Abstract
Aging is a complex process accompanied by a decreased capacity of cells to cope with random damages induced by reactive oxygen species, the natural by-products of energy metabolism, leading to protein aggregation in various components of the cell. Chaperones are important players in the aging process as they prevent protein misfolding and aggregation. Small chaperones, such as small heat shock proteins, are involved in the refolding and/or disposal of protein aggregates, a feature of many age-associated diseases. In Drosophila melanogaster, mitochondrial Hsp22 (DmHsp22), is localized in the mitochondrial matrix and is preferentially up-regulated during aging. Its overexpression results in an extension of life span (>30%) (Morrow, G., Samson, M., Michaud, S., and Tanguay, R. M. (2004) FASEB J. 18, 598–599 and Morrow, G., Battistini, S., Zhang, P., and Tanguay, R. M. (2004) J. Biol. Chem. 279, 43382–43385). Long lived flies expressing Hsp22 also have an increased resistance to oxidative stress and maintain locomotor activity longer. In the present study, the cross-species effects of Hsp22 expression were tested. DmHsp22 was found to be functionally active in human cells. It extended the life span of normal fibroblasts, slowing the aging process as evidenced by a lower level of the senescence associated β-galactosidase. DmHsp22 expression in human cancer cells increased their malignant properties including anchorage-independent growth, tumor formation in nude mice, and resistance to a variety of anticancer drugs. We report that the DmHsp22 interacts and inactivates wild type tumor suppressor protein p53, which may be one possible way of its functioning in human cells.
Keywords: Aging, Chaperones/Heat Shock, Organisms/Drosophila, Protein/Heat Shock, Tumor/Suppressor/p53, Viruses/Reovirus
Introduction
The small heat shock proteins (sHsps)5 are members of a superfamily of proteins that share a highly conserved ∼80-amino acid α-crystallin domain in their C-terminal domain. Like other heat shock proteins, sHsps exhibit a highly dynamic structure forming both homo- and hetero-oligomeric complexes, possess chaperone-like activity, and are involved in a plethora of functions ranging from protein folding to apoptosis and stress resistance. In human cells, there are 11 members in the family: Hsp27 (HspB1, Hsp25), myotonic dystrophy protein kinase-binding protein (HspB2), HspB3, αA-crystallin (HspB4), αB-crystallin (HspB5), Hsp20 (p20, HspB6), cardiovascular heat shock protein (HspB7), Hsp22 (Hsp22/HspB8/H11) and HspB9 (1–4). Although very similar in structure, the different sHsps show specificity in terms of response to stress, in tissue, and developmental expression, suggesting that they have different functions. sHsps have been reported to act at various steps of the apoptotic process, but the structure/functions of these small proteins remain poorly understood. The tissue and developmental stage-specific pattern of sHsp expression has suggested their role in specific tissue functions, maintenance, and modulation of tissue integrity, dynamics of structural elements, spatial organization, and apoptosis that play an important role in development, control of cell proliferation, and carcinogenesis (1, 5–7). Specific point mutations in a number of small Hsps can destabilize the protein structure and have been reported to be associated with a number of neurodegenerative disorders such as distal hereditary motor neuropathy and/or Charcot-Marie-Tooth neuromuscular diseases (2, 3, 8). Interestingly, K141N and K141E mutations that induced structural destabilization of the protein also reduced its chaperone activity, enhanced the formation of homo- and heterodimers, and increased the formation of intracellular aggregates (9–13). Indeed, wild type Hsp22 was shown to block the accumulation of polyglutamine protein (Htt43Q) in inclusion bodies, suggesting its molecular chaperone function keeps Htt43Q in a soluble and rapidly degradable state.
In Drosophila melanogaster, in which the sHsps were first reported, there are four main members that show distinct intracellular localization (14), One of these, D. melanogaster Hsp22 (DmHsp22) localizes distinctly in the mitochondrial matrix (15) and shows heat and oxidative stress-inducible features and chaperone-like activity (16). DmHsp22 is up-regulated during Drosophila aging (17). Flies selected for increased longevity were also found to display an earlier onset of Hsp22 and Hsp23 transcription (18). Ubiquitous or targeted expression of Hsp22 within motor neurons resulted in a 30% increase in Drosophila mean life span and beneficial effects such as, increased locomotor activity, resistance to oxidative injuries, and thermal stress (19). On the other hand, flies compromised for Hsp22 expression showed a 40% decrease in life span and displayed a 30% decrease in locomotor activity and higher sensitivity to mild stress (20). Histone deacetylase inhibitor trichostatin A was shown to promote hsp22 gene transcription and extend the life span of D. melanogaster (21, 22). The selected long lived Drosophila lines showed higher level of Hsp22 expression, responded more rapidly to heat, and were more tolerant to high temperature (15, 18, 22).
In the present study, we investigated the cross-species effects of Hsp22. We report that the DmHsp22 is functionally active in human cells. Indeed, the expression of DmHsp22 extends the life span of normal human fibroblasts and increased the malignant properties of human cancer cells. Moreover, DmHsp22-expressing cancer cells formed aggressive tumors and acquired a drug-resistant phenotype. We further report that DmHsp22 interacts with and inactivate wild type tumor suppressor p53 in human cancer cells, suggesting that this interaction may represent one of the mechanisms of its proproliferative function.
MATERIALS AND METHODS
Cell Culture
Human normal fibroblasts (TIG-1) and cancerous cells, osteosarcoma (U2OS), breast carcinoma (MCF7), and lung carcinoma (A549), were propagated in low glucose Dulbecco's modified Eagle's minimal essential medium supplemented with 10% fetal bovine serum and 1% antibiotics-antimycotics (Sigma). DmHsp22-overexpressing cells were generated by retrovirus-driven expression. The cDNA encoding D. melanogaster Hsp22 protein was cloned into the HindIII site of the vector CX4neo (pCX4neo, provided by Dr. Akagi, Osaka, Japan). For production of retroviruses, Plat-E, an ecotropic murine leukemia virus-packaging cell line was transfected with the pVPack-GP (expressing gag and pol) and pVPack VSVG (expressing env) vectors (Stratagene) along with either pCXneo retroviral vector or pCX4neo/DmHsp22 using FuGENE6 (Roche Applied Science). After 48 h, culture supernatants were collected and filtered through 0.45-μm filters. The filtered supernatants (viral stock) were used for cell infection without freeze thawing. Cells (2 × 105/well in 6-well dishes) were treated with 8 μg/ml polybrene at 37 °C for 1 h, after which they were incubated with 200–300 μl of filtered viral stock for 1 h followed by addition of 2 ml of Dulbecco's modified Eagle's minimal essential medium and continued incubation at 37 °C for further 48 h. The infected cells were selected in medium containing G418 (100–200 μg/ml for normal and 500 μg/ml for cancer cells) until stable expressing cell lines were obtained. The stably infected cells lines were examined for expression of DmHsp22 by Western blotting and immunostaining with specific antibody (15) and were maintained in 100 μg/ml G418-supplemented medium. Normal control and DmHsp22-expressing cells were serially passaged in parallel. The passage ratio was kept at 1:16 for young cultures and was decreased to 1:8 or 1:4 as they aged. Serial passaging was continued until cells stopped dividing and entered senescence. The nondividing senescent cells were maintained for >4 weeks before termination of the experiment.
Western Blotting and Immunoprecipitation
Control and DmHsp22-expressing cells were trypsinized and solubilized in Nonidet P-40 lysis buffer. The lysates were centrifuged at 13,000 rpm for 10 min at 4 °C to remove cell debris. The protein concentrations were determined by Bradford protein assay following the manufacturer's instructions. Lysates containing 20–30 μg protein were boiled in SDS sample buffer for 5 min, subjected to SDS-PAGE under reducing conditions and then transferred onto Immobilon-P membranes (Millipore) using a semidry transfer apparatus (ATTO). After washing with Tris-buffered saline-Triton X-100, the membrane was incubated with the first and second (horseradish peroxidase-conjugated) antibodies as indicated. The membrane was washed thrice in Tris-buffered saline-Triton X-100 and once in Tris-buffered saline, then subjected to an enhanced chemiluminescence-mediated visualization (Amersham Biosciences) using an Lumino Image Analyzer equipped with a CCD camera (LAS3000-mini, FujiFilm). Mitochondrial fractions were prepared using the Qproteome mitochondria isolation kit (Qiagen) following the manufacturer's instructions. Washed cells were suspended with lysis buffer for disrupting the plasma membrane. After incubation for 10 min, lysate were centrifuged at 1,000 × g for 10 min at 4 °C, and then the supernatant was harvested as a cytosolic fraction. Pellet containing nuclei, cell debris, and unbroken cells was resuspended with ice-cold disruption buffer. Resuspended pellet was centrifuged at 1,000 × g for 10 min at 4 °C, and then the supernatant was harvested as a microsomal fraction. The resulting pellet was centrifuged at 8,000 × g for 10 min at 4 °C for mitochondrial fraction by density gradient purification. After centrifugation, the band toward the bottom of the tube was harvested as a mitochondrial fraction. For in vivo coimmunoprecipitation, cell lysates (300 μg protein) in 300 μl of Nonidet P-40 lysis buffer were incubated at 4 °C for 1–2 h with anti-DmHsp22 antibody. Immunocomplexes were separated by incubation with Protein A/G-Sepharose, and Western blotting was performed with the indicated antibodies using the procedure described above.
Immunostaining
Cells grown on coverslips were fixed in methanol:acetone (1:1), permeabilized with 0.2% Triton X-100 and blocked with 5% bovine serum albumin in phosphate-buffered saline (PBS). Cells were probed with primary antibody (as indicated) for 1–2 h. Staining was visualized by Alexa Fluor 594 (rabbit)-conjugated secondary antibody. The cells were examined on a Carl Zeiss microscope attached with either a Photometrics Synsys monochrome CCD (charge-coupled device) or an AxioCam HRC color camera (Zeiss).
Senescence-associated β-Galactosidase Assay
Senescent cells were detected using the standard protocol for senescence-associated β-galactosidase staining assay. In brief, cells were washed with PBS, fixed with 2% formaldehyde/0.2% glutaraldehyde in PBS for 10 min, and washed again with PBS, followed by an overnight incubation in staining solution (citric acid/phosphate buffer, pH 6.0, 5 mm K3Fe (CN)6, 5 mm K4Fe(CN)6, 2 mm MgCl2, 150 mm NaCl, supplemented with 1 mg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) at 37 °C. Stained cells were observed under the microscope and photodocumented with a NIKON camera.
Cell Transformation Assays
Cell transformation assay was performed by using the Cell Transformation Detection kit (CHEMICON) following the manufacturer's instructions. Base agar solution (0.8% agar in culture media) and top agar solution (0.4% agar in culture media) were prepared and preincubated at 37 °C. Plates (24-well) were coated with base agar solution and stored at 4 °C until use. Prior to use, plates were warmed up for 5 min by placing at 37 °C incubator. An equal number of MCF7, U2OS, A549, and each Hsp22-expressing cells (1250 cells/well of 24-well plate) was mixed with top agar solution. The mixture of cells and top agar solution was then placed on the top of base agar layer. Cells were fed with cell culture media twice a week and incubated at 37 °C with 5% CO2. On day 28, colonies were stained with cell stain solution. Colony-forming activity is expressed as the mean number of colonies in five fields from three independent experiments.
Chemotaxis Assay
Chemotaxis assays were performed on control and DmHsp22-transduced cells. Cells at 60–70% confluency were washed with cold PBS, trypsinized, and resuspended in Dulbecco's modified Eagle's minimal essential medium supplemented with 0.5% bovine serum albumin (Sigma) at 2 × 105 cells/ml. 2 × 104 cells were plated in Transwell inserts (12 mm-pore, Costar), and the invasion assay was performed following the manufacturer's instructions. Fibronectin from human plasma (Sigma) was used as chemoattractant. Cells that moved through the transwell were fixed with 4% formaldehyde in PBS and stained with crystal violet (0.2% in PBS).
Nude Mice Assay
Balb/c nude mice (4 weeks old, female) were bought from Nihon Clea. Control and DmHsp22-transduced cells (1 × 106 suspended in 0.3 ml of growth medium) were injected subcutaneously into the flank of nude mice (two sites per mouse). Tumor formation was monitored for the next month. The assay was regarded as positive if tumors appeared and grew progressively.
In Vitro Cell Invasion Assay
Control and DmHsp22-expressing cells were grown in the monolayer. A wound was made in the monolayer of cells by completely scratching the cells in a line with a pipette tip. Cells were washed a few times with PBS to remove cell debris and fed with fresh medium. The time of scratching wound was designated as time 0. Cells were allowed to proliferate and migrate into the wound during the next 48 h. Migration of cells into the wound was recorded under a phase contrast microscope with a 10× phase objective.
Cell Viability and Drug Resistance Assay
Equal number of control and DmHsp22-expressing MCF7 cells were seeded on a 96-well plate in a medium containing 5% FBS at 1 × 104 cells per well. When the cells reached subconfluence, cells were treated with campothecin, nocodazol, etoposide, or taxol at different doses. After a 48-h incubation, cultured media containing drug was removed, and 5% alamarBlue in culture media was added to culture wells. After a 4-h incubation at 37 °C, culture plates were read on a microplate reader at 595 nm. All assays were performed in triplicate. The number of living cells was calculated relative to the cells with nontreatment of drug, cultured, and treated with 5% alamarBlue in culture media under the same conditions, as were the experimental groups.
Statistical Analysis
The data obtained by three or more independent experiments were analyzed by Mann-Whitney test (nonparametric rank sum test) using StatView software (Abacus Concepts, Inc., Berkeley, CA). Differences were considered significant when p < 0.05.
RESULTS AND DISCUSSION
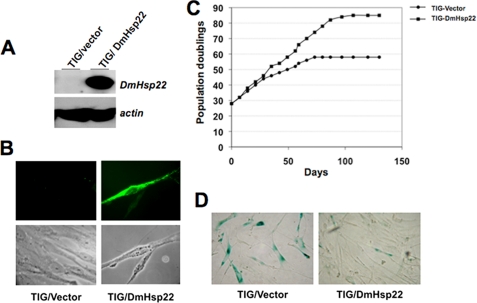
Drosophila mitochondrial Hsp22 was cloned into a retroviral expression vector as described in “Material and Methods” and was expressed in normal human fibroblasts (TIG-1). The cells were infected with vector (control) or DmHsp22-expressing retroviral vector at population doubling (PD) 24; the expression of DmHsp22 was confirmed by Western blotting and immunostaining assays with an antibody that specifically detected DmHsp22 (Fig. 1, A and B; data not shown). Of note, no cross-reactivity of the antibody was observed with any of the other human proteins. Control and DmHsp22-expressing cells were serially passaged. Whereas control cells entered senescence at 45 PD, as marked by their morphology and slow growth, the DmHsp22-expressing cells showed younger morphology and continued dividing until ∼65 PD (Fig. 1C). Control cells underwent 58 PD, whereas DmHsp22-expressing cells divided for 84 PD. Upon examination of senescence-specific β-galactosidase staining in control and DmHsp22 cells at parallel PD (48–50 PD), we found significant lower level of β-galactosidase expression in DmHsp22-expressing cells (Fig. 1D), suggesting that these cells have extended life span and slowed the rate of the aging process. Of note, the parallel experiments in which the mitochondrial chaperone mortalin/mtHsp70 was infected into TIG-1 cells gave a similar life span extension as reported earlier (23, 24).
FIGURE 1.
Normal human fibroblasts expressing DmHsp22 underwent extended PDs in culture. Whereas control cells divided for 58 PDs, the DmHsp22-expressing cells underwent 84 PDs. Expression of DmHsp22 was detected by Western blotting (A) and immunostaining (B) using anti-DmHsp22 antibody. In vitro serial passaging of the control and DmHsp22-infected cells was performed in parallel, and the senescent cells were maintained for at least 3 weeks (C). Cells were stained for senescence-associated β-galactosidase; representative images at ∼45 PD are shown (D).
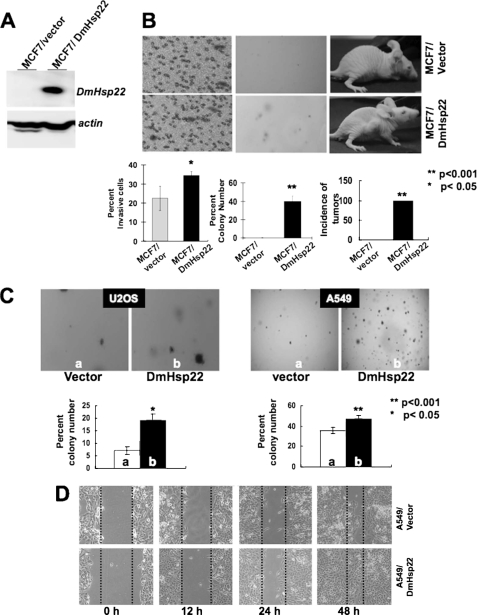
Up-regulation of mortalin has been reported in a variety of tumors, and it was shown to contribute to human carcinogenesis (25–27). Based on our above findings that the small mitochondrial Hsp22 of D. melanogaster caused life span extension of human fibroblasts, we next investigated the effects of DmHsp22 expression on human cancer cells. Human breast cancer cells (MCF7) infected with DmHsp22 virus were examined for their malignant properties in comparison to the control cells (Fig. 2). Cell transformation, cell migration, and nude mice assays in control (vector-transfected) and DmHsp22-expressing cells (Fig. 2, A and B) revealed that the expression of DmHsp22 caused malignant transformation of breast cancer cells. Control cells neither showed migration in chemotaxis assay, nor anchorage-independent growth (colony formation in soft agar), and nor formed tumors in nude mice tumor. In contrast, DmHsp22-expressing cells showed migration in the chemotaxis assay, anchorage-independent growth, and formed tumors in nude mice (Fig. 2B). Similar to the MCF7 cells, human lung carcinoma (A549) and osteosarcoma (U2OS) cells showed enhanced malignant properties as a consequence to the expression of DmHsp22. Anchorage-independent growth was increased in both cell types as a consequence of DmHsp22 expression (Fig. 2C). In wound scratch/cell invasion assays, DmHsp22-expressing cells showed enhanced motility (Fig. 2D). These data demonstrated that the DmHsp22 is functional in human cells and has effects that match with the overexpression of mortalin.
FIGURE 2.
DmHsp22 expression in human breast (MCF7) and lung (A549) cancer cells with wild type p53 caused their malignant transformation. Expression of DmHsp22 was detected by Western blotting with anti-Hsp22 antibody (A). Vector and DmHsp22-infected cells were assayed for invasion by chemotaxis assay (B, left panel), colony forming capacity (B, middle panel), and tumor formation in nude mice (B, right panel). Colony-forming efficiency of the vector and DmHsp22-expressing A549 and U2OS (osteosarcoma) cells is shown in C. Invasive capacity of A549 cells as investigated by wound-scratch assay is shown in D. Differences were considered statistically significant when p < 0.05.
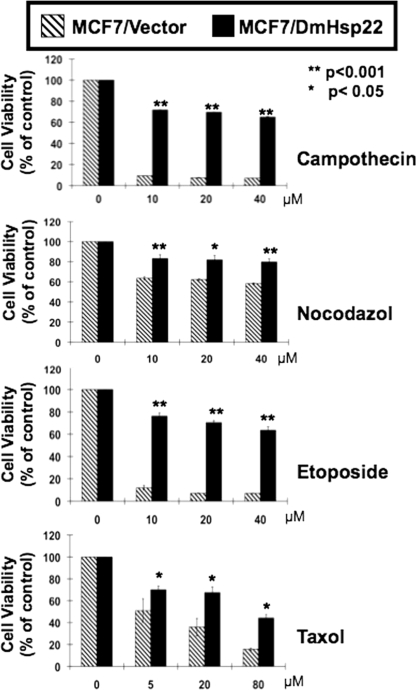
Drug resistance is a common feature of aggressive cancers. In particular, estrogen receptor-positive breast cancers that are treated with endocrine therapy meet only a limited success rate because of the drug-resistant phenomenon. For example, a large number of originally tamoxifen-sensitive tumors develop resistance after several months of treatment while still expressing the estrogen receptor (28). Mechanism(s) of estrogen response plays a critical role in the etiology, progression, drug resistance, and treatment of breast cancers and are not clearly understood. Furthermore, cadmium (metalloestrogen and environmental pollutant) that has been implicated in breast cancer resulted in an induction of human HspB8, suggesting that this sHsp may mediate estrogenic response in estrogen receptor-positive breast cancer cells that often develop drug resistance (29–31). In view of these reports and our findings described above, we next examined whether the mitochondrial DmHsp22, besides increasing the malignant properties of cancer cells, could make them resistant to drugs. The response to the four commonly used anti-cancer drugs was examined by cell viability assays. As shown in Fig. 3, the DmHsp22-expressing cells showed resistance to killing by each of the four drugs tested. Small Hsps have also been shown to form small molecular mass oligomers and interacts with biological membranes and proteins (3). Therefore, it is possible that such interactions of Hsp22 with membrane elements may regulate the drug intake and metabolism and hence modulate the signal transduction that induces growth arrest or killing.
FIGURE 3.
Drug response of vector and DmHsp22-infected cells was determined by cell viability assay. DmHsp22-expressing cells showed drug resistance. Representative response to camptothecin, nocodazol, etoposide, and taxol in three independent experiments are shown. Differences were considered statistically significant when p < 0.05.
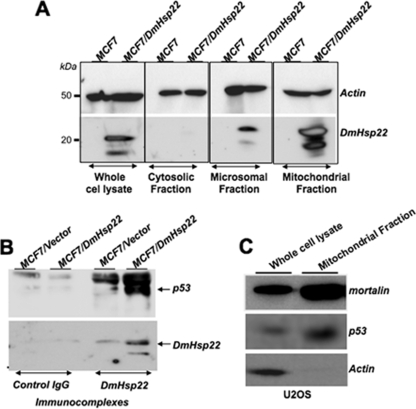
The tumor suppressor p53 pathway has a definitive role in cellular senescence and is inactivated in a large variety of cancers. Mortalin was shown to interact with p53 and inactivates its transcriptional activation function (32–35) that contributes to its proproliferative impact. To get insights to the mechanism(s) by which DmHsp22 induced proproliferative phenotypes in human cells, we examined its effect on the p53 pathway. We first examined the subcellular localization of DmHsp22 by cell fractionation and detection with anti-DmHsp22 antibody. As expected and shown in Fig. 4A, DmHsp22 was localized predominantly in the mitochondria, although there was some amount detected in the microsomal fraction as well. Immunoprecipitation of DmHsp22 and subsequent detection of p53 in the DmHsp22 immunocomplexes revealed interactions between the two proteins (Fig. 4B). An increasing number of recent studies has shown that p53 is localized in mitochondria and contributes to apoptosis independent to its transcriptional activation function in the nucleus (36–39). Although MCF7 cells could not be used for this assay due to extremely low levels of wild type p53 in these cells, we detected mitochondrial localization of p53 in U2OS cells (Fig. 4C).
FIGURE 4.
Cellular localization of DmHsp22 and its interaction with p53 protein. Subcellular localization of DmHsp22 in MCF7 cells was determined by cellular fractionation into cytosolic, microsomal, and mitochondrial fractions and detection of the protein by Western blotting with anti-DmHsp22 antibody. As shown, DmHsp22 localized in the mitochondria and microsomal fractions (A). DmHsp22 was immunoprecipitated with anti-Hsp22 antibody, and the presence of p53 in immunocomplexes was determined by Western blotting with anti-p53 antibody. p53 was detected in the DmHsp22 complexes (B) in MCF7 cells and also mitochondrial fraction of U2OS cells (C).
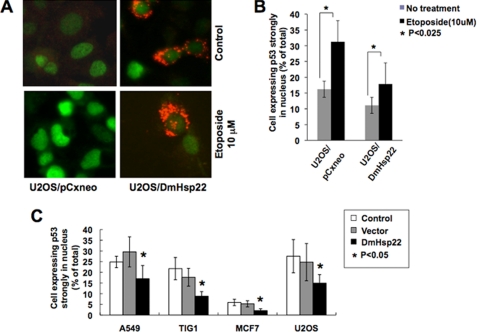
We next hypothesized that, similar to mortalin, DmHsp22 may cause functional inactivation of p53 by its cytoplasmic sequestration (32). We tested this by undertaking double immunostaining assays for p53 (green) and DmHsp22 (red). We found that, whereas control U2OS cells had 67% of cells showing nuclear p53, DmHsp22-expressing cells showed only 22% cells with nuclear p53. Furthermore, the intensity of nuclear p53 staining in DmHsp22-expressing cells was lower as compared with the control cells (Fig. 5). Furthermore, in DmHsp22-expressing U2OS cells, we found that there was a significant decrease in DNA damage-induced activation of p53 pathway. As shown in Fig. 5A, control cells showed strong nuclear p53 staining subsequent to the treatment with etoposide, whereas in DmHsp22-expressing cells, the nuclear p53 was significantly abrogated and was particularly noticeable in cells that expressed high levels of DmHsp22, suggesting that the DmHsp22 inactivated tumor suppressor p53 by interacting with and inhibiting its nuclear transport (Fig. 5, A and B). Quantitation of number of cells with very strong nuclear p53 staining subsequent to etoposide treatment in control and DmHsp22-expressing cells revealed that the inactivation of p53 by DmHsp22 in U2OS, A549, and MCF7 cells was statistically significant (Fig. 5B and data not shown). We further examined if the similar inactivation of p53 by DmHsp22 occurred in normal human cells and had resulted in their lifespan extension as shown in Fig. 1C. Immunostaining of p53 in control and DmHsp22-expressing TIG-1, A549, MCF7, and U2OS cells revealed that in each case DmHsp22 expression caused ∼40% decrease in number of cells showing nuclear staining for p53 (Fig. 5C). Taken together, these data suggested that the inactivation of p53 by DmHsp22 is not cell line-specific, and it may be responsible for proproliferative effects of DmHsp22 (delayed aging of normal cells and increased malignant properties of cancer cells) seen in human normal and cancer cells.
FIGURE 5.
Inactivation of wild type p53 by DmHsp22. p53 was activated by exposure of cells to etoposide that caused nuclear translocation of p53, seen as bright nuclear staining (green). In many DmHsp22-expressing cells, etoposide-induced nuclear translocation of p53 was abrogated (A). Quantitation of the data obtained from three independent experiments and the statistical significance are shown (B). Reduction in number of cells with nuclear p53 in DmHsp22-expressing cells was observed in four human cells, including A549-lung carcinoma, MCF7-breast carcinoma, U2OS-bone carcinoma, and TIG-1-normal fibroblasts. Quantitation of the data obtained from three experiments and statistical significance are shown (C). Differences were considered statistically significant when p < 0.05.
At the present time, there are no known mammalian homologs of DmHsp22. Whereas DmHsp22 is localized in the matrix of mitochondria in mammalian cells, the other mammalian sHsps are mainly localized in the cytosol and nucleus; some cytosolic sHsps, for example HspB2, have been reported to interact with the outer membrane of the mitochondria (40). In this regard, DmHsp22 appears to be similar to mammalian mtHsp70/mortalin and Hsp60 that are found predominantly in mitochondria (41). Besides the mitochondrial localization that is observed for mortalin, Hsp60, and DmHsp22, they have molecular chaperone function in vitro and in vivo (15, 16, 42). Of note, whereas mortalin and DmHsp22 caused life span extension of normal human cells, Hsp60 was not seen to have such function in parallel assays, suggesting that Dmhsp22 functions like mortalin. The mechanism by which it increases life span warrants further studies. The cross-species effects would be of particular interest in understanding the molecular mechanisms by which DmHsp22 regulates life span and thus help to unveil the in vivo role of a mitochondrial small Hsp.
Acknowledgments
We thank Hoang Thi Hang and Tomoko Yaguchi for assistance.
This work was supported in part by grants from the Canadian Institutes of Health Research (MOP-77796), New Energy and Industrial Technology Development Organization (Japan), the Ministry of Commerce Industry and Energy (10030051), and the Korea Science and Engineering Foundation (R01-2006-000-10084-0, R15-2004-024-02001-0, M10416130002-04N1613-00210).
- sHsp
- small heat shock protein
- DmHsp22
- Drosophila mitochondrial Hsp22
- PD
- population doubling
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Dierick I., Irobi J., De Jonghe P., Timmerman V. (2005) Ann. Med. 37, 413–422 [DOI] [PubMed] [Google Scholar]
- 2.Hu Z., Chen L., Zhang J., Li T., Tang J., Xu N., Wang X. (2007) J. Neurosci. Res. 85, 2071–2079 [DOI] [PubMed] [Google Scholar]
- 3.Shemetov A. A., Seit-Nebi A. S., Gusev N. B. (2008) J. Neurosci. Res. 86, 264–269 [DOI] [PubMed] [Google Scholar]
- 4.Kampinga H. H., Hageman J., Vos M. J., Kubota H., Tanguay R. M., Bruford E. A., Cheetham M. E., Chen B., Hightower L. E. (2009) Cell Stress Chaperones 14, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benndorf R., Sun X., Gilmont R. R., Biederman K. J., Molloy M. P., Goodmurphy C. W., Cheng H., Andrews P. C., Welsh M. J. (2001) J. Biol. Chem. 276, 26753–26761 [DOI] [PubMed] [Google Scholar]
- 6.Fontaine J. M., Sun X., Benndorf R., Welsh M. J. (2005) Biochem. Biophys. Res. Commun. 337, 1006–1011 [DOI] [PubMed] [Google Scholar]
- 7.Michaud S., Marin R., Tanguay R. M. (1997) Cell Mol. Life Sci. 53, 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrow G., Tanguay R. M. (2008) Biotechnol. J. 3, 728–739 [DOI] [PubMed] [Google Scholar]
- 9.Irobi J., Van Impe K., Seeman P., Jordanova A., Dierick I., Verpoorten N., Michalik A., De Vriendt E., Jacobs A., Van Gerwen V., Vennekens K., Mazanec R., Tournev I., Hilton-Jones D., Talbot K., Kremensky I., Van Den Bosch L., Robberecht W., Van Vandekerckhove J., Van Broeckhoven C., Gettemans J., De Jonghe P., Timmerman V. (2004) Nat. Genet. 36, 597–601 [DOI] [PubMed] [Google Scholar]
- 10.Carra S., Sivilotti M., Chávez Zobel A. T., Lambert H., Landry J. (2005) Hum. Mol. Genet. 14, 1659–1669 [DOI] [PubMed] [Google Scholar]
- 11.Fontaine J. M., Sun X., Hoppe A. D., Simon S., Vicart P., Welsh M. J., Benndorf R. (2006) FASEB J. 20, 2168–2170 [DOI] [PubMed] [Google Scholar]
- 12.Kim M. V., Kasakov A. S., Seit-Nebi A. S., Marston S. B., Gusev N. B. (2006) Arch. Biochem. Biophys. 454, 32–41 [DOI] [PubMed] [Google Scholar]
- 13.Simon S., Fontaine J. M., Martin J. L., Sun X., Hoppe A. D., Welsh M. J., Benndorf R., Vicart P. (2007) J. Biol. Chem. 282, 34276–34287 [DOI] [PubMed] [Google Scholar]
- 14.Michaud S., Morrow G., Marchand J., Tanguay R. M. (2002) Prog. Mol. Subcell. Biol. 28, 79–101 [DOI] [PubMed] [Google Scholar]
- 15.Morrow G., Inaguma Y., Kato K., Tanguay R. M. (2000) J. Biol. Chem. 275, 31204–31210 [DOI] [PubMed] [Google Scholar]
- 16.Morrow G., Heikkila J. J., Tanguay R. M. (2006) Cell Stress Chaperones 11, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King V., Tower J. (1999) Dev. Biol. 207, 107–118 [DOI] [PubMed] [Google Scholar]
- 18.Kurapati R., Passananti H. B., Rose M. R., Tower J. (2000) J. Gerontol. A Biol. Sci. Med. Sci. 55, B552–559 [DOI] [PubMed] [Google Scholar]
- 19.Morrow G., Samson M., Michaud S., Tanguay R. M. (2004) FASEB J. 18, 598–599 [DOI] [PubMed] [Google Scholar]
- 20.Morrow G., Battistini S., Zhang P., Tanguay R. M. (2004) J. Biol. Chem. 279, 43382–43385 [DOI] [PubMed] [Google Scholar]
- 21.Tao D., Lu J., Sun H., Zhao Y. M., Yuan Z. G., Li X. X., Huang B. Q. (2004) Acta Biochim. Biophys. Sin. 36, 618–622 [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y., Sun H., Lu J., Li X., Chen X., Tao D., Huang W., Huang B. (2005) J. Exp. Biol. 208, 697–705 [DOI] [PubMed] [Google Scholar]
- 23.Kaul S. C., Yaguchi T., Taira K., Reddel R. R., Wadhwa R. (2003) Exp. Cell Res. 286, 96–101 [DOI] [PubMed] [Google Scholar]
- 24.Kaula S. C., Reddelb R. R., Sugiharac T., Mitsuia Y., Wadhwac R. (2000) FEBS Lett. 474, 159–164 [DOI] [PubMed] [Google Scholar]
- 25.Dundas S. R., Lawrie L. C., Rooney P. H., Murray G. I. (2005) J. Pathol. 205, 74–81 [DOI] [PubMed] [Google Scholar]
- 26.Yi X., Luk J. M., Lee N. P., Peng J., Leng X., Guan X. Y., Lau G. K., Beretta L., Fan S. T. (2008) Mol. Cell Proteomics 7, 315–325 [DOI] [PubMed] [Google Scholar]
- 27.Wadhwa R., Takano S., Kaur K., Deocaris C. C., Pereira-Smith O. M., Reddel R. R., Kaul S. C. (2006) Int. J. Cancer 118, 2973–2980 [DOI] [PubMed] [Google Scholar]
- 28.Robertson J. F., Cannon P. M., Nicholson R. I., Blamey R. W. (1996) Int. J. Biol. Markers 11, 29–35 [DOI] [PubMed] [Google Scholar]
- 29.Sun X., Fontaine J. M., Bartl I., Behnam B., Welsh M. J., Benndorf R. (2007) Cell Stress Chaperones 12, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besada V., Diaz M., Becker M., Ramos Y., Castellanos-Serra L., Fichtner I. (2006) Proteomics 6, 1038–1048 [DOI] [PubMed] [Google Scholar]
- 31.Fichtner I., Becker M., Zeisig R., Sommer A. (2004) Eur. J. Cancer 40, 845–851 [DOI] [PubMed] [Google Scholar]
- 32.Wadhwa R., Takano S., Robert M., Yoshida A., Nomura H., Reddel R. R., Mitsui Y., Kaul S. C. (1998) J. Biol. Chem. 273, 29586–29591 [DOI] [PubMed] [Google Scholar]
- 33.Kaul S. C., Aida S., Yaguchi T., Kaur K., Wadhwa R. (2005) J. Biol. Chem. 280, 39373–39379 [DOI] [PubMed] [Google Scholar]
- 34.Ma Z., Izumi H., Kanai M., Kabuyama Y., Ahn N. G., Fukasawa K. (2006) Oncogene 25, 5377–5390 [DOI] [PubMed] [Google Scholar]
- 35.Walker C., Böttger S., Low B. (2006) Am. J. Pathol. 168, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U. M. (2003) Mol. Cell 11, 577–590 [DOI] [PubMed] [Google Scholar]
- 37.Mihara M., Moll U. M. (2003) Methods Mol. Biol. 234, 203–209 [DOI] [PubMed] [Google Scholar]
- 38.Morselli E., Galluzzi L., Kepp O., Kroemer G. (2009) Cell Cycle 8, 1647–1648 [PubMed] [Google Scholar]
- 39.Vaseva A. V., Marchenko N. D., Moll U. M. (2009) Cell Cycle 8, 1711–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa M., Tsujimoto N., Nakagawa H., Iwaki T., Fukumaki Y., Iwaki A. (2001) Exp. Cell Res. 271, 161–168 [DOI] [PubMed] [Google Scholar]
- 41.Deocaris C. C., Kaul S. C., Wadhwa R. (2006) Cell Stress Chaperones 11, 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heikkila J. J., Kaldis A., Morrow G., Tanguay R. M. (2007) Biotechnol. Adv. 25, 385–395 [DOI] [PubMed] [Google Scholar]