Abstract
Sus1 is a central component of the yeast gene gating machinery, the process by which actively transcribing genes such as GAL1 become associated with nuclear pore complexes. Sus1 is a component of both the SAGA transcriptional co-activator complex and the TREX-2 complex that binds to nuclear pore complexes. TREX-2 contains two Sus1 chains that have an articulated helical hairpin fold, enabling them to wrap around an extended α-helix in Sac3, following a helical hydrophobic stripe. In SAGA, Sus1 binds to Sgf11 and has been proposed to provide a link between SAGA and TREX-2. We present here the crystal structure of the complex between Sus1 and the N-terminal region of Sgf11 that forms an extended α-helix around which Sus1 wraps in a manner that shares some similarities with the Sus1-Sac3 interface in TREX-2. However, the Sus1-binding site on Sgf11 is somewhat shorter than on Sac3 and is based on a narrower hydrophobic stripe. Engineered mutants that disrupt the Sgf11-Sus1 interaction in vitro confirm the importance of the hydrophobic helical stripe in molecular recognition. Helix α1 of the Sus1-articulated hairpin does not bind directly to Sgf11 and adopts a wide range of conformations within and between crystal forms, consistent with the presence of a flexible hinge and also with results from previous extensive mutagenesis studies (Klöckner, C., Schneider, M., Lutz, S., Jani, D., Kressler, D., Stewart, M., Hurt, E., and Köhler, A. (2009) J. Biol. Chem. 284, 12049–12056). A single Sus1 molecule cannot bind Sgf11 and Sac3 simultaneously and this, combined with the structure of the Sus1-Sgf11 complex, indicates that Sus1 forms separate subcomplexes within SAGA and TREX-2.
Keywords: Nucleus/Nuclear Export, Nucleus/Nuclear Pore, Protein/Protein-Protein Interactions, Protein/Structure, Transcription, Integration Gene Expression
Introduction
There is an emerging consensus that the different steps of the gene expression pathway are tightly coupled and show a high degree of interdependence (2–5). Integration of the steps that lead from transcription to mRNA nuclear export (including splicing, 5′ cap addition, and polyadenylation) relies on a complex network of interactions between activated genes, processing factors and the nuclear pore complex (NPC).3 For a subset of genes, this integration is achieved by tethering actively transcribed genes to the NPC, a process known as “gene gating” (6), in which the small nuclear protein, Sus1, is a central component (7–10). Sus1 is part of both the 2-MDa Spt-Ada-Gcn5 acetyltransferase (SAGA) complex, that is a co-activator for transcription by RNA polymerase II, and the TREX-2 (transcription and export-2) complex that tethers SAGA to NPCs (reviewed in Ref. 11). The SAGA complex possesses histone acetyltransferase and deubiquitination activities and is bound to chromatin during active transcription (12). The TREX-2 complex contains Sac3, Thp1, Sem1, and Cdc31 in addition to Sus1 and has roles in both nuclear mRNA export and transcription elongation (8, 11, 13–16). The TREX-2 complex binds the mRNA export factors Mex67 and Mtr2 as well as NPCs (13) and, by interacting with both SAGA and NPCs, facilitates gene gating (9). It has been proposed that gated genes may benefit from enhanced or optimal rates of messenger synthesis compared with genes residing in an intranuclear position (17), and this may be of particular importance in producing a rapid cellular response to stress, consistent with the involvement of a significant proportion of SAGA regulated genes in stress response (18).
Yeast SAGA is a large 2-MDa complex that functions as a transcriptional co-activator for a number of RNA polymerase II dependent genes (12, 19). The Gcn5 histone-acetyltransferase forms a submodule with multiple Ada proteins that regulate gene expression by acetylation of histones H3 and H2B (20–22). The SAGA complex also contains a second enzymatic activity through the deubiquitinating (DUB) module formed by the ubiquitin-specific protease, Ubp8, together with SAGA subunits Sgf73, Sgf11, and Sus1 (23, 24). The complex deubiquitinates Lys123 on histone H2B that has been shown to have downstream effects on the methylation state of histone H3 and a regulatory role in gene activation (25, 26). Within the yeast SAGA complex, Sgf11 binds Sus1 directly, forming a stable dimer (23). However, both the Sgf11/Sus1 heterodimer and Ubp8 are required for all three proteins to bind to Sgf73 (23, 24). Analogous interactions are found between Sgf11, Sus1, and Ubp8 homologues in both Drosophila and human cells (27–29). Sus1 is also associated with mRNA biogenesis during transcription elongation, being observed at coding regions in a SAGA-dependent manner (30). This is consistent with the presence of SAGA subunits within both open reading frames and at promoter regions (31). Histone deubiquitination and Ubp8 catalytic activity appear to be required for the association of actively transcribing genes with NPCs during gene gating (24). Although the precise details of the arrangement of the individual proteins within the SAGA DUB module remains to be established, the interaction between Sgf11 and Sus1 is necessary for its integrity (11, 23, 29).
Within the TREX-2 complex, residues 723–805 of Sac3 (the “CID” domain) bind two Sus1 chains and one chain of the calmodulin-like centrin, Cdc31. The crystal structure of this complex (10) shows that the Sac3 CID region adopts an extended α-helical conformation about which Sus1 and Cdc31 wrap. Both Sus1 chains have an articulated helical hairpin fold that is based on five α-helical segments linked by putative hinges. This conformation has been proposed to enable them to wrap around the Sac3 helix, like fingers gripping a thin rod (10). The binding interface contains few hydrogen bonds or polar interactions and is based on a hydrophobic stripe that winds around the Sac3 helix. Both Sus1 molecules (designated Sus1A and Sus1B) bind to stripes within the Sac3 CID region. In each case, the hydrophobic stripe is generated by a four-residue sequence repeat in which the first two residues are Phe, Tyr, Ile, Leu, or Met or have a side chain containing a considerable hydrophobic portion, such as Arg or Glu. In both Sus1-binding sites on Sac3, the binding motif was ∼25-residues-long. Both Sus1 chains and Cdc31 are required for optimal NPC association of TREX-2 (10, 24).
Because Sus1 is found in both SAGA and TREX-2, it was initially proposed that Sus1 may physically bridge the two complexes (7). More recently, a more dynamic role for Sus1 in linking the two complexes has been proposed, whereby the competitive exchange of Sus1 molecules between SAGA and TREX-2 would serve to both physically link the complexes and modulate their function (1). Alternatively, the presence of Sus1 in both complexes may be coincidental, and this protein may have mechanistically separate roles in SAGA and TREX-2. Other proteins may also be important in linking the two complexes and, for example, deletion of the SAGA component Sgf73 alters the association of Sus1 with the TREX-2 complex (24, 30). Sgf73 also plays a role in recruiting Sac3 and Thp1 to SAGA, and it has been suggested that Sgf73 may alter the TREX-2 component Sac3 allowing for efficient TREX-2 assembly (24). Distinguishing between these different putative functions of Sgf11 and Sus1 has been difficult because it was not known how Sus1 binds to Sgf11 (11, 32). Extensive mutagenesis studies of Sus1 showed that, although it was possible to generate mutants in which Sac3 binding was lost while Sgf11 binding was retained, it was not possible to generate mutants in which Sgf11 binding was lost while Sac3 binding was retained (1). These mutagenesis results indicated that Sus1 binding to these two partners was in some way different but did not indicate what these differences were.
Here, we present the crystal structure of the N-terminal region of Sgf11 bound to Sus1, its direct binding partner in the SAGA complex. Sgf11 forms an extended α-helix around which Sus1 wraps in a manner that has some similarities to the way in which it binds to Sac3 in the Sac3-Cdc31-Sus1 complex. However, the Sus1-binding site on Sgf11 is somewhat shorter than on Sac3 and is based on a narrower hydrophobic stripe. As a consequence, helix α1 of the Sus1-articulated hairpin fold does not bind directly to Sgf11 and, consistent with the presence of a flexible hinge between it and the next helix (α2), takes up a remarkably wide range of conformations within and between crystal forms. We have engineered mutants based on the structure of the complex that disrupt the Sgf11-Sus1 interaction and have used these to confirm the importance of the hydrophobic stripe in molecular recognition.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Full-length and truncated Sgf11 variants were generated by PCR using yeast genomic DNA (Novagen; Beston, UK) and cloned into pGEXTEV (33), a modified version of pGEX-4T-1 (GE Healthcare) in which the thrombin site has been replaced with a TEV protease site. Sus1 cDNA was cloned into an untagged expression vector, pNMTK, a modified pOPT vector (34) with kanamycin resistance. Sgf11 Ala insertion mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene). The cloning of Cdc31 and Sac3 variants has been described elsewhere (10).
For crystallography, GST-Sgf11 constructs (amino acids 1–33 and 7–33) were coexpressed with untagged Sus1 in BL21 (DE3) CodonPlus RIL cells in ZYM-5052 autoinducing medium (35) at 20 °C. Cells were lysed by high pressure cavitation (10–15k pounds per square inch) in 50 mm Tris-HCl (pH 8.0), 25% w/v sucrose, 1 mm EGTA, and 1 mm phenylmethanesulphonyl fluoride. Complete EDTA-free protease inhibitor mixture (Roche) was added to the lysed cells. Cells were clarified by centrifugation, filtered through a 0.45-μm membrane, and bound to glutathione-Sepharose 4B resin (GE Healthcare) for 1 h at 4 °C. The resin was washed with 500 ml of 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol to remove nonspecifically bound proteins, and the Sgf11-Sus1 complex released from the GST tag by overnight incubation with 100 μg of His-TEV protease (S219V mutant; (36)). The complex was further purified on a HiLoad Superdex 75 26/60 prep-grade column (GE Healthcare) in 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol.
Crystallization and Data Collection
Crystals were grown at 19 °C by hanging drop vapor diffusion in either 3.2 m sodium formate (Sgf11-(7–33)-Sus1) or 1.1 m NaH2PO4 and 20 mm K2HPO4 (Sgf11-(1–33)-Sus1). Crystals of Sgf11-(7–33)-Sus1 required no cryoprotectant; however, crystals of Sgf11-(1–33)-Sus1 were exposed to mother liquor containing 30% (w/v) sucrose before flash cooling in liquid nitrogen prior to data collection. Crystals of Sgf11-(1–33)-Sus1 were also dehydrated by vapor diffusion, by adding increasing amounts of sucrose into the mother liquor for up to a week before flash freezing. Crystallographic data were collected either in-house using a Rigaku FR-E+ SuperBright generator equipped with Osmic mirrors and a Mar DTB camera or at the Diamond Synchrotron (Oxford, UK) (Table 1).
TABLE 1.
Crystal data
| Crystals | ||
| Sgf11 fragment | 7–33 | 1–33 |
| Space group | P61 | P61 |
| Unit cell dimensions (Å) | ||
| a | 75.15 | 68.67 |
| b | 75.15 | 68.67 |
| c | 197.37 | 232.86 |
| Data collection | ||
| Wavelength (Å) | 1.54059 | 1.0719 |
| Resolution range (Å)a | 25–2.1 (2.21–2.1) | 59.4–2.7 (2.85–2.7) |
| Total observationsa | 503,521 (37,147) | 382,872 (56,409) |
| Unique observationsa | 36,176 (5,136) | 17,082 (2,485) |
| Completeness (%)a | 98.3 (95.3) | 100 (100) |
| Multiplicity | 13.9 (7.2) | 22.4 (22.6) |
| Rpim (%)a | 1.5 (29.2) | 2.4 (29.7) |
| Rmerge (%)a | 5.9 (75.7) | 11.2 (138.6) |
| Mean I/σ(I)a | 26.2 (2.4) | 18.1 (2.7) |
| Refinement | ||
| Rcryst/Rfree (%) | 19.0/23.7 | 21.3/25.3 |
| Bond length r.m.s.d. (Å)b | 0.002 | 0.002 |
| Bond angle r.m.s.d. | 0.6° | 0.62° |
| MolProbity score/percentile | 1.41/99 | 2.10/97 |
| Ramachandran plot (%) | ||
| Favoured | 99.5 | 98.4 |
| Allowed | 0.5 | 1.6 |
| Forbidden | 0 | 0 |
| PDB accession codec | 3KIK | 3KJL |
a Parentheses refer to final resolution shell.
b r.m.s.d., root mean square deviation.
c PDB, Protein Data Bank.
Structure Solution and Refinement
Although both crystal forms appeared to have P6122 symmetry, subsequent analysis indicated that each had P61 symmetry and were almost perfectly merohedrally twinned. The crystals obtained with Sgf11 (residues 7–33) diffracted isotropically to a resolution of 2.11 Å, whereas those using residues 1–33 gave very anisotropic diffraction extending to 2.4 Å resolution along the c axis, but to only 2.7 Å resolution along the a and b axes. Molecular replacement using residues 21–90 of Sus1 from the Sac3-Sus1-Cdc31 complex (10) in PHASER (CCP4) gave a solution for four chains in the asymmetric unit of the Sgf11-(7–33)-Sus1 crystals, which was then refined using twinned refinement option in PHENIX (37) employing the twinning operator (h, -h-k, -l) and with Rfree flags set appropriately. Alternating cycles of refinement and rebuilding in COOT generated a model with an R-factor of 19.0% (Rfree = 23.7%), excellent geometry (Table 1) and a MolProbity score of 1.41 (99th percentile). Because of the presence of merohedral symmetry, stringent geometric constraints were applied, and these were then optimized to minimize the Rfree value during refinement. TLS parameters were obtained from the TLS website (38). The model obtained for the Sgf11-(7–33)-Sus1 crystals was then used to solve the Sgf11-(1–33)-Sus1 crystals using molecular replacement, which, after iterative cycles of refinement and rebuilding, gave a model with an R-factor of 21.3% (Rfree = 25.3%) and excellent geometry (Table 1).
In Vitro Binding Studies
Binding assays were carried out by immobilizing GST fusion protein containing complexes on glutathione 4B resin from clarified bacterial lysates. Sgf11 fragments and mutants were coexpressed with untagged Sus1, and the cell lysates were incubated with resin at 4 °C for 1 h. Resin from the 1–33, 7–33, and 1–44 Sgf11 fragments was washed with 20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol. Whereas resin from the full-length Sgf11 variants was washed with 20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 10 μm ZnCl2, and 5 mm β-mercaptoethanol. Samples were analyzed by SDS-PAGE.
Binding studies demonstrating the mutually exclusive binding of Sus1 to Sac3 or Sgf11 involved immobilizing coexpressed GST-Sac3-Cdc31 complexes onto glutathione 4B resin from clarified bacterial lysates by incubation at 4 °C for 1 h. The resin was washed with 20 mm Tris-HCl (pH 8.0), 200 mm NaCl, and 1 mm dithiothreitol. The full-length Sgf11-Sus1 complex was purified essentially as described for Sgf11-(1–33) and Sgf11-(7–33) complexes, except buffers did not contain EDTA or EGTA. Instead, the lysis buffer contained 100 μm ZnCl2, GST wash buffer and size exclusion chromatography buffers contained 10 μm ZnCl2 and 5 mm β-mercaptoethanol. The purified Sgf11-Sus1 complex was added to resin-bound GST-Sac3-Cdc31 complexes and incubated at 4 °C for 1 h. The resin was washed with 20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 10 μm ZnCl2, and 5 mm β-mercaptoethanol. Unbound and resin bound material was analyzed by SDS-PAGE.
RESULTS AND DISCUSSION
Sus1 Wraps around a Helix Formed by Residues 8–30 of Sgf11
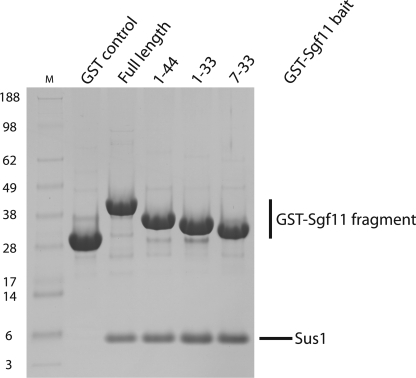
In contrast to Sac3, each Sgf11 chain binds only a single Sus1 chain (10). Inspection of the Sgf11 sequence (24, 29) indicated the presence of a putative CCCH zinc finger domain at its C terminus (residues ∼65–99). Deletion mutants indicated that Sus1 binding involved primarily the N terminus of Sgf11 (residues 1–33; Fig. 1). Crystals suitable for high resolution structure determination were obtained with Sgf11 fragments comprising residues 1–33 and 7–33. The crystals containing Sgf11 residues 1–33 showed anisotropic diffraction, whereas those containing residues 7–33 diffracted to 2.1 Å resolution and were solved first using molecular replacement based on a model of residues 21–90 of Sus1 from the Cdc31-Sac3-Sus1 complex (10). The structure obtained in this way was then used to obtain a structural model for the complex between Sus1 and Sgf11 residues 1–33. Both crystal forms had P61 symmetry with near prefect merohedral twinning and the asymmetric unit of each contained four copies of the Sgf11-Sus1 complex. In each case, there was an approximate 2-fold rotation axis relating one pair of complexes to the other pair. Although the relation between these pairs was different in the two crystal forms, the arrangement of chains within a pair was preserved.
FIGURE 1.
An N-terminal region of Sgf11 comprising residues 7–33 is sufficient for binding to Sus1. GST fusions of Sgf11 and a series of fragments were coexpressed with Sus1 in Escherichia coli, and complexes were immobilized on glutathione-Sepharose 4B resin from clarified lysates. After extensive washing, bound material was analyzed by SDS-PAGE.
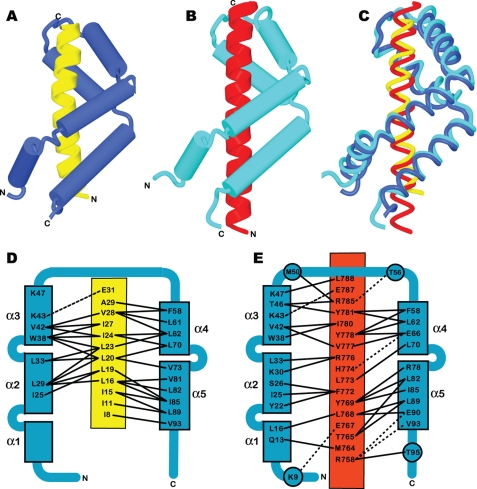
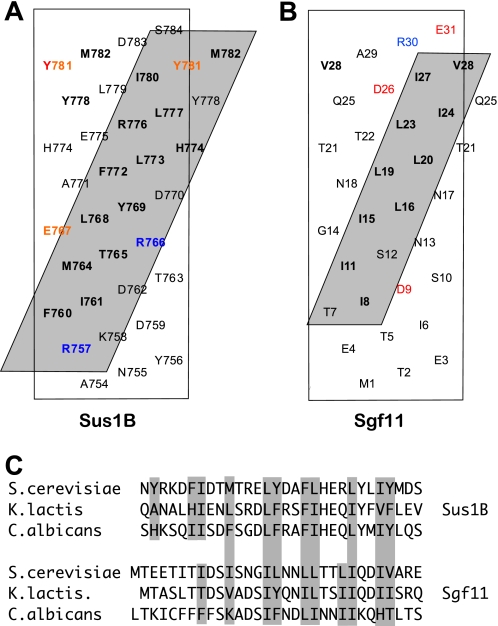
Fig. 2A and supplemental Movie S1 illustrate the structure of the Sgf11-Sus1 complex. Sus1 retained the articulated α-helical hairpin fold observed in crystals of complexes with Sac3 (Fig. 2, B and C, and supplemental Movie S2) (10). This fold is based on five α-helices (α1–5) connected by flexible linkers that enable Sus1 to wrap around an isolated α-helix such as that observed for the Sgf11 N-terminal region (residues 8–30). The helical conformation was lost for residues 2–6 and 31–33 of Sgf11. The Sgf11-Sus1 interface had a buried surface area 1,078 Å2, which was slightly lower than the 1,324–1,546 Å2 observed for the two Sus1-Sac3 interfaces in the Sac3 CID complex (10). Molecular recognition in the Sgf11-Sus1 complex was based on the hydrophobic helical stripe on the Sgf11 helix derived from an ∼four-residue-repeating sequence motif (Fig. 3). A similar pattern of hydrophobic residues was seen in the sequences of Sgf11 from Kluyveromyces lactis and Candida albicans (Fig. 3C). Although the helical stripe motif on Sgf11 was similar in some respects to that observed for the Sac3-Sus1 interfaces, it was narrower and shorter than in Sac3. The interaction interface involved residues 8–32 of Sgf11 and was primarily hydrophobic with few H-bonds or salt bridges (Figs. 2D and 3B). The Sgf11 interaction interface on Sus1 was based primarily on the inner surface of helices α2–5, with little contact with helix α1 and was thus less extensive than in Sac3 (Figs. 2 and 3). However, in the DUB complex, Sus1 helix α1 may be involved with interactions with other components such as Sgf73 and/or Ubp8. In contrast to the interaction with Sac3, Sus1 does not encircle the Sgf11 helix completely and instead leaves one side exposed.
FIGURE 2.
Overview of the structure of the Sgf11-Sus1 complex (yellow, Sgf11 residues 8–31; blue, Sus1) (A) and comparison with Sus1 bound to Sac3 (red, Sac3 residues 755–788; cyan, Sus1) (B). C, the structures (represented as worm format in the same orientations as A and B) of the two complexes superimposed. Sus1 takes up an articulated helical hairpin fold based on five helices separated by flexible hinges that enables it to bend around the Sgf11 helix. D, the principal interactions between Sus1 (blue) and Sgf11 (yellow), which are somewhat different to those observed between Sus1 (blue) and Sac3 (red) (E, see Ref. 10).
FIGURE 3.
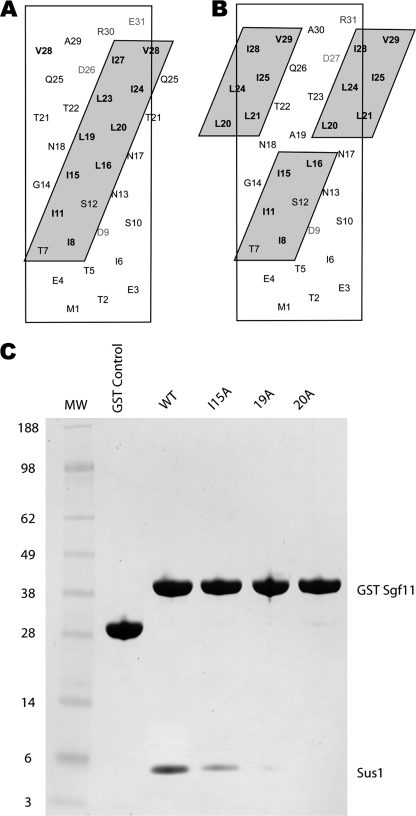
A and B, helical nets showing the hydrophobic stripe on Sac3 to which Sus1 binds (A; see Ref. 10) and comparing it with the stripe on Sgf11 (B). The Sus1-interaction interface on Sgf11 is clearly less extensive than that seen on Sac3. Charged residues are red (positive) and blue (negative). C, the hydrophobic stripe on Saccharomyces cerevisiae Sgf11 is generated by an ∼four-residue sequence repeat of hydrophobic side chains. Similar four-residue sequence repeat motifs were present in Sgf11 from K. lactis and C. albicans.
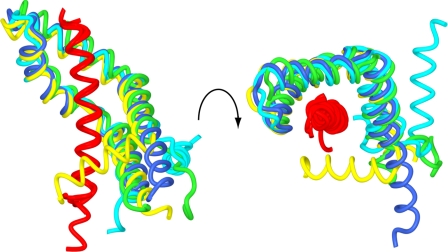
There was a striking variation in the position of Sus1 helix α1 between Sgf11-Sus1 complexes within and between crystals (Fig. 4 and supplemental Movie S3) so that in the most extreme case, this helix had been rotated by almost 180° between different copies. Moreover, the structural integrity of this region of Sus1 was reduced in the Sgf11 complex, and often, the density in this region was less clear than in other regions of the structure. This remarkable structural plasticity provided direct evidence for the flexibility of the hinge between helices α1 and -2.
FIGURE 4.
Structural plasticity of Sus1. Worm traces of the different copies of the complex in the crystals superimposed to illustrate that although the relative positions of helices α2–5 were well conserved between different copies of the Sgf11-Sus1 complex, the position of helix α1 was very variable. The Sgf11 helix is shown in red; two chains from the complex with Sgf11 residues 7–33 are shown in blue and yellow, and two chains from the complex with Sgf11 residues 1–33 are shown in cyan and green.
The structure of the Sgf11-Sus1 interface and comparison with Sus1 bound to Sac3 is consistent with the results obtained from a mutagenesis study of Sus1 (1), in which it was possible to obtain mutants that retained affinity for Sgf11, whereas affinity for Sac3 was lost. The predominantly hydrophobic nature of the Sgf11-Sus1 interface was consistent with the failure of mutagenesis of charged residues in Sus1 (mutants sus1-1 to sus1-9) to impair the interaction with either Sac3 or Sgf11 (1). The lack of contact between Sgf11 and the N and C termini of Sus1 seen in the crystal structures of the complex was consistent with the failure of deletions of Sus1 residues 1–10 or 91–96 to alter its binding to Sgf11. Mutations of Gly37 and Trp38 to Ala (sus1-11) impaired binding to both Sac3 and Sgf11. Trp38 is strongly conserved in Sus1 (10), and Gly37 forms the hinge between helices α2 and α3, and so these mutations would be expected to cause major disruption to the molecular architecture of the Sus1 molecule, consistent with this mutant binding neither Sgf11 nor Sac3. Mutations within the hinge between Sus1 helices α1 and α2 (sus1-10) had little effect on the interaction with Sgf11 but did impair binding to Sac3, consistent with helix α1 making considerable contact in the Sus1-Sac3 complexes. Finally, mutation in the hinge between Sus1 helices α4 and α5 (sus1–12 V73A/D75A) caused a less severe phenotype for Sus1 binding but did not alter the Sgf11 interaction, probably because these residues are not intimately involved in the Sgf11-Sus1 interface (Fig. 2, D and E). As illustrated in Figs. 2 and 3, the Sus1-binding interface on Sac3 was more extensive than the Sgf11-binding interface and overlapped it completely, accounting for the inability to engineer Sus1 mutants in which Sac3 binding was retained, whereas Sgf11 binding was impaired.
Mutations That Interfere with the Sus1-Sgf11 Interaction
Previous studies (10, 39) highlighted the difficulty of interfering with interactions of the type observed between Sgf11 and Sus1 using simple point mutations. The interaction interface was primarily hydrophobic, and so mutations based on charge repulsion could not be employed, whereas dramatic mutations, involving substituting large hydrophobic residues or substituting charged for hydrophobic residues, would increase the likelihood of introducing major conformational changes. In analogous studies on the binding of calmodulin to helical peptides, Ala substitutions sometimes even enhanced affinity (39). We therefore used an alternative strategy to disrupt the interaction, based on inserting an Ala residue in the Sgf11 helix, so that the hydrophobic stripe to which Sus1 binds was disrupted. As illustrated in Fig. 5, A and B, insertion of a single residue in an α-helix results in a rotation of ∼100°. Insertion of a single Ala after Sgf11 residues 18 or 19 (lanes 19A and 20A in Fig. 5C) resulted in a dramatic decrease in the affinity of Sgf11 for Sus1 in in vitro pulldown assays (Fig. 5C). The effectiveness of this mutagenic approach is highlighted by comparison with a point mutation in the hydrophobic stripe at Ile15 (I15A) that had a less dramatic effect on Sus1 binding. In summary, the results obtained with these mutants confirm the importance of the hydrophobic stripe on Sgf11 in its interaction with Sus1.
FIGURE 5.
Ala insertions in the Sgf11 helix disrupt binding to Sus1. A, helical nets show the hydrophobic stripe on Sgf11 to which Sus1 binds. B, shown is an illustration of how the stripe is disrupted by insertion of an Ala after residue 18. C, binding assays demonstrate the reduced affinity of Sus1 for Sgf11 Ala variants. GST control (lane 2); GST-Sgf11 wild-type (lane 3), I15A (Ala substitution for Ile15, lane 4), 19A (Ala inserted after residue 18, lane 5), and 20A (Ala inserted after residue 19, lane 6) were coexpressed with full-length Sus1 and affinity purified from cell lysates. Proteins were visualized by SDS-PAGE.
Sus1 Binding to Sac3 and Sgf11 Is Mutually Exclusive
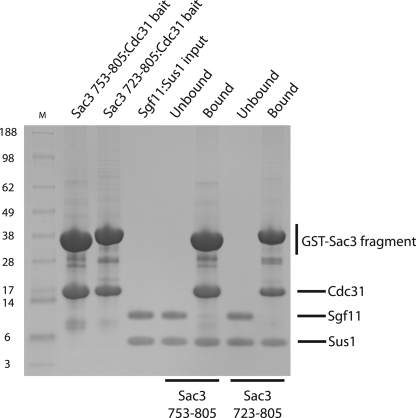
The observation that Sus1 wrapped around an α-helix in a similar way when complexed to either Sgf11 or Sac3, indicated that its binding to these two partners should be mutually exclusive. We tested this hypothesis by challenging two Sac3-Cdc31 complexes with the Sgf11-Sus1 complex (Fig. 6). The two Sac3 complexes, containing the Sus1B and Sus1A/B-binding sites, were unable to bind the Sgf11-Sus1 complex. These results indicated that a Sus1 chain binds to Sac3 and Sgf11 in a mutually exclusive manner because Sgf11 is not observed in the bound material, whereas Sus1 is evident. Interestingly, in vitro Sac3 can extract Sus1 from a Sus1/Sgf11 heterodimer (Fig. 6) and this may reflect Sus1 equilibrating between the two complexes.
FIGURE 6.
Sus1 binds to Sac3 and Sgf11 in a mutually exclusive manner. GST-Sac3 753–805 (containing the Sus1B-binding site) and GST-Sac3 723–805 (containing the Sus1A and Sus1B-binding sites) were co-expressed with Cdc31 and immobilized on glutathione Sepharose 4B resin from clarified lysates. After resin washing, purified Sgf11-Sus1 complex was added and incubated. The unbound fraction and washed resin was analyzed by SDS-PAGE. Sgf11 is not observed in the bound material whereas Sus1 is present indicating the inability of Sus1 to bind Sac3 and Sgf11 simultaneously.
It was initially suggested that Sus1 could function to physically bridge the SAGA and TREX-2 complexes directly (7). However, the current study and previous work tend to support a more complex model for SAGA and TREX-2 interaction (1, 24). The similarity of the Sus1-Sgf11 and Sus1-Sac3 interaction interfaces (Figs. 2 and 3), combined with the indication that the binding of these two partners to Sus1 was mutually exclusive (Fig. 6), would make it unlikely that a single Sus1 molecule could link the SAGA and TREX-2 complexes. In principle, the Sus1 chains in each complex could interact with one another or with other components of the complexes to form an interface. We have not observed any indication of dimerization of either the Sgf11-Sus1 or Sac3-Sus1-Cdc31 complexes that would be consistent with a Sus1-Sus1 interaction. However, our present data cannot exclude the possibility that Sus1 may bridge the two complexes with a low affinity interaction that could, for example, be between the Sus1 chains of each complex (perhaps mediated by an additional factor) or the C-terminal zinc finger of Sgf11 interacting with the Sac3 CID region of the TREX-2 complex. Alternatively, it has been proposed that Sus1 could link the SAGA and TREX-2 complexes functionally by a dynamic exchange mechanism in which the competitive capture of Sus1 from TREX-2 by the DUB complex could dissociate TREX-2 from NPCs and up-regulate histone deubiquitination (1). This regulation would be reversed upon the reciprocal exchange of Sus1 from the DUB to TREX-2 and thereby could synchronize the NPC binding of TREX-2 to transcriptional events (1). It has also been suggested that post-translational modifications of Sus1 or other components of TREX-2 or SAGA could play a role in regulating gene gating (1). For example, Lys9 of Sus1 forms a salt bridge in the Sus1A-Sac3 and Sus1B-Sac3 interface but does not participate in the Sgf11-Sus1 interface (Figs. 2 and 3), and so post-translational modification of this residue could potentially contribute to switching the binding preference of Sus1 from Sac3 (TREX-2) to Sgf11 (SAGA).
In summary, we have determined the structure of the Sgf11-Sus1 complex and shown that Sus1 functions in this complex to wrap around the extended helix formed by Sgf11 residues 8–31. The structure of this second Sus1 complex indicates that this molecule probably has a common function in each complex that is based on stabilizing extended single α-helices that would normally be anticipated to have very low stability when isolated in solution. However, the hydrophobic stripe that formed the Sus1-binding interface on Sgf11 was less extensive than that observed on Sac3 and provided a structural explanation for how it was possible to produce Sus1 mutants, in which binding to Sgf11 was retained while binding to Sac3 was lost (1), but how it was not possible to impair Sac3 binding without also losing Sgf11 binding. Overall, these structural results also favor a model in which Sus1 forms a component of separate subcomplexes in SAGA and TREX-2 rather than a single Sus1 chain linking the two complexes. Although further work will be required to establish precisely how Sus1 and Sgf11 participate in the linkage of SAGA to TREX-2, the structural and biochemical data obtained here identify the stabilizing of long α-helices in both complexes as an important function that contributes to their acting as interaction platforms that promote integration of transcriptional and export events in the gene expression machinery.
Supplementary Material
Acknowledgments
We are most grateful for the comments and criticisms of our colleagues in Cambridge and Heidelberg and to Neil Marshall for assistance. We also thank James Sandy for assistance at Diamond beamline I02.
This work was supported in part by a Wellcome Trust Program Grant (to M. S.) and by grants from the Deutsche Forschungsgemeinschaft (SBF638, B3 to E. H.).
The atomic coordinates and structure factors (codes 3KIK and 3KJL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies S1–S3.
- NPC
- nuclear pore complex
- SAGA
- Spt- Ada-Gcn5 acetyltransferase
- DUB
- deubiquitinating
- GST
- glutathione S-transferase.
REFERENCES
- 1.Klöckner C., Schneider M., Lutz S., Jani D., Kressler D., Stewart M., Hurt E., Köhler A. (2009) J. Biol. Chem. 284, 12049–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler A., Hurt E. (2007) Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 3.Brown C. R., Silver P. A. (2007) Curr. Opin. Genet. Dev. 17, 100–106 [DOI] [PubMed] [Google Scholar]
- 4.Brown C. R., Kennedy C. J., Delmar V. A., Forbes D. J., Silver P. A. (2008) Genes Dev. 22, 627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luna R., Gonzalez-Aguilera C., Aguilera A. (2009) RNA Biol. 6, 145–148 [DOI] [PubMed] [Google Scholar]
- 6.Blobel G. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Navarro S., Fischer T., Luo M. J., Antúnez O., Brettschneider S., Lechner J., Pérez-Ortín J. E., Reed R., Hurt E. (2004) Cell 116, 75–86 [DOI] [PubMed] [Google Scholar]
- 8.Fischer T., Rodríguez-Navarro S., Pereira G., Rácz A., Schiebel E., Hurt E. (2004) Nat. Cell Biol. 6, 840–848 [DOI] [PubMed] [Google Scholar]
- 9.Cabal G. G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J. C., Hurt E. C., Nehrbass U. (2006) Nature 441, 770–773 [DOI] [PubMed] [Google Scholar]
- 10.Jani D., Lutz S., Marshall N. J., Fischer T., Köhler A., Ellisdon A. M., Hurt E., Stewart M. (2009) Mol. Cell 33, 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Navarro S. (2009) EMBO Rep. 10, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmers H. T., Tora L. (2005) Trends Biochem. Sci. 30, 7–10 [DOI] [PubMed] [Google Scholar]
- 13.Fischer T., Strässer K., Rácz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. (2002) EMBO J. 21, 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilmes G. M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., Cagney G., Collins S. R., Whitworth G. B., Kress T. L., Weissman J. S., Ideker T., Guthrie C., Krogan N. J. (2008) Mol. Cell 32, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Aguilera C., Tous C., Gómez-González B., Huertas P., Luna R., Aguilera A. (2008) Mol. Biol. Cell 19, 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faza M. B., Kemmler S., Jimeno S., González-Aguilera C., Aguilera A., Hurt E., Panse V. G. (2009) J. Cell Biol. 184, 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., Widom J., Brickner J. H. (2007) PLoS Biol. 5, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huisinga K. L., Pugh B. F. (2004) Mol. Cell 13, 573–585 [DOI] [PubMed] [Google Scholar]
- 19.Grant P. A., Duggan L., Côté J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F., Berger S. L., Workman J. L. (1997) Genes Dev. 11, 1640–1650 [DOI] [PubMed] [Google Scholar]
- 20.Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., Workman J. L. (1999) J. Biol. Chem. 274, 5895–5900 [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A., Tan S. (2002) J. Biol. Chem. 277, 7989–7995 [DOI] [PubMed] [Google Scholar]
- 22.Sanders S. L., Jennings J., Canutescu A., Link A. J., Weil P. A. (2002) Mol. Cell Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler A., Pascual-García P., Llopis A., Zapater M., Posas F., Hurt E., Rodríguez-Navarro S. (2006) Mol. Biol. Cell 17, 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köhler A., Schneider M., Cabal G. G., Nehrbass U., Hurt E. (2008) Nat. Cell Biol. 10, 707–715 [DOI] [PubMed] [Google Scholar]
- 25.Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. (2003) Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel J. A., Torok M. S., Sun Z. W., Schieltz D., Allis C. D., Yates J. R., 3rd, Grant P. A. (2004) J. Biol. Chem. 279, 1867–1871 [DOI] [PubMed] [Google Scholar]
- 27.Kurshakova M., Maksimenko O., Golovnin A., Pulina M., Georgieva S., Georgiev P., Krasnov A. (2007) Mol. Cell 27, 332–338 [DOI] [PubMed] [Google Scholar]
- 28.Kurshakova M. M., Krasnov A. N., Kopytova D. V., Shidlovskii Y. V., Nikolenko J. V., Nabirochkina E. N., Spehner D., Schultz P., Tora L., Georgieva S. G. (2007) EMBO J. 26, 4956–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H. G., Krasnov A., Georgieva S. G., Schüle R., Takeyama K., Kato S., Tora L., Devys D. (2008) Mol. Cell 29, 92–101 [DOI] [PubMed] [Google Scholar]
- 30.Pascual-García P., Govind C. K., Queralt E., Cuenca-Bono B., Llopis A., Chavez S., Hinnebusch A. G., Rodríguez-Navarro S. (2008) Genes Dev. 22, 2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker S. P., Grant P. A. (2007) Oncogene 26, 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmes G. M., Guthrie C. (2009) Mol. Cell 33, 671–672 [DOI] [PubMed] [Google Scholar]
- 33.Matsuura Y., Stewart M. (2004) Nature 432, 872–877 [DOI] [PubMed] [Google Scholar]
- 34.Teo H., Gill D. J., Sun J., Perisic O., Veprintsev D. B., Vallis Y., Emr S. D., Williams R. L. (2006) Cell 125, 99–111 [DOI] [PubMed] [Google Scholar]
- 35.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 36.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 37.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 38.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 39.Montigiani S., Neri G., Neri P., Neri D. (1996) J. Mol. Biol. 258, 6–13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.