Abstract
Nitric oxide (NO) plays a pivotal role in tumorigenesis, particularly with relation to cancer cell invasion and metastasis. NO can reversibly couple to cysteine thiols to form an S-nitrosothiol, which regulates the enzymatic activities of target proteins. c-Src is a tyrosine kinase that promotes cancer cell invasion and metastasis. Interestingly, c-Src can be activated by NO stimulation. However, mechanisms by which NO stimulates Src kinase activity have not been elucidated. We report here that NO causes S-nitrosylation of c-Src at cysteine 498 (Cys498) to stimulate its kinase activity. Cys498 is conserved among Src family kinases, and Cys506 of c-Yes, which corresponds to Cys498 of c-Src, was also important for the NO-mediated activation of c-Yes. Estrogens may work synergistically with NO to induce the proliferation and migration of many kinds of breast cancer cells. For example, β-estradiol induces the expression of endothelial nitric synthase and production of NO in MCF7 cells. We found that activation of c-Src in MCF7 cells by β-estradiol stimulation was mediated by the S-nitrosylation of Cys498. In addition, we report that disruption of E-cadherin junctions and enhancement of cell invasion by β-estradiol stimulation was mediated by NO-dependent activation of c-Src. These results identify a novel signaling pathway that links NO and Src family kinases to cancer cell invasion and metastasis.
Keywords: Cell/Migration, Signal Transduction, Signal Transduction/Phosphotyrosine, Signal Transduction/Protein Kinases/Tyrosine, Breast Cancer, S-nitrosylation, Nitric Oxide, Src
Introduction
Nitric oxide (NO) is a free radical gas that regulates numerous biological processes such as neurotransmission, vasodilatation, and macrophage-mediated immunity. NO can react with superoxide to produce secondary intermediates such as peroxynitrite and nitrogen dioxide, which have cytotoxic effects and post-translational modification of proteins (1). NO is synthesized from l-arginine by three major isoforms of NO synthase (NOS)3: endothelial NOS (eNOS), neuronal NOS, and inducible NOS (2). Numerous biological processes described for NO results from binding of NO to the heme iron of guanylate cyclase, which results in an increase of cGMP production and activation of downstream signaling pathways. NO also contributes to cell signaling by inducing post-translational modifications of proteins. S-Nitrosylation is the reversible coupling of NO moiety to a reactive cysteine thiol to form an S-nitrosothiol (3, 4). Accumulating evidence has shown that S-nitrosylation is an important post-translational modification that regulates a large variety of cellular functions and signaling events including tumorigenesis (5). Many proteins have been reported to be S-nitrosylated, and it is becoming evident that S-nitrosylation is one of the major mechanisms to convey NO-based cell signals (6).
c-Src is a non-receptor tyrosine kinase that plays pivotal roles in numerous cellular processes such as proliferation, migration, and transformation. c-Src consists of distinct functional regions (7). The N-terminal region is unique with low conservation through Src family kinases and contains a myristoylation signal (8, 9). Following this region are SH3 and SH2 domains, which are protein binding domains and play important roles for cell signals (10, 11). Catalytic domain is responsible for tyrosine kinase activity and is linked to a short C-terminal tail with a regulatory tyrosine residue (Tyr527) (12). Upon phosphorylation of Tyr527, the regulatory tail interacts with the SH2 domain, causing a closed conformation that inactivates the kinase (13). Dephosphorylation of Tyr527 disrupts the intramolecular interaction, which enables SH2 and SH3 domains to interact with downstream proteins and catalytic domain with substrates (14). Autophosphorylation of Tyr416 located in the catalytic domain is also required for the full activation of the kinase (15). v-Src is a viral protein encoded by the avian oncogene of Rous sarcoma virus, which is deleted of the C-terminal regulatory region of c-Src and thus is constitutively active and able to transform cells. In addition to the phosphorylation-based regulation of c-Src, recent studies have indicated a possible role of cysteine modification for the regulation of the kinase (16, 17). Our previous studies have indicated the importance of cysteine residues located in the C terminus of the catalytic domain (18, 19). Four cysteines, namely Cys483, Cys487, Cys496, and Cys498 are clustered in the C terminus of c-Src, which we referred to as the cysteine-clustered motif (CC motif) (20). Substitution of these cysteine residues rendered the kinase refractory to the inactivation by SH-alkylating agents and the activation by mercuric ions that have high affinity for thiols of cysteines (19–21). Recent studies have also indicated that cysteine oxidation of c-Src is important for the regulation of the kinase after integrin-mediated cell adhesion (17).
We have previously reported the activation of c-Src by NO stimulation through cysteine modification (22). In this report, we further studied the mechanism of NO-mediated activation of c-Src and demonstrated that S-nitrosylation of Cys498 is important for the activation of the kinase. Furthermore, we show that c-Src activation by β-estradiol stimulation was dependent on the production of NO, which was critical for the disruption of cell-cell contact and induction of cell invasion.
MATERIALS AND METHODS
Cells, Chemicals, and Antibodies
SYF, SYF/WT, and SYF/C498A cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% of fetal bovine serum. MCF7 cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% of fetal bovine serum and 0.01 mg/ml of insulin. SNAP was obtained from Cayman Chemicals (Ann Arbor, MI). Other reagents were obtained from the following manufacturers: β-estrogen, S-methyl methanethiosulfonate (MMTS), biotin HRP, PP2, and L-NMMA (Funakoshi, Tokyo, Japan); HRP-conjugated streptavidin (Takara, Tokyo, Japan); streptavidin-agarose beads (Sigma); Micro Bio-Spin 6 column (Bio-Rad); nickel-nitrilotriacetic acid (Ni-NTA; Qiagen). Anti-Src monoclonal antibody, monoclonal antibody 327, was kindly provided by Dr. J. S. Brugge. Anti-phospho-Src (Tyr416), anti-phospho-FAK (Tyr397), and anti-phospho-FAK (Tyr925) antibodies were purchased from Cell Signal Technologies (Tokyo, Japan). Anti-FAK and anti-eNOS were obtained from BD Biosciences.
Construction of Mutant c-Src
Full-length avian c-src was amplified by PCR and then ligated into pcDNA3.1 vector (Invitrogen) between BamHI and EcoRI sites. Substitution of amino acids was performed as described previously (18), and each mutation was confirmed by sequencing.
Immunoblot
Samples were electrophoresed on SDS-polyacrylamide gels and transferred to the polyvinylidene difluoride membrane (Immobilon, Millipore, Billerica, MA). The membrane was blocked with 1% skim milk for 1 h, incubated with primary antibodies for 1 h, washed with TBS-T for 15 min, and incubated with HRP-labeled secondary antibodies for 1 h. Signals were detected with the ECL system (Nakalai, Tokyo, Japan). Signal intensities were measured using Light Capture II equipped with CS analyzer (ATTO Corp., Tokyo, Japan).
Detection of S-Nitrosylated c-Src
Biotin switch technique (BST) was performed to detect S-nitrosylation of c-Src as described previously (23). To detect S-nitrosylation of c-Src in cells after SNAP treatment, cells were cultured without serum for 2 h, treated with SNAP for 90 min, and then lysed with HEN buffer (250 mm HEPES, 1 mm EDTA, 0.1 mm neocuproine, pH 7.7). Cell debris was removed by centrifugation, and SDS and MMTS were added to the final concentrations of 2.5 and 0.1%, respectively. Following frequent vortex and incubation at 50 °C for 30 min, lysates were precipitated with 3 volumes of acetone at −20 °C for 1 h. Proteins were centrifuged at 15,000 rpm for 20 min, and the pellets were suspended in 250 μl of HEN buffer with 1% SDS. Samples were further incubated with 0.4 mm biotin-HPDP in the presence or absence of 5 mm ascorbate for 1 h in the dark. After acetone precipitation, proteins were resuspended in 250 μl of HEN buffer with 1% SDS, followed by addition of 750 μl of neutralization buffer (20 mm HEPES, 0.1 m NaCl, 1 mm EDTA, 0.5% Triton X-100, pH 7.7). 50 μl of streptavidin-agarose beads were added to each sample and incubated for 1 h at room temperature. Beads were washed with washing buffer (neutralization buffer with 500 mm NaCl) five times, followed by one neutralization buffer wash. To detect total endogenous S-nitrosylated proteins, 50 μl of 2× SDS sample buffer without 2-mercaptoethanol was added and immunoblotted with HRP-labeled streptavidin. To detect S-nitrosylation of c-Src, 50 μl of 2× SDS sample buffer was added to the beads and immunoblotted with anti-Src antibody.
For the detection of S-nitrosylation in vitro, either C-terminally His-tagged c-Src or C498ASrc was immunoprecipitated with Ni-NTA and incubated with SNAP for 20 min. Immunoprecipitates were suspended in 100 μl of HEN buffer with 2% SDS and 0.1 mm MMTS and incubated for 30 min at 50 °C with frequent vortex. MMTS was removed using a Micro Bio-Spin 6 column. 8 μl of 2 mm biotin-HPDP and 4 μl of 100 mm ascorbate were added to the 68 μl of the eluate. 5× SDS sample buffer without 2-mercaptoethanol was added to the eluate and immunoblotted with HRP-labeled streptavidin.
Wound Healing Assay
Confluent monolayers of cells were serum-starved overnight and then scratched with a 10-μl tip. Cells were stimulated with SNAP and incubated for 18 h. Photographs were taken, and distance of migration was measured using measurement tools in Adobe Photoshop. Five randomly selected fields were evaluated for cell migration, and three independent experiments were performed.
Cell Aggregation Assay
MCF7 cells were stimulated with β-estradiol in the presence or absence of either L-NMMA or PP2 for 24 h, and then cell aggregation assay was performed. Briefly, cells were treated with 0.01% crystallized trypsin in Ca2+- and Mg2+-free HEPES-buffered saline (HCMF, pH 7.4) or HCMF with 1 mm CaCl2 (HMF) at 37 °C for 20 min and then washed with HCMF to obtain single cell suspensions. The cells were then washed with 0.05% soybean trypsin inhibitor in HCMF and incubated at 37 °C for 20 min with gyratory shaking in HMF with 1% bovine serum albumin. The data were plotted as the percentage of single cells relative to total single cells at zero time and are presented as the average of determination from three different experiments.
Invasion Assay
MCF7 cells were assayed for their invasiveness using a modified Boyden chamber. MCF7 cells were seeded onto Matrigel-coated filters with or without β-estradiol in the presence or absence of either L-NMMA or PP2. 18 h later, cells that invaded the lower surface of the filters were fixed, stained, and quantified by counting three randomly selected fields under the microscope. To determine whether Cys498 was essential for the β-estradiol induced invasion of MCF7 cells, cells were transfected with siRNA-targeting 3′UTR of human c-Src together with either expression vector encoding wild type avian c-Src or C498A Src using Lipofectamine 2000 according to manufacturer's protocol (Invitrogen). To visualize transfected cells, an expression vector encoding green fluorescent protein was also transfected. Two days after transfection, cells were subjected to invasion assay. The number of invaded cells that expressed green fluorescent protein was counted and normalized by the transfection efficiency. Experiments were performed three times, and average values and S.D. were determined. The sequence of siRNA we used to knockdown c-Src is 5′-GCAUUCGAGAUGGCAGAUUTT-3′.
In Vitro Kinase Assay
c-Src and mutants were expressed in SYF cells, and 48 h later, cells were lysed with RIPA buffer (20 mm Tris-HCl, 150 mm NaCl, 0.1% SDS, 0.5% Triton X-100, 0.5% sodium deoxycholate, pH 7.4) and immunoprecipitated with anti-Src antibody. Immunoprecipitates were washed and incubated in kinase buffer (10 mm Tris-HCl, 5 mm MgCl2, pH 7.4) with [γ32-P]ATP and 1 μg of casein at 30 °C for 20 min in the presence or absence of SNAP. SDS sample buffer was added to the reaction mixtures and then subjected to electrophoresis and analyzed by the BAS 2000 system.
RESULTS
S-Nitrosylation of c-Src by SNAP Stimulation
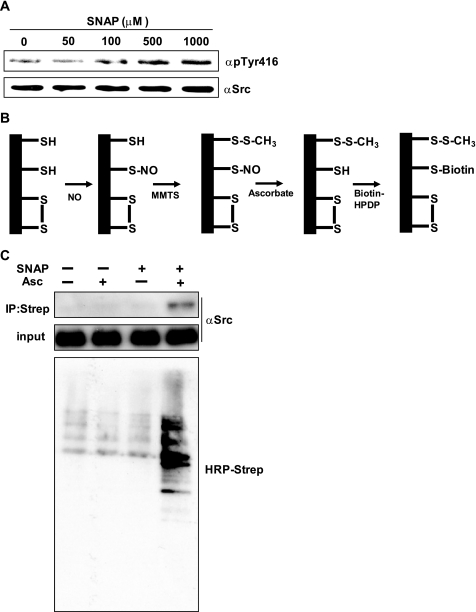
We previously reported that the catalytic activity of c-Src was activated by nitric oxide (22); therefore, we examined whether c-Src was S-nitrosylated by NO stimulation in cells. SYF cells are fibroblast cells derived from c-Src, c-Yes, and Fyn knock-out mice (24). We transfected wild type avian c-Src to SYF cells and established a cell line that constitutively expressed c-Src (SYF/WT). SYF/WT cells were treated with SNAP, a NO-generating reagent, at different concentrations for 90 min, after which cells were lysed and phosphorylation of c-Src was examined by Western blot. Using an antibody that specifically detects the phosphorylation of Tyr416 of c-Src, which is increased when the kinase is active, we found that SNAP treatment increased the phosphorylation of Tyr416 in a dose-dependent manner (Fig. 1A). We next examined S-nitrosylation of c-Src by BST, which detects S-nitrosothiols of proteins. Fig. 1B is the schematic diagram of the BST. After S-nitrosylation of proteins, free thiols of proteins were blocked with MMTS, followed by reduction of S-nitrosothiols with ascorbate. Biotin-HPDP then binds to the reduced thiols, after which biotinylated proteins are immunoprecipitated with streptavidin beads. SYF/WT cells were treated with 500 μm SNAP for 90 min, and then the BST was performed with or without ascorbate to show the specificity of the assay (25). As shown in Fig. 1C, S-nitrosylation of endogenous proteins and c-Src was elevated after SNAP treatment.
FIGURE 1.
Activation and S-nitrosylation of c-Src by SNAP treatment. A, SYF/WT cells were serum-starved for 2 h and then stimulated with the indicated concentrations of SNAP for 90 min. Cells were lysed, and phosphorylation of Tyr416 was examined by Western blot. B, a schematic diagram of BST is shown. After S-nitrosylation of proteins, free thiols of proteins are blocked with MMTS, followed by reduction of S-nitrosothiols to free thiols by ascorbate. HPDP-biotin binds to the free thiols, and then the proteins are immunoprecipitated (IP) by streptavidin beads, followed by detection of the proteins by HRP-labeled streptavidin or anti-Src antibody. C, SYF/WT cells were serum-starved for 2 h, stimulated with 500 μm SNAP, and then subjected to the BST as described under “Materials and Methods.” The upper panel shows S-nitrosylated c-Src. The middle panel shows the amount of input protein. The lower panel shows the S-nitrosylated endogenous proteins.
S-Nitrosylation of Cys498 Is Critical for the Activation of c-Src by SNAP in Vitro
Next, we attempted to determine the critical cysteine residues involved in the activation of c-Src by NO. There are nine cysteine residues in c-Src. Three cysteine residues are in the SH2 domain, and six cysteine residues are in the kinase domain. We previously reported the activation of c-Src by mercuric chloride, which was dependent on the modification of cysteine residues located in the short C-terminal region of the kinase domain. There are four cysteine residues in this region, which we previously named the cysteine-clustered motif (CC motif) (Fig. 2A). We focused on these four cysteines and substituted each residue to alanine by site-directed mutagenesis. C483A-, C487A-, C496A-, and C498A-Src were transiently expressed in SYF cells and immunoprecipitated with anti-Src antibody. Immunoprecipitates were incubated with or without SNAP, and catalytic activities were examined. Although C487A-Src was partially resistant to the SNAP-mediated activation, C498A-Src was completely resistant (Fig. 2B). Because Cys498 is conserved among Src family kinases, we substituted Cys506 of c-Yes, which corresponds to Cys498 of c-Src, and examined its activation by SNAP treatment. Wild type c-Yes was activated by SNAP; however, the activation of C506A-Yes was remarkably inhibited (Fig. 2C). We next examined S-nitrosylation of both wild type and C498A-Src in vitro. Wild type c-Src and C498A-Src were immunoprecipitated, treated with SNAP for 20 min, and then subjected to the BST. We found that S-nitrosylation of C498A-Src was significantly reduced compared with that of wild type (Fig. 2D), indicating that S-nitrosylation of Cys498 is crucial for the NO-mediated activation of c-Src.
FIGURE 2.
Cys498 is critical for the activation of c-Src by SNAP in cells. A, alignment of amino acid sequence of CC motif of Src family kinases and FAK. B, in vitro kinase assay of wild type c-Src and mutants in the presence or absence of 5 μm SNAP. Lower panels are immunoblot analyses of expression of Src kinases. C, in vitro kinase assay of wild type c-Yes and C506A-Yes. D, Wild type c-Src and C498A-Src were immunoprecipitated, incubated with 5 μm SNAP for 20 min, and then subjected to the BST as described under “Materials and Methods.” The upper panel indicates the S-nitrosylated proteins.
S-Nitrosylation of Cys498 Is Critical for the Enhanced Cell Migration by SNAP
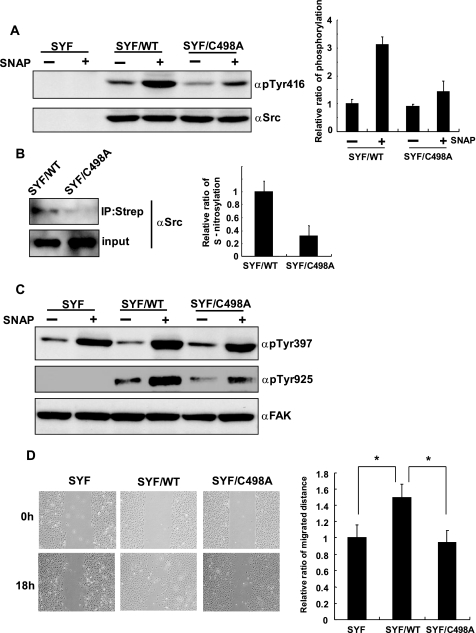
To address the physiological role of S-nitrosylation of c-Src, we established a cell line that constitutively expressed C498A-Src (SYF/C498A). SYF, SYF/WT, and SYF/C498A cells were stimulated with 500 μm SNAP for 90 min, and activation of wild type and C498A-Src was examined by Western blot. Although C498A-Src was activated by SNAP treatment, the level of activation of C498A-Src was significantly reduced compared with that of wild type c-Src (Fig. 3A). Consistent with this result, the level of S-nitrosylation of C498A was less than that of wild type c-Src after SNAP treatment (Fig. 3B). Furthermore, we also investigated the phosphorylation of FAK, which is one of the substrates of c-Src, to evaluate the activation of c-Src after SNAP stimulation. Tyr397 of FAK is autophosphorylated by various extracellular stimuli and correlates with the activity of the kinase. Phosphorylated Tyr397 has high affinity for the SH2 domain of Src family kinases. Once c-Src is recruited to the phosphorylated Tyr397, it phosphorylates Tyr925 of FAK to create a binding site for the Grb-SOS complex. SNAP treatment of cells increased phosphorylation of Tyr397 of FAK in every cell line (Fig. 3C). Because FAK has the cysteine residue that corresponds to Cys498 of Src (Fig. 2A), we assume that FAK could also be activated by S-nitrosylation of the cysteine residue. In contrast to the phosphorylation of Tyr397, the phosphorylation of Tyr925, which is dependent on the activation of c-Src, following SNAP stimulation was clearly reduced in SYF/C498A cells compared with that of SYF/WT cells, suggesting the pivotal role of Cys498 for the NO-mediated activation of c-Src (Fig. 3C).
FIGURE 3.
S-Nitrosylation of Cys498 mediates the activation of c-Src and cell migration by SNAP. A, SYF, SYF/WT, and SYF/C498A cells were serum-starved for 2 h and then treated with 500 μm SNAP for 90 min. Cells were lysed, and phosphorylation of c-Src was examined by Western blot. Three independent experiments were performed, and the relative ratio of phosphorylation is indicated. B, SYF/WT and SYF/C498A cells were serum-starved for 2 h, treated with SNAP for 90 min, and then subjected to BST. Three independent experiments were performed, and the relative ratio of S-nitrosylation is indicated. C, SYF, SYF/WT, and SYF/C498A cells were serum-starved for 2 h, treated with 500 μm SNAP, and then cells were lysed, and phosphorylation of FAK was examined by Western blot. D, SYF, SYF/WT, and SYF/C498A cells were grown to confluence and serum-starved for 2 h. Scratch was made by a 10-μl tip and treated with 500 μm SNAP for 18 h. Distance of migration was measured in five randomly selected fields, and three independent experiments were performed. *, p < 0.01. IP, immunoprecipitation. Error bars indicate S.D.
Src and FAK are critical signaling molecules for cell migration; and thus, we performed a wound healing assay to assess the role of NO-dependent Src activation in cell migration. Cells were grown to confluence and wound was made by scratching with a 10-μl tip and stimulated with 500 μm SNAP for 18 h. Strikingly, SYF and SYF/C498A cells exhibited a decreased cell migration into the wound area as compared with the SYF/WT cells (Fig. 3D). These results, together with the aforementioned observations, suggest that the enhanced cell migration by NO was mediated by the activation of c-Src, whose activity was dependent on the S-nitrosylation of Cys498.
Activation of c-Src by β-Estradiol Is Dependent on the S-Nitrosylation of Cys498
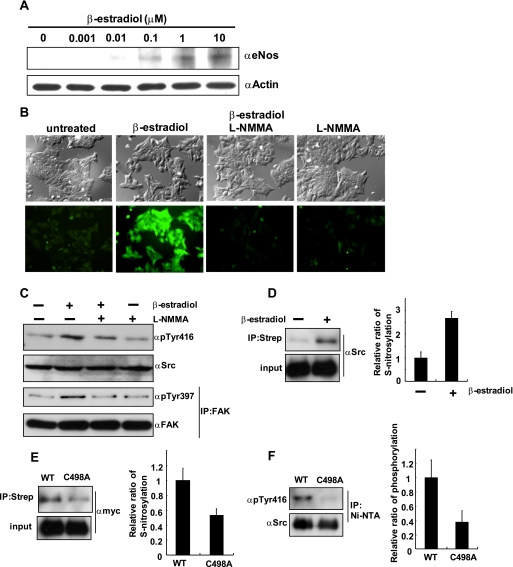
Because NO is known to activate cancer cell proliferation, invasion, and metastasis (26), we examined the effects of NO-dependent activation of c-Src in MCF7 cells. MCF7 cells are breast cancer derived cells that have estrogen receptor with minor invasive property, and stimulation of MCF7 cells with β-estradiol was reported to induce cell invasion (27). β-estradiol stimulation induced the expression of eNOS in a dose-dependent manner (Fig. 4A), which is consistent with the previous report (28). Next, we examined the NO production using DAF-2T, a NO-sensitive fluorescent dye, in cells cultured under similar conditions. Fluorescent microscopic images revealed that β-estradiol treatment induced production of NO, which was drastically suppressed by the addition of L-NMMA, a known NOS inhibitor (Fig. 4B). We then determined the levels of activation of c-Src and FAK by β-estradiol stimulation using phospho-specific antibodies. As shown in Fig. 4C, both c-Src and FAK were activated by β-estradiol stimulation, which was abolished by the addition of L-NMMA, indicating that the activation of c-Src and FAK by β-estradiol was mediated by NO. S-Nitrosylation of c-Src was also increased by β-estradiol stimulation (Fig. 4D). To compare the levels of S-nitrosylation between wild type c-Src and C498A-Src, C-terminally Myc-tagged either wild type c-Src or C498A-Src was transiently expressed in MCF7 cells and stimulated with β-estradiol for 24 h and then subjected to BST. As expected, S-nitrosylation of C498A was less than that of wild type c-Src (Fig. 4E). Finally, we checked the phosphorylation levels of wild type c-Src and C498A-Src after β-estradiol stimulation. C-terminally His-tagged wild type c-Src and C498A-Src were expressed in SYF cells, stimulated with β-estradiol for 24 h, immunoprecipitated with Ni-NTA, and then phosphorylation of Tyr416 of c-Src was examined. As shown in Fig. 4F, phosphorylation of C498A-Src was decreased compared with that of wild type c-Src.
FIGURE 4.
Activation of c-Src after β-estradiol stimulation in MCF7 cells is mediated by NO. A, MCF7 cells were stimulated with the indicated concentrations of β-estradiol for 24 h, and expression of eNOS was examined by Western blot. B, after 24 h of β-estradiol (100 nm) stimulation, MCF7 cells were incubated with DAF2 for 1 h and analyzed by fluorescent microscopy. C, MCF7 cells were stimulated with β-estradiol (100 nm) for 24 h with or without L-NMMA (50 μm), and cell lysates were analyzed for the phosphorylation of c-Src and FAK. D, MCF7 cells were treated with β-estradiol (100 nm) or left untreated for 24 h, and then the BST was performed to detect S-nitrosylation of c-Src. Three independent experiments were performed, and the relative ratio of S-nitrosylation is indicated. E, C-terminally Myc-tagged c-Src or C498A-Src was expressed in MCF7 cells, treated with β-estradiol (100 nm) for 24 h, and then subjected to the BST. Three independent experiments were performed, and the relative ratio of S-nitrosylation is indicated. F, C-terminally His-tagged c-Src or C498A-Src was expressed in MCF7 cells and stimulated with β-estradiol (100 nm) for 24 h. Cells were lysed and immunoprecipitated (IP) with Ni-NTA, and then the phosphorylation was examined by Western blot. Three independent experiments were performed, and the relative ratio of phosphorylation is indicated. Strep, streptavidin.
NO-mediated c-Src Activation Modulates E-cadherin Expression, Cell-Cell Adhesion, and Cell Invasion
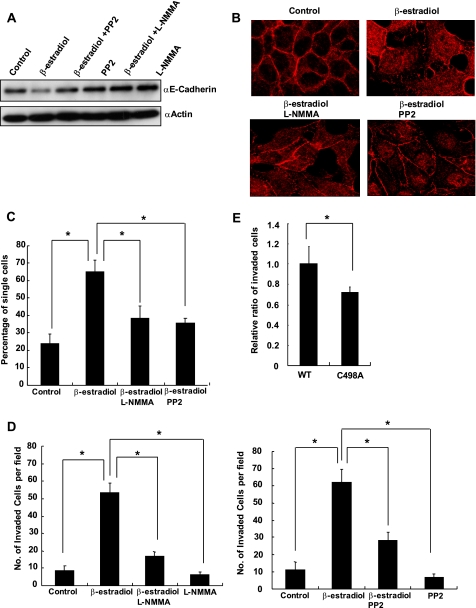
The expression of E-cadherin has been previously reported to be suppressed in the presence of β-estradiol (29). E-cadherin is a critical mediator of cell-cell adhesion, and disruption of E-cadherin is associated with the dissemination of cancer cells to metastasize (30). To determine whether the suppression of E-cadherin was dependent on the production of NO and the activation of c-Src, we attempted to address this issue by treating MCF7 cells with β-estradiol in the presence or absence of either L-NMMA or Src inhibitor, PP2. We found that the inhibition of either NO production or c-Src activation abolished the β-estradiol-mediated suppression of E-cadherin (Fig. 5A), indicating that the reduced expression of E-cadherin was dependent on the NO-mediated Src activation. We then performed an immunocytochemistry analysis to assess the effect of β-estradiol on cell-cell contact. MCF7 cells were fixed and immunostained for E-cadherin after β-estradiol stimulation in the absence or presence of either L-NMMA or PP2. As shown in Fig. 5B, E-cadherin immunostaining showed that β-estradiol disrupted cell-cell adhesion, which was partially restored by the addition of either L-NMMA or PP2. To further investigate the effect of c-Src activation on cell-cell adhesion, we performed a cell aggregation assay. Results indicated that the disrupted cell-cell adhesion induced by β-estradiol was recovered by the addition of either L-NMMA or PP2 (Fig. 5C). Next, we carried out a cell invasion test using the modified Boyden chamber method to study the involvement of NO-mediated Src activation in β-estradiol dependent invasion of MCF7 cells. MCF7 cells were loaded onto the upper chamber and stimulated with β-estradiol for 18 h. Cells that invaded the lower surface of the filter were fixed, stained and counted. As shown in Fig. 5D, β-estradiol up-regulated the cell invasion, whereas either L-NMMA or PP2 treatment suppressed cell invasion. Finally, to confirm that S-nitrosylation of Cys498 is required for the β-estradiol-induced cell invasion, we transfected siRNA targeting 3′-UTR of human c-Src and an expression vector encoding green fluorescent protein together with either an expression vector encoding wild type avian c-Src or C498A Src. The siRNA suppressed expression of c-Src (data not shown). Two days after transfection, cells were subjected to the invasion assay with the presence of β-estradiol. Fig. 5E shows cell invasion was reduced when C498A-Src was expressed, indicating that S-nitrosylation of Cys498 is involved in the β-estradiol-induced cell invasion.
FIGURE 5.
Disruption of cell-cell adhesion and induction of cell invasion after β-estradiol stimulation was mediated by NO and c-Src. A, MCF7 cells were stimulated with β-estradiol (100 nm) in the presence or absence of either L-NMMA (50 μm) or PP2 (25 μm) for 24 h, and expression of E-cadherin was examined by Western blot. B, MCF7 cells were stimulated with β-estradiol (100 nm) in the presence or absence of either L-NMMA (50 μm) or PP2 (25 μm) for 24 h and immunostained for E-cadherin. C, MCF7 cells were stimulated with β-estradiol (100 nm) in the presence or absence of either L-NMMA (50 μm) or PP2 (25 μm) for 24 h, and cell aggregation assay was performed. Percentages of single cells were counted, and mean averages + S.D. from three independent experiments are indicated. *, p < 0.01. D, MCF7 cells were loaded onto Matrigel-coated filters with or without β-estradiol (100 nm) in the presence or absence of either L-NMMA (50 μm) or PP2 (25 μm). Cells that invaded to the lower surface of the filters were fixed, stained, and quantified by counting three randomly selected fields under the microscope. Mean averages + S.D. from three independent experiments are indicated. *, p < 0.01. E, MCF7 cells were transfected with siRNA targeting 3′-UTR of human c-Src and a vector encoding green fluorescent protein together with either a vector encoding wild type avian c-Src or C498A Src. Two days after transfection, cells were subjected to the invasion assay in the presence of β-estradiol (100 nm). The number of cells that invaded to the lower surface was counted and normalized by transfection efficiency. Three independent experiments were performed, and the relative ratio of migrated cells is indicated. *, p < 0.01. Error bars indicate S.D.
DISCUSSION
In this report, we showed evidence that Cys498 of c-Src, which is conserved among Src family kinases, plays a pivotal role in the activation of c-Src by NO. Accumulating evidence has indicated that S-nitrosylation regulates enzymatic activity of various proteins, which consequently modulate signaling pathways and gene expression. Using the biotin switch technique, we found that S-nitrosylation of C498A-Src was reduced compared with that of wild type after SNAP treatment in vitro, indicating that the activation of c-Src by SNAP was dependent on the S-nitrosylation of Cys498. Moreover, using cells that stably express either wild type c-Src or C498A-Src, we demonstrated that S-nitrosylation of Cys498 was critical for the NO-mediated activation of c-Src in cells.
c-Src is a non-receptor tyrosine kinase, and its catalytic activity is mainly regulated by the phosphorylation of Tyr527. However, recent evidence has shown the importance of cysteine modification in the regulation of the kinase. Reactive oxygen species, such as superoxide and hydrogen peroxide, serve as secondary messengers to regulate signaling pathways of certain growth factors and cell adhesion. A number of studies have suggested that cysteine modification of c-Src plays an important role for the reactive oxygen species-mediated signal transductions; however, whether c-Src is activated or inactivated by oxidation remains unclear (31). Upon cell adhesion, c-Src is activated by the formation of an intramolecular S–S bond between Cys245 and Cys487, which are highly conserved residues among the Src family kinases and are localized in the SH2 domain and kinase domain, respectively (17). On the other hand, Kemble et al. (31) reported the direct inactivation of c-Src by oxidation. They showed that the inactivation was caused by oxidation of Cys277, which resulted in homodimerization of c-Src linked by a disulfide bridge. We previously reported the critical role of cysteine residues in the CC motif for the regulation of catalytic activity of c-Src (18). SH-alkylating agents such as BIPM and NAM are known to inhibit kinase activity of v-Src by directly binding to cysteine residues. Mutation of cysteine residues in the CC motif abolished the inactivation of the kinase by these agents (19). On the other hand, mercuric ions that have high affinity for thiol side chain of cysteine residues activate c-Src, which is dependent on the modification of Cys498 (20). These previous studies and our present study strongly suggest the importance of cysteine modification for the regulation of c-Src for diverse intracellular signaling pathways.
A number of studies have indicated that both NO and c-Src are involved in the progression of tumor formation. NOS activity has been detected in various tumors and has been associated with tumor grade and proliferation rate. A recent study showed that eNOS-mediated Ras activation by S-nitrosylation was important for initiation and maintenance of tumor (32). c-Src is known to be activated in several cancers, and there is convincing evidence that increased Src activity is associated with more invasive and aggressive phenotypes (33). Our results indicate a possible role of NO-mediated c-Src activation for tumorigenesis and invasion.
Breast cancer is one of the most common malignancies, and estrogens are major contributors to the development of the cancer. Since β-estradiol stimulation of MCF7 cells has been reported to induce eNOS expression, we treated MCF7 cells and examined activation of c-Src and its physiological effect. We found that c-Src was activated by β-estradiol stimulation, which was dependent on the production of NO. Furthermore, S-nitrosylation of c-Src was increased by β-estradiol stimulation, and the phosphorylation and S-nitrosylation of C498A-Src after β-estradiol stimulation were reduced compared with those of wild type c-Src, indicating that S-nitrosylation of Cys498 is critical for the NO-mediated activation of c-Src. E-cadherin is a calcium-dependent cell-cell adhesion glycoprotein forming the core of the epithelial adherens junction. Loss of E-cadherin is a hallmark of many invasive tumors, and increased Src activity is associated with a disruption of E-cadherin-mediated cell-cell contacts in tumor cells (34). We found NO-mediated c-Src activation was critical for the suppression of E-cadherin expression and disruption of cell-cell contacts after β-estradiol stimulation. Loss of E-cadherin not only disrupts cell-cell adhesion but also activates multiple transcriptional pathways to induce cell invasion (30). Stimulation of MCF7 cells with β-estradiol induced cell invasion that was dependent on the production of NO and activation of c-Src. Our experimental results suggest that NO-mediated activation of c-Src is critical for cancer cell dissemination and invasion, which could be mediated by suppressing E-cadherin expression.
NO exerts diverse physiological functions; however, precise mechanisms are still unknown. Recent evidence has clearly indicated that regulation of enzymatic activities of proteins by S-nitrosylation plays critical roles for various physiological functions (5). Because Cys498 is conserved in Src family kinases, our result suggests that some physiological phenomenon elicited by NO may be mediated by the activation of Src family kinases. Further studies about NO-mediated activation of tyrosine kinases may advance our understanding of physiological functions of NO.
Acknowledgments
We thank members of Division of Cancer Biology for technical assistance and helpful discussion.
This work was supported by grants from the Ministry of Education, Science, Culture and Technology of Japan.
- NOS
- nitric-oxide synthase
- eNOS
- endothelial NOS
- WT
- wild type
- CC
- cysteine-clustered
- HRP
- horseradish peroxidase
- Ni-NTA
- nickel-nitrilotriacetic acid
- BST
- biotin switch technique
- MMTS
- S-methyl methanethiosulfonate
- SH
- Src homology
- SNAP
- S-nitroso-n-acetyl-d,l-penicillamine
- FAK
- focal adhesion kinase.
REFERENCES
- 1.Stamler J. S., Singel D. J., Loscalzo J. (1992) Science 258, 1898–1902 [DOI] [PubMed] [Google Scholar]
- 2.Nathan C., Xie Q. W. (1994) Cell 78, 915–918 [DOI] [PubMed] [Google Scholar]
- 3.Stamler J. S., Jaraki O., Osborne J., Simon D. I., Keaney J., Vita J., Singel D., Valeri C. R., Loscalzo J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7674–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., Singel D. J., Loscalzo J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 6.Stamler J. S., Lamas S., Fang F. C. (2001) Cell 106, 675–683 [DOI] [PubMed] [Google Scholar]
- 7.Jove R., Kornbluth S., Hanafusa H. (1987) Cell 50, 937–943 [DOI] [PubMed] [Google Scholar]
- 8.Pellman D., Garber E. A., Cross F. R., Hanafusa H. (1985) Nature 314, 374–377 [DOI] [PubMed] [Google Scholar]
- 9.Cross F. R., Garber E. A., Pellman D., Hanafusa H. (1984) Mol. Cell. Biol. 4, 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pawson T. (1995) Nature 373, 573–580 [DOI] [PubMed] [Google Scholar]
- 11.Mayer B. J., Gupta R. (1998) Curr. Top. Microbiol. Immunol. 228, 1–22 [DOI] [PubMed] [Google Scholar]
- 12.Cartwright C. A., Eckhart W., Simon S., Kaplan P. L. (1987) Cell 49, 83–91 [DOI] [PubMed] [Google Scholar]
- 13.Xu W., Harrison S. C., Eck M. J. (1997) Nature 385, 595–602 [DOI] [PubMed] [Google Scholar]
- 14.Brown M. T., Cooper J. A. (1996) Biochim. Biophys. Acta. 1287, 121–149 [DOI] [PubMed] [Google Scholar]
- 15.Jove R., Hanafusa T., Hamaguchi M., Hanafusa H. (1989) Oncogene. Res. 5, 49–60 [PubMed] [Google Scholar]
- 16.Chiarugi P. (2008) Mol. Cells. 26, 329–337 [PubMed] [Google Scholar]
- 17.Giannoni E., Buricchi F., Raugei G., Ramponi G., Chiarugi P. (2005) Mol. Cell. Biol. 25, 6391–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senga T., Miyazaki K., Machida K., Iwata H., Matsuda S., Nakashima I., Hamaguchi M. (2000) Oncogene 19, 273–279 [DOI] [PubMed] [Google Scholar]
- 19.Oo M. L., Senga T., Thant A. A., Amin A. R., Huang P., Mon N. N., Hamaguchi M. (2003) Oncogene 22, 1411–1417 [DOI] [PubMed] [Google Scholar]
- 20.Senga T., Hasegawa H., Tanaka M., Rahman M. A., Ito S., Hamaguchi M. (2008) Cancer Sci. 99, 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman M. A., Senga T., Oo M. L., Hasegawa H., Biswas M. H., Mon N. N., Huang P., Ito S., Yamamoto T., Hamaguchi M. (2008) Oncol. Rep. 19, 975–980 [PubMed] [Google Scholar]
- 22.Akhand A. A., Pu M., Senga T., Kato M., Suzuki H., Miyata T., Hamaguchi M., Nakashima I. (1999) J. Biol. Chem. 274, 25821–25826 [DOI] [PubMed] [Google Scholar]
- 23.Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Nat. Cell. Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 24.Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO. J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrester M. T., Foster M. W., Stamler J. S. (2007) J. Biol. Chem. 282, 13977–13983 [DOI] [PubMed] [Google Scholar]
- 26.Ying L., Marino J., Hussain S. P., Khan M. A., You S., Hofseth A. B., Trivers G. E., Dixon D. A., Harris C. C., Hofseth L. J. (2005) Cancer Res. 65, 9132–9136 [DOI] [PubMed] [Google Scholar]
- 27.Albini A., Graf J., Kitten G. T., Kleinman H. K., Martin G. R., Veillette A., Lippman M. E. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8182–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loibl S., Bratengeier J., Farines V., von Minckwitz G., Spänkuch B., Schini-Kerth V., Nepveu F., Strebhardt K., Kaufmann M. (2006) Pathol. Res. Pract. 202, 1–7 [DOI] [PubMed] [Google Scholar]
- 29.Oesterreich S., Deng W., Jiang S., Cui X., Ivanova M., Schiff R., Kang K., Hadsell D. L., Behrens J., Lee A. V. (2003) Cancer. Res. 63, 5203–5208 [PubMed] [Google Scholar]
- 30.Onder T. T., Gupta P. B., Mani S. A., Yang J., Lander E. S., Weinberg R. A. (2008) Cancer Res. 68, 3645–3654 [DOI] [PubMed] [Google Scholar]
- 31.Kemble D. J., Sun G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5070–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim K. H., Ancrile B. B., Kashatus D. F., Counter C. M. (2008) Nature 452, 646–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frame M. C. (2002) Biochim. Biophys. Acta. 1602, 114–130 [DOI] [PubMed] [Google Scholar]
- 34.Frame M. C., Fincham V. J., Carragher N. O., Wyke J. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 233–245 [DOI] [PubMed] [Google Scholar]