A. G. (2006) Nat. Immunol. 7, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 7.Horng T., Barton G. M., Flavell R. A., Medzhitov R. (2002) Nature 420, 329–333 [DOI] [PubMed] [Google Scholar]
- 8.Horng T., Barton G. M., Medzhitov R. (2001) Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 10.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 11.Oshiumi H., Sasai M., Shida K., Fujita T., Matsumoto M., Seya T. (2003) J. Biol. Chem. 278, 49751–49762 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., Takeda K., Akira S. (2002) Nature 420, 324–329 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., Akira S. (2003) Nat. Immunol. 4, 1144–1150 [DOI] [PubMed] [Google Scholar]
- 15.Kagan J. C., Medzhitov R. (2006) Cell 125, 943–955 [DOI] [PubMed] [Google Scholar]
- 16.Kagan J. C., Su T., Horng T., Chow A., Akira S., Medzhitov R. (2008) Nat. Immunol. 9, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson R. A., Boronenkov I. V., Doughman S. D., Kunz J., Loijens J. C. (1999) J. Biol. Chem. 274, 9907–9910 [DOI] [PubMed] [Google Scholar]
- 18.Donaldson J. G. (2003) J. Biol. Chem. 278, 41573–41576 [DOI] [PubMed] [Google Scholar]
- 19.D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 20.Zhang H., Zhang R., Luo Y., D'Alessio A., Pober J. S., Min W. (2004) J. Biol. Chem. 279, 44955–44965 [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., Yin G., Surapisitchat J., Berk B. C., Min W. (2001) J. Clin. Invest. 107, 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda A., Nogami M., Yokozeki T., Yamazaki M., Nakamura H., Watanabe H., Kawamoto K., Nakayama K., Morris A. J., Frohman M. A., Kanaho Y. (1999) Cell 99, 521–532 [DOI] [PubMed] [Google Scholar]
- 23.Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. (2000) J. Cell Biol. 149, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCudden C. R., Hains M. D., Kimple R. J., Siderovski D. P., Willard F. S. (2005) Cell. Mol. Life Sci. 62, 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radhakrishna H., Donaldson J. G. (1997) J. Cell Biol. 139, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z., Tseng C. P., Pong R. C., Chen H., McConnell J. D., Navone N., Hsieh J. T. (2002) J. Biol. Chem. 277, 12622–12631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.