Abstract
A change of cytosolic pH 7 to 4 opens the bacterial potassium channel KcsA. However, the overall gating mechanism leading to channel opening, especially the contribution of the cytoplasmic domain, remains unsolved. Here we report that deletion of the cytoplasmic domain resulted in changes in channel conductance and gating behavior at pH 4 without channel opening at pH 7. To probe for rearrangements in the cytoplasmic domain during channel opening, amino acid residues were substituted with cysteines and labeled with a fluorophore (tetramethylrhodamine maleimide) that exhibits increased fluorescence intensity upon transfer from a hydrophilic to hydrophobic environment. In all cases channel open probability (Po) was ∼1 at pH 4 and ∼0 at pH 7. Major increases in fluorescence intensity were observed for tetramethylrhodamine maleimide-labeled residues in the cytoplasmic domain as pH changed from 7 to 4, which suggests the fluorophores shifted from a hydrophilic to hydrophobic environment. Dipicrylamide, a lipid soluble quencher, reduced the fluorescence intensities of labeled residues in the cytosolic domain at pH 4. These results reveal that a decrease in pH introduces major conformational rearrangements associated with channel opening in the KcsA cytoplasmic domain.
Keywords: Ion Channels, Liposomes, Potassium Channels, Protein Conformation, Protein Structure
Introduction
Ion-channels are membrane proteins that regulate cell function by controlling the membrane ion permeability (1). Many ion channels are regulated by stimuli at their cytoplasmic domain. For example, BK, a large conductance calcium-activated potassium channel, is activated when calcium binds to its intracellular binding site (2). Other channels like the hyperpolarization-activated cyclic nucleotide-gated cation channel and the cyclic nucleotide-gated channel are activated by the direct binding of cyclic nucleotides to their cytoplasmic binding domains (3). These bindings give rise to rearrangements in the cytoplasmic domains and stabilize the channels in their open states. In the case of the inward rectifier K+-channel KirBac1.1, gating is proposed to be controlled by the interaction between the cytoplasmic domains and the membrane (4). Thus, the cytoplasmic domains of these channels are important in regulating their functions. Furthermore, in each case it is thought that changes in the cytoplasmic domain are transmitted through a linker to the channel core region, which includes the channel pore and leads to gate opening.
KcsA2 is a minimal potassium channel from Streptomyces lividans. It consists of a transmembrane pore, an α-helix at the N terminus, and a cytoplasmic domain made of 35 amino acids at the C terminus of each subunit. It is a tetramer with four homologous subunits, each of which has two transmembrane segments, TM1 and TM2 (5, 6). The closed structure of the KcsA has been solved (7), whereas the open structure has not. In general, although the molecular mechanism for ion conduction has been revealed (8), that for gating is still unknown. The KcsA channel gating mechanism is of particular importance because this channel is a relatively simple potassium channel, and therefore, its mechanism can likely be used as a model for more complicated channels. Jiang et al. (9) have proposed a model by combining the closed structure of KcsA and the open structure of MthK, which is another potassium channel whose open structure is known. According to their model, which is currently the most accepted model regarding KcsA gating, the bundle crossing, which is composed of four TM2 segments, forms the gate.
KcsA is referred to as a pH-gated channel because its open probability is higher at acidic pH on the intracellular side of the membrane than at neutral pH. Acidic pH increases the open probability indirectly by destabilizing the closed state (10). Because the intracellular pH of S. lividans remains near neutral during opening, the physiological stimulus for opening the KcsA channel is unknown (11). To understand the entire gating mechanism, we need to understand the changes in structure associated with gating and the physiological stimulus. It has been reported from NMR spectroscopy (12) and surface plasmon resonance studies (13) that there are several parts of the channel that rearrange dramatically during channel gating. These studies showed that there are at least two different conformations that respond to changes in pH and that large rearrangements occur around specific regions including the selectivity filter (Glu-71—Asn-80), the C terminus of TM2 (Ala-98—Gly-116), and the cytoplasmic domain.
In this study we measured conformational changes in a KcsA channel by detecting changes in fluorescence intensity to determine the position change of amino acid residues associated with gating in the cytoplasmic domain. As a result, the part of the channel ranging from the C terminus of TM2 to the adjacent part of the cytoplasmic domain was found to be in a hydrophobic environment when the channel was activated at pH 4, whereas at pH 7, when the channel was closed, it was in a hydrophilic environment. We also measured the fluorescence resonance energy transfer between fluorophores at the cytoplasmic domain in the presence of dipicrylamide (DPA), a membrane-localized quencher (14) that lowers fluorescence intensities at acidic pH. These results suggest that the region from the C terminus of TM2 to the adjacent part of the cytoplasmic domain moves largely toward the membrane when the gate opens.
EXPERIMENTAL PROCEDURES
Constructs and Mutants
KcsA cloned into pQE-30 vectors including an N-terminal hexahistidine tag were kindly provided by Dr. Kubo of the National Institute of Advanced Industrial Science and Technology. KcsAΔ125–160, KcsAΔ135–160, and KcsAΔ145–160 were obtained by introducing a stop codon at positions 125, 135, and 145, respectively. Mutations were performed with the QuikChangeTM site-directed mutagenesis kit (Stratagene). In addition, all mutated channels had an E71A mutation that prevented gate inactivation (15).
Protein Expression and Purification
pQE-30 plasmids containing the KcsA sequence were transformed into Escherichia coli XL1-Blue and overexpressed by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm. Expressed channels were extracted from membrane fractions by 10 mm n-dodecyl β-d-maltoside (Dojin). Co2+ affinity gel beads (TALON Metal Affinity Resins, Clontech) equilibrated with normal buffer (5 mm n-dodecyl β-d-maltoside, 20 mm Tris-HCl, pH 7.6, 100 mm KCl) were added to the extracted channel protein solution and incubated for 30 min at 4 °C to bind them to histidine tags. Nonspecific bound proteins were removed with wash buffer (normal buffer containing 20 mm imidazole). The channel proteins were eluted with elution buffer (normal buffer containing 400 mm imidazole).
Fluorescence Labeling
Mutant channels were fluorescently labeled by mixing 2.5 μΜ proteins with an equimolecular amount of tetramethylrhodamine (TMR) maleimide followed by incubation for 12–16 h on ice. Excess dyes were removed by a Co2+ affinity gel column. Labeled channels were eluted with elution buffer.
Reconstitution into Liposomes
The phospholipid mixture (POPE:POPG = 3:1) in chloroform was evaporated and suspended in S buffer (450 mm KCl, 4 mm NMG, 20 mm HEPES, pH 7.2) to a concentration of 10 mg lipid/ml followed by sonication. CHAPS was added to the suspension to a final concentration of 34 mm and incubated for 2 h. Then all proteins eluted from the affinity column (0.5 nmol) and lipid suspension were mixed together and incubated for 20 min. This mixture was dialyzed against S buffer containing BIO-BEADs for 3 days to remove detergents and reconstitute it into liposomes. The liposome suspension was centrifuged at 100,000 × g for 20 min at 4 °C. The precipitation was resuspended with 250 μl of a solution containing 100 mm KCl, 300 mm sucrose, and 10 mm Tris-Hepes, pH 7.2. Labeling with TMR was confirmed by SDS-PAGE and, following fluorescence measurements, with Molecular Imager (Bio-Rad). The liposome suspension was stored at −80 °C.
Channel Current Recordings
Channel currents were measured by the planar bilayer method (16, 17). Bilayers were made by painting a lipid solution (POPE:POPG = 3:1 in n-decane) across a small hole on a thin plastic sheet. Currents were recorded in a symmetrical solution containing 200 mm KCl and 10 mm Tris-MES, pH 4.0. The bath solution was held at virtual ground such that voltage at the upper solution connected to a patch clamp amplifier by an Ag-AgCl electrode-defined membrane potential.
Fluorescence Measurements
A liposome suspension containing fluorescently labeled proteins (20 μl) was diluted to 200 μl with 200 mm KCl with 10 mm Tris-MES at pH 4 or 7 and centrifuged at 100,000 × g for 20 min at 4 °C. The precipitation was suspended with the same solution (200 μl) and sonicated to equilibrate internal and external solution. Fluorophores were excited at 532 nm, and emission spectra were obtained with a fluorescence spectrometer (RF-5300PC, Shimadzu). To measure DPA quenching effects, 0.2 mm DPA was added before the centrifugation.
RESULTS
Effects of Deleting the Cytoplasmic Domain on Activity
To investigate the effect of the cytoplasmic domain on KcsA channel gating, we made three cytoplasmic domain deletion mutants, KcsAΔ125–160, KcsAΔ135–160, and KcsAΔ145–160, and investigated their properties. Each channel had an E71A mutation, which made the channel non-inactivating. Wild type KcsA channels have a very low open probability even at acidic pH because of gate inactivation (15). However, it is known that removing electric charges from residue Glu-71 results in shortening the inactivation time and increasing the open probability (15, 18).
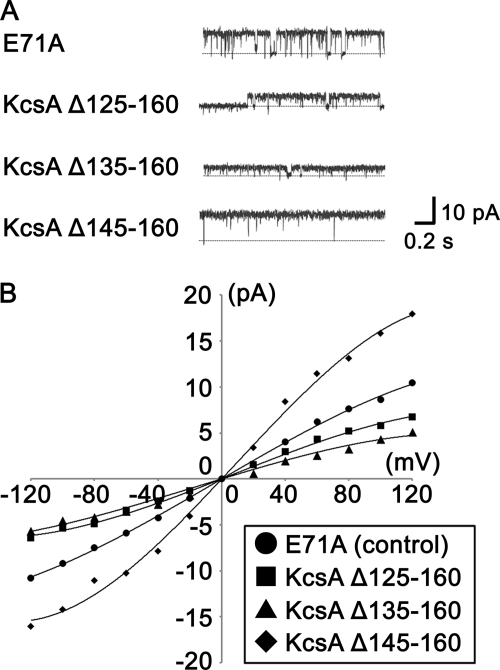
To measure the activities of cytoplasmic domain deletion mutants, the mutants were reconstituted into liposomes, inserted into planar bilayers, and had their single channel currents measured. Single channel records were taken in a symmetrical solution of 200 mm KCl at pH 4.0, which is a condition that promotes the opening of the wild type channel. Fig. 1A shows typical single channel records for the cytoplasmic domain deletion mutants at 60 mV. Fig. 1B is the single channel I-V relationships for the mutants. The lowest conductance of the six measurements is shown. As summarized in Table 1, the chord conductance at 100 mV of KcsAΔ125–160 was much smaller than that of full-length KcsA (E71A), whereas the chord conductance of KcsAΔ145–160 was larger. KcsAΔ135–160 had various conductances ranging between 43 and 90 pS. In addition, the open probability of KcsAΔ125–160 was lower. These changes in channel properties indicate that the cytoplasmic domain has an important role in regulating function.
FIGURE 1.
Current measurement of cytoplasmic domain deletion KcsA mutants. Single channel currents of mutant channels reconstituted into an artificial planar lipid bilayer were recorded in a symmetrical solution containing 200 mm KCl and 10 mm Tris-MES, pH 4.0. A, representative current records of KcsA E71A (control), KcsAΔ125–160, KcsAΔ135–160, and KcsAΔ145–160 at 60 mV are shown. B, I-V relationships of cytoplasmic domain deletion mutants are shown. ●, KcsA E71A; ■, KcsAΔ125–160; ▴, KcsAΔ135–160; ♦, KcsAΔ145–160.
TABLE 1.
Channel properties of cytoplasmic domain deletion KcsA mutants
Chord conductances and open probabilities of full-length KcsA(E71A) and cytoplasmic domain deletion KcsA mutants were measured at 100 mV in a symmetrical solution containing 200 mm KCl and 10 mm Tris-MES, pH 4.0 (mean ± S.D., n = 3–9).
| Chord conductances | Open probability | |
|---|---|---|
| pS | ||
| E71A (control) | 92 ± 8 | 0.91 ± 0.06 |
| KcsA Δ125–160 | 50 ± 11 | 0.68 ± 0.13 |
| KcsA Δ135–160 | 43–90 | 0.84 ± 0.10 |
| KcsA Δ145–160 | 144 ± 18 | 0.97 ± 0.04 |
Site-specific Mutagenesis and Fluorescent Labeling
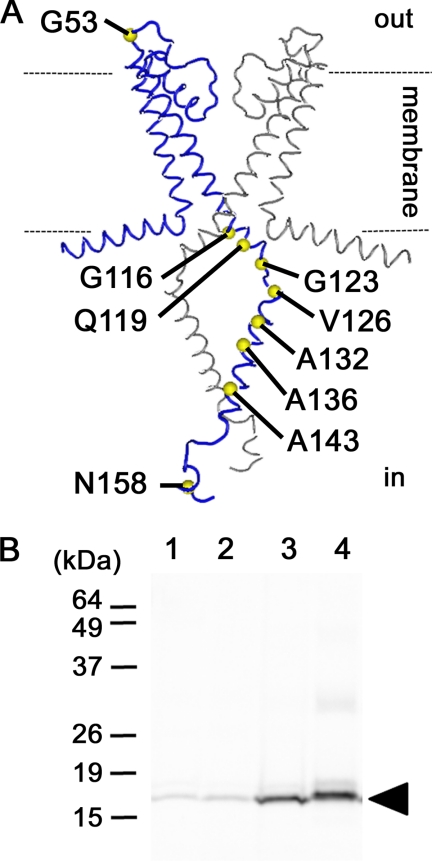
To examine the mechanism that regulates gating, rearrangements of residues in the cytoplasmic domain were investigated. Therefore, certain amino acid residues were fluorescently labeled with TMR-maleimide, and the changes in fluorescence intensities that accompanied these changes in pH were measured. Single cysteine mutants were made in which one amino acid residue was substituted for cysteine to achieve specific labeling with TMR-maleimide. Each channel had an E71A mutation, which made the channel non-inactivating. All single cysteine mutants were successfully expressed in E. coli and purified through a His affinity column. Nine of 13 single cysteine mutants were labeled with TMR-maleimide. Six mutants were labeled in the cytoplasmic domain (G123C, V126C, A132C, A136C, A143C, and N158C), 2 at the interface between the transmembrane and cytoplasmic domains (G116C and Q119C) and 1 in the extracellular loop between the 2 trans-membrane domains (G53C) (Fig. 2A). Fig. 2B shows a fluorography for wild type (lane 1), E71A (lane 2), G53C (lane 3), A132C (lane 4) after each was labeled with TMR-maleimide and reconstituted into liposomes. We confirmed that the single cysteine mutants G53C and A132C were specifically labeled.
FIGURE 2.
Deduced structure of KscA and attachment sites for TMR-maleimide. A, tested mutations are shown on a structure deduced from EPR measurements (Protein Data Bank code 1F6G). Two of four subunits are presented in this figure. Positions where a cysteine was introduced are indicated with a yellow circle. Dotted lines show membrane/solution interfaces. B, purified mutants were labeled with TMR-maleimide and reconstituted into liposomes. Proteins were analyzed by fluorography (Molecular Imager, Bio-Rad) after SDS-PAGE. The arrowhead indicates the KcsA monomer. Lane 1, wild type; lane 2, E71A; lane 3, G53C; lane 4, A132C.
Reconstitution of Labeled Mutants into Liposomes and Channel Activity Measurements
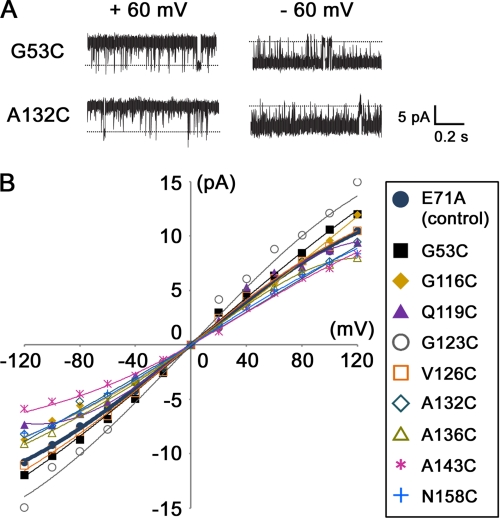
To measure the activities of fluorescently labeled channels, their single channel currents were measured. Fig. 3A shows typical single channel records for channel mutants labeled with TMR, whereas Fig. 3B shows the single channel I-V relationships for various mutants. As a result, all mutants had pH-dependent channel activity. The chord conductance at 100 mV ranged between 70 and 121 pS, whereas the values for wild type and single E71A mutants were 84 and 92 pS, respectively. The open probability at 100 mV ranged between 0.89 and 0.99, which approximated the E71A mutant (0.93). These results indicate that labeling had no significant effect on channel activity for all TMR-labeled mutants.
FIGURE 3.
Current measurement of TMR-labeled KcsA mutants. Single channel currents of mutant channels reconstituted into an artificial planar lipid bilayer were recorded in a symmetrical solution containing 200 mm KCl and 10 mm Tris-MES, pH 4.0. A, representative current records of mutant channels (G53C and A132C) are shown. B, I-V relationships of TMR-labeled KcsA mutants are shown.
Correlation between Open Probability and Fluorescence
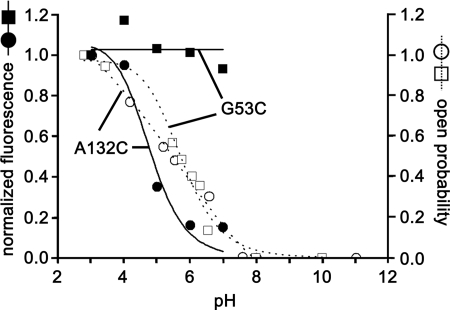
To compare fluorescence with open channel probability, we plotted the pH dependence of fluorescence along with the open probability obtained from channel current recordings. As shown in Fig. 4, the fluorescence of A132C increased with a decrease in pH and correlated very well with the open probability of the channel. Also, the pKa of the fluorescence change (pKa = 4.8) corresponded to the pKa of the KcsA proton sensor (pKa = 5.4). This strongly indicates that fluorophores with high intensity corresponded to channels in the open state, whereas fluorophores with low intensity corresponded to the closed state. We know that fluorescence intensity from TMR is higher in a hydrophobic environment than in a hydrophilic environment (19). Therefore, the change in A132C fluorescence indicates that TMR molecules moved between hydrophilic and hydrophobic environments. In contrast, the fluorescence of G53C did not change significantly, showing even if this site moved during gating, it remained in the same environment.
FIGURE 4.
Correlation between fluorescence and channel open probability. Representative examples of the pH-dependent fluorescence change in mutants G53C and A132C are plotted along the open probability obtained from their channel current recordings. The pKa of the A132C fluorescence change was 4.8. The pKa of the A132C open probability was 5.4.
Fluorescence Measurements at pH 4 and 7
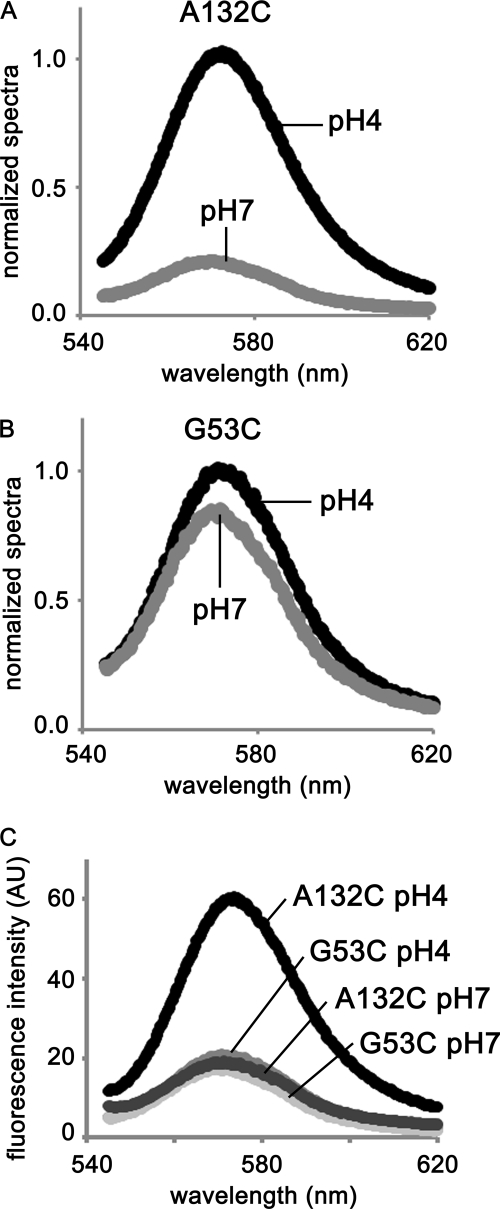
For all tested mutants, the open probability was ∼1.0 in an acidic solution but was 0 in a neutral one. We measured fluorescence at pH 4 and 7 from TMR attached to substituted cysteines. Fig. 5, A and B, show representative results of the emission spectra for A132C and G53C. The fluorescence of A132C was higher at pH 4 than at pH 7, almost 5 times so at 570 nm (Fig. 5A). This result indicates that Cys-132 was in a more hydrophobic environment when the gate was open than when the gate was closed. On the other hand, we did not see any significant differences for G53C between pH 4 and 7 (Fig. 5B). As seen in Fig. 5C, the fluorescence intensity of G53C was, even at pH 4, much lower than that of A132C, indicating that G53C was continuously in a hydrophilic environment.
FIGURE 5.
Emission spectra of TMR-A132C and TMR-G53C. We measured fluorescence intensities of the TMR-labeled KcsA mutants A132C (A) and G53C (B) after reconstituting them into liposomes. The excitation wave length was 532 nm. The data were normalized at 570 nm and pH 4. C, spectra of A132C and G53C are superimposed.
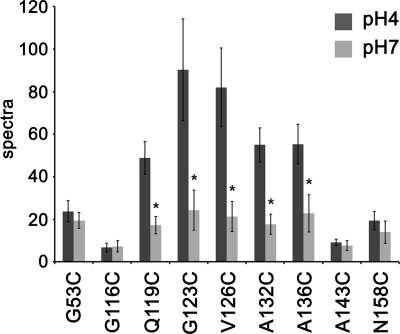
For the other seven TMR-labeled mutants, the fluorescence intensity results are summarized in Fig. 6. These data were normalized for protein concentration. The fluorescence intensities from TMRs located in the cytoplasmic domain (Q119C, G123C, V126C, A132C, and A136C) changed drastically with a change in pH, which shows that these residues were in a hydrophobic environment when the gate opened but switched to a hydrophilic environment when the gate closed. On the contrary, TMR fluorescence at pH 7 for G53C, which faces the extracellular space, for G116C, which locates at the interface between the membrane and cytoplasmic space, and for A143C and N158C, which are in the cytoplasmic domain, were not significantly different from that measured at pH 4. Thus, the environmental change for each of these residues in response to gating and change in pH were smaller compared with the above five residues.
FIGURE 6.
Fluorescence intensities of TMR-labeled KcsA mutants at pH 4 and 7. Fluorescence intensities were measured at 570 nm for nine different mutants at pH 4 and 7. The histogram shows results normalized for protein concentration (mean ± S.D., n = 3–6). *, p < 0.05 compared with values measured at pH 4 for each mutant.
The Cytoplasmic Domain Is in a Hydrophobic Environment When the Gate Opens
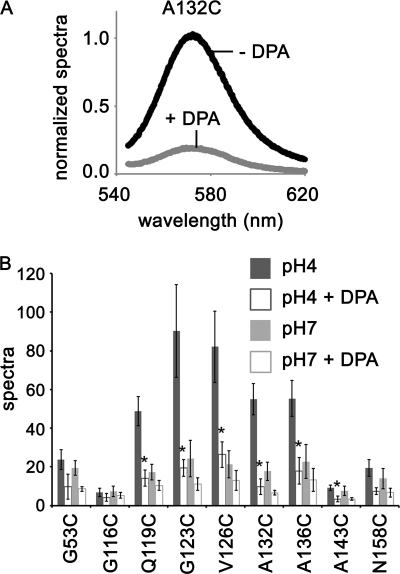
We found that the C terminus of TM2 and the adjacent part of the cytoplasmic domain, in which the aforementioned five residues locate, are in a hydrophobic environment while in the open state. This result implies that either these regions were packed by the four cytoplasmic domains or are located in a hydrophobic environment of the membrane during the open state. To identify which, we measured TMR fluorescence in the presence of the membrane localized quencher DPA. Fig. 7A shows representative results of the emission spectra for A132C at pH 4 in the presence and absence of DPA. The fluorescence of A132C was higher in the absence of DPA than in the presence of DPA. The fluorescence intensity results for the other TMR-labeled mutants are summarized in Fig. 7B. These data were normalized for protein concentration. Quenching effects were also observed at pH 7 even at the end of the C termini (N158C), which is known from crystallography to locate farthest from the membrane (7). Larger quenching effects were detected in the presence of DPA at pH 4 for five mutants (TMR-Q119C, G123C, V126C, A132C, and A136C), each having high intensities in the absence of DPA at pH 4. The fluorescence intensities were not only quenched by DPA but also enhanced by the hydrophobic environment. Therefore, it is likely that quenching effects were underestimated. This indicates that these residues moved into the membrane or in the vicinity of the membrane at pH 4.
FIGURE 7.
Effect of a membrane-localized quencher DPA on TMR-labeled KcsA mutants at pH 4 and pH 7. A, shown are emission spectra for A132C in the presence and absence of DPA. The data were normalized at 570 nm in the absence of DPA. B, fluorescence intensities in the presence and absence of DPA at pH 4 and pH 7 at 570 nm are shown. The histogram shows results normalized for protein concentration (mean ± S.D., n = 3–6). *, p < 0.05 compared with quenching ratio by DPA at pH 7.
DISCUSSION
Although the physiological stimulus for opening the KcsA channel is still unknown, it is likely that intracellular factors are involved. It has been reported that intracellular low pH destabilizes the closed structure, which increases the open probability (10), which suggests pH change regulates gating even if in some cases intracellular pH levels are tightly kept at neutral pH like it is in S. lividans.
To understand the gating mechanism of KcsA physically, structural information regarding full-length KcsA, especially at the cytoplasmic C termini domains, during open and closed states is needed because it is most likely that the cytoplasmic domain regulates gating and likely does so in several other channels too. For example, in the case of the MthK channel, conformational changes in the cytoplasmic domain involving Ca2+ binding sites induced by Ca2+ binding are thought to exert a force on the pore through inner helix extensions (20). Additionally, exchanging the cytoplasmic cyclic nucleotide binding domain of the cyclic nucleotide-gated channel for the cytoplasmic domain of KcsA resulted in a response to cAMP.
Our electrophysiological results indicate that the cytoplasmic domain regulates gating. pH dependences of cytoplasmic domain deletion KcsA mutants were not different from wild type KcsA. However, channel properties such as conductance and open probability changed with a larger deleted region showing a lower conductance (Fig. 1). In addition, KcsAΔ125–160 had a lower open probability. From these changes, we concluded that the cytoplasmic domain does not act as a pH sensor but plays an important role for gate regulation.
Because the fluorescence intensity of the fluorophore TMR is higher in a hydrophobic environment than in a hydrophilic environment, we labeled KcsA with TMR and observed its conformational rearrangements, which allowed us to identify the environment in which TMR-labeled residues resided. KcsA sites were labeled mainly in the cytoplasmic domain and had their fluorescence measured at pH 4 (activated condition) and pH 7 (non-activated condition). At pH 4, KcsA mutants labeled at the C terminus of TM2 (Q119C) or at the adjacent part of the cytoplasmic domain (G123C, V126C, A132C, and A136C) showed high fluorescence intensities. On the other hand, at pH 7 their intensities were low. These results show that these regions were located in a hydrophobic environment in the open state and in a hydrophilic environment in the closed state.
We also determined that the hydrophobic environment included the membrane by observing the quenching of TMR fluorescence by using the membrane-localized quencher DPA. It has been reported that when the gate opens, intersubunit interactions become weak in the cytoplasmic domain, destabilizing the oligomer (13). It is, therefore, unlikely that TMRs were packed into a hydrophobic environment by the four cytoplasmic domains when the gate opened. There is, however, still a possibility that the observed change in fluorescence did not arise from the interaction between TMR and the membrane but from an interaction between TMR and the surrounding amino acid residues in the different subunits. For example, Trp, Tyr, Phe, and His all show quenching effects on TMR fluorescence (21, 22). To ensure that the observed changes in fluorescence corresponding to changes in pH were due to TMR moving from the hydrophilic solution to the hydrophobic membrane, we measured fluorescence in the presence of DPA (Fig. 7). Larger quenching effects were detected in the presence of DPA at pH 4 for five mutants (TMR-Q119C, G123C, V126C, A132C, and A136C), each having high intensities in the absence of DPA at pH 4. When the distance between TMR and DPA or the distance between TMR and the membrane becomes sufficiently close, TMR fluorescence decreases because of fluorescence resonance energy transfer from the TMR to the DPA (14). Hence, quenching of the TMR fluorescence shows that the region discussed above was inside or very close to the membrane. We also detected larger quenching effects for A143C, which did not show the significant difference between fluorescence intensities at pH 4 and 7 (Fig. 6), indicating that the distance between Ala-143 and the membrane was closer at pH 4 than at pH 7 and that this site remained in the same environment. Therefore, it seems that when the channel opened, even if the site moved toward the membrane, it never entered the membrane.
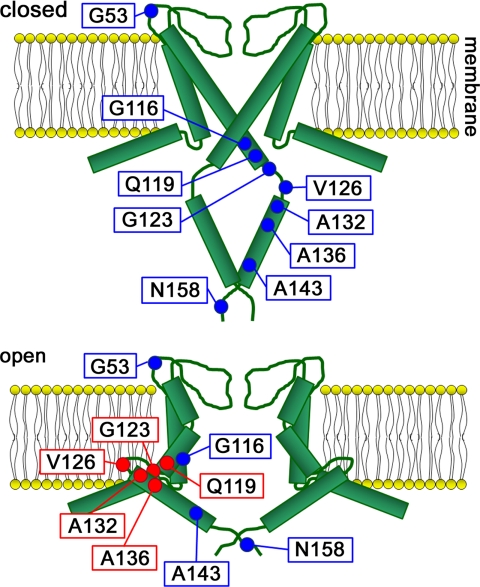
From these results and previous studies, we propose a conformation model (Fig. 8). In the closed state a pH sensing site near the bundle crossing (C terminus of TM2) forms a complex network that stabilizes the closed state (10), and the four cytoplasmic domains form a stable oligomer. In this state the nine labeled residues we observed in this paper are in a hydrophilic environment. When intracellular pH becomes low, the network at the pH-sensing site disrupts (10), and the TM2 helices bend at a hinge point around Gly-99, causing them to splay open (9). This results in the oligomer of the cytoplasmic domains destabilizing (13) and each cytoplasmic domain moving toward the membrane, which widens the gate. Eventually a part of the cytoplasmic domain is located in the membrane or near the membrane to regulate gating. Therefore, the environment around the C terminus of TM2 (Gln-119) and part of the cytoplasmic domain (Ala-123, Val-126, Ala-132, and Ala-136) switch from a hydrophilic to hydrophobic environment.
FIGURE 8.
Model accounting for rearrangements. The four cytoplasmic domains of the subunits form an oligomer in the closed state (pH 7). The oligomer is destabilized in the open state (pH 4) causing the cytoplasmic domains to swing such that they come close to or enter the membrane.
This model can explain the electrophysiological data of the cytoplasmic domain deletion mutants. The cytoplasmic domain plays an important role for pulling the gate wider; therefore, deletions in this region should prevent the gate from easily opening. Consistent with this, the conductance of the cytoplasmic domain deletion mutants KcsAΔ125–160 and KcsAΔ135–160 was smaller. The open probability for KcsAΔ125–160 was also lower. Therefore, the cytoplasmic region may be essential to make the gate open, leading to the characteristics of KcsA gating. However, we did not observe a decrease in the conductance or open probability of KcsAΔ145–160. In fact, the conductance increased. This result suggests that the C termini of the cytoplasmic domain determine the domain position, which would then influence channel activity.
In this study we showed a conformational change occurs at the Gln-119 site during the gating of KcsA. It has been said that the C terminus of TM2, which includes Gly-116 and Gln-119, is an important region for gating. X-ray analysis has indicated that four TM2 helices forming a bundle in the closed state splay open to create a wide entryway in the channel (5, 9). However, we could not detect any change at the Gly-116 site because TMR detects only environmental differences. In other words, Gly-116 remained in the same environment regardless of pH.
To clarify protein function, one needs to do more than solve the protein structure. The protein dynamic behavior must also be studied. This is because flexible regions in the protein are likely very important for its function but are also the most difficult to resolve in structural studies. Here, we used ion channels as a model to demonstrate this point. The large channel rearrangement observed in this study occurred in the cytoplasmic domain, whose structure has not yet been determined at acidic pH because of its flexible nature. In fact, single molecule studies have shown that KcsA channel gating results in large motions that do not synchronize with the channel functions (23), implying that the channel structure cannot uniquely determine when the gate opens, although we do know that the C termini of the cytoplasmic domain are in or in the vicinity of the membrane. Thus, to further clarify the mechanism behind KcsA gating, studies focusing on other factors, especially at the single molecule level, are required.
Acknowledgments
We thank Dr. Tai Kubo (Advanced Industrial Science and Technology, Tukuba, Japan) for sending us the plasmid pQE-30/KcsA. We also thank Dr. Peter Karagiannis for carefully revising the manuscript.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Molecular and System Life Science and Innovative Nanoscience of Supermolecular Motor Proteins Working in Biomembranes) and the Hayashi Memorial Foundation for Female Natural Scientists.
- KcsA
- potassium channels from S. lividans
- DPA
- dipicrylamine
- TMR
- tetramethylrhodamine
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio] propane sulfonate
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]
- MES
- 4-morpholineethanesulfonic acid
- TM
- transmembrane
- pS
- picosiemens.
REFERENCES
- 1.Hille B. (ed) (2001) Ion Channels of Excitable Membranes, Sinauer, Sunderland, MA [Google Scholar]
- 2.Magleby K. L. (2003) J. Gen. Physiol. 121, 81–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craven K. B., Zagotta W. N. (2006) Annu. Rev. Physiol. 68, 375–401 [DOI] [PubMed] [Google Scholar]
- 4.Enkvetchakul D., Jeliazkova I., Bhattacharyya J., Nichols C. G. (2007) J. Gen. Physiol. 130, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 6.Cortes D. M., Cuello L. G., Perozo E. (2001) J. Gen. Physiol. 117, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uysal S., Vásquez V., Tereshko V., Esaki K., Fellouse F. A., Sidhu S. S., Koide S., Perozo E., Kossiakoff A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6644–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morais-Cabral J. H., Zhou Y., MacKinnon R. (2001) Nature 414, 37–42 [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Nature 417, 523–526 [DOI] [PubMed] [Google Scholar]
- 10.Thompson A. N., Posson D. J., Parsa P. V., Nimigean C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6900–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corvini P. F., Gautier H., Rondags E., Vivier H., Goergen J. L., Germain P. (2000) Microbiology 146, 2671–2678 [DOI] [PubMed] [Google Scholar]
- 12.Baker K. A., Tzitzilonis C., Kwiatkowski W., Choe S., Riek R. (2007) Nat. Struct. Mol. Biol. 14, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamoto M., Shimizu H., Inoue F., Konno T., Sasaki Y. C., Oiki S. (2006) J. Biol. Chem. 281, 28379–28386 [DOI] [PubMed] [Google Scholar]
- 14.Taraska J. W., Zagotta W. N. (2007) Nat. Struct. Mol. Biol. 14, 854–860 [DOI] [PubMed] [Google Scholar]
- 15.Cordero-Morales J. F., Cuello L. G., Zhao Y., Jogini V., Cortes D. M., Roux B., Perozo E. (2006) Nat. Struct. Mol. Biol. 13, 311–318 [DOI] [PubMed] [Google Scholar]
- 16.Ide T., Yanagida T. (1999) Biochem. Biophys. Res. Commun. 265, 595–599 [DOI] [PubMed] [Google Scholar]
- 17.Ide T., Takeuchi Y., Yanagida T. (2002) Single Molecules 1, 33–42 [Google Scholar]
- 18.Cordero-Morales J. F., Jogini V., Lewis A., Vásquez V., Cortes D. M., Roux B., Perozo E. (2007) Nat. Struct. Mol. Biol. 14, 1062–1069 [DOI] [PubMed] [Google Scholar]
- 19.Cha A., Bezanilla F. (1997) Neuron 19, 1127–1140 [DOI] [PubMed] [Google Scholar]
- 20.Chakrapani S., Perozo E. (2007) Nat. Struct. Mol. Biol. 14, 180–182 [DOI] [PubMed] [Google Scholar]
- 21.Eftink M. R. (1994) Biophys. J. 66, 482–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Barkley M. D. (1998) Biochemistry 37, 9976–9982 [DOI] [PubMed] [Google Scholar]
- 23.Shimizu H., Iwamoto M., Konno T., Nihei A., Sasaki Y. C., Oiki S. (2008) Cell 132, 67–78 [DOI] [PubMed] [Google Scholar]