Abstract
COX-2 (cyclooxygenase-2) is a pivotal player in inflammatory processes, and ultraviolet radiation is a known stimulus for COX-2 expression in skin cells. Here, an induction of COX-2 expression in HaCaT human keratinocytes was observed only upon exposure of cells to UVB (280–320 nm) but not to UVA radiation (320–400 nm), as demonstrated by reverse transcription-PCR and Western blotting. Prostaglandin E2 levels were elevated in cell culture supernatants of HaCaT cells exposed to UVB. COX-2 mRNA stability was dramatically increased by UVB irradiation. Both the stabilization of COX-2 mRNA and the enhancement of COX-2 steady-state mRNA and protein levels caused by UVB were prevented both by inhibition and small interfering RNA-induced depletion of p38MAPK, a kinase strongly activated upon exposure to UVB, suggesting p38MAPK-dependent mRNA stabilization as a mechanism of UVB-induced COX-2 expression. A dramatic decrease in COX-2 expression induced by UVB was elicited by small interfering RNA-based depletion of a stress-responsive mRNA stabilizing protein regulated by p38MAPK, i.e. HuR; UVB-induced elevation of COX-2 mRNA and protein levels coincided with an accumulation of HuR in the cytoplasm and was attenuated in cells depleted of HuR. Moreover, UVB-induced generation of prostaglandin E2 by HaCaT cells was blunted by HuR depletion, suggesting that stress kinases (such as p38MAPK) as well as HuR are excellent targets for approaches aiming at interfering with induction of COX-2 expression by UVB.
Keywords: Cell/Epithelial, Eicosanoids/Cyclooxygenase Pathway, Phosphorylation/Kinases/Tyrosine, RNA/Turnover, Signal Transduction/Protein Kinases/MAP, Tissue/Organ Systems/Skin
Introduction
Cyclooxygenases catalyze the rate-limiting step in prostaglandin biosynthesis, i.e. the conversion of arachidonic acid to prostaglandin (PG)3 H2, which in turn is converted by various synthases to different prostaglandins or thromboxane A2, important mediators in inflammatory processes. Two genes coding for isoforms of cyclooxygenase (COX-1 and COX-2) are known (1). Although COX-1 and a COX-1 variant, termed COX-3 (2), are constitutively expressed, expression of COX-2 is strongly inducible by growth factors, cytokines, and other stimuli, resulting in the production of prostaglandins during inflammatory processes. One such potent stimulus for COX-2 induction is UV radiation. Both UVB (280–320 nm) (3) and UVA (320–400 nm) (4) were reported previously to enhance the expression of COX-2 in human keratinocytes, followed by an increased production of the inflammatory mediator PGE2, a major prostaglandin in skin. Analysis of the relative contributions of UV ranges to the effects of solar light on COX-2 levels demonstrated that UVB is a far more efficient inducer of COX-2 expression; for example, UVB and UVA-2 (320–350 nm) but not UVA-1 (350–400 nm) contributed to COX-2 induction by simulated solar light in artificial human epidermis (5). Several lines of evidence link COX-2 and PGE2 to the development of UV-induced skin cancer, such as the findings that COX-2 and PGE2 levels are elevated in skin cancer versus normal tissue, that PGE2 is a promoting factor in skin carcinogenesis, and that depletion or inhibition of COX-2 attenuates skin carcinogenesis in various models of induced carcinogenesis (6).
The induction of COX-2 expression by UVB has been demonstrated to be mediated by both transcriptional and post-transcriptional mechanisms. Isoforms of the MAPK family member p38MAPK play an essential role in these processes and were found to mediate UVB-induced elevation of COX-2 promotor activity in human keratinocytes by phosphorylating cAMP-responsive element-binding protein and ATF-1 (activating transcription factor), which interact with the COX-2 promoter (7, 8). Moreover, the aryl hydrocarbon receptor was recently demonstrated to be involved in transcriptional control of COX-2 expression in response to UVB (9). Although it has been known for some time that exposure to UVB affects RNA stability and post-transcriptional regulation of gene expression (10, 11), this was recently also demonstrated for COX-2 whose mRNA was stabilized in HaCaT keratinocytes exposed to UVB (12).
HuR is an mRNA-stabilizing protein related to the Drosophila embryonic lethal abnormal vision family of proteins (13) known to be modulated by mitogenic and stress-causing agents, including UV radiation (14, 15). Stress-induced modulation of HuR activity may be achieved by phosphorylation, such as by protein kinase C isoforms, resulting in its translocation to the cytoplasm (16, 17). Furthermore, p38MAPK has been shown to stimulate transfer of HuR to the cytosol and to affect HuR mRNA stabilizing activity (15). It was recently demonstrated in human keratinocytes that COX-2 mRNA coprecipitates with endogenous HuR, suggesting that HuR binds to COX-2 mRNA in a constitutive manner; moreover, forced overexpression of an HuR-GFP construct stabilizes COX-2 mRNA in unstimulated HaCaT cells (12).
In this study, we demonstrate that the stress-responsive kinase p38MAPK and the RNA-stabilizing protein HuR are involved in UVB-induced COX-2 expression in HaCaT cells and further demonstrate that both stress kinases and HuR are excellent targets for a pharmacological approach that interferes with COX-2 expression and its induction by UVB.
EXPERIMENTAL PROCEDURES
Cell Culture, UV Irradiation, and siRNA Transfections
HaCaT human immortalized keratinocytes (18) were a kind gift from Prof. P. Boukamp, Heidelberg, Germany. Cells were held at 37 °C in a humidified atmosphere with 5% (v/v) CO2 and cultured in Dulbecco's modified Eagle's medium (PAA, Pasching, Austria) supplemented with (final concentrations) 9% (v/v) fetal calf serum (PAA), 2 mm Glutamax (Invitrogen), and penicillin/streptomycin (100 units/ml and 0.1 mg/ml, respectively; PAA). For mRNA stability measurements and UVA irradiations, cells were cultured in RPMI 1640 medium (Sigma). Exposure of cells to UVA was performed with a UVA700 irradiation device (Waldmann, Villingen-Schwenningen, Germany) emitting in the UVA spectral region (320–400 nm) with a maximum emission at 365 nm. For exposure to UVB, TL20W/12RS-UV bulbs (Philips, Eindhoven, The Netherlands) were used emitting most of the energy in the UVB spectral region (280–320 nm) with a maximum emission at 310 nm. Irradiations were at an intensity of 49 milliwatts/cm2 (UVA) and 0.60–0.85 milliwatt/cm2 (UVB), respectively. Cell viability after exposure to UVB was determined using the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide to the corresponding blue formazan by viable cells. For irradiations, cells were grown to ∼90% confluence and held in serum-free media for 24 h prior to exposure to UV, washed once with PBS, and covered with PBS during exposure to UV or during sham irradiation. Inhibitors of signaling molecules or transporters (SB202190 was from Sigma, and staurosporine, bisindolylmaleimide I, rottlerin, and leptomycin B were from Calbiochem) or the respective solvent controls (DMSO) were added to media 30 min prior to exposure of cells to UV and were also present throughout post-irradiation incubations. siRNAs targeting the coding sequence (HuR1, sense GAGGCAAUUACCAGUUUCA) and the 3′-untranslated region (HuR2, UCUUAAGUUUCGUAAGUUA) of human HuR mRNA and a nontargeting control siRNA (UUCUCCGAACGUGUCACGUUU) were used at 150 nm (final concentration) and were purchased from Qiagen (Hilden, Germany). Knockdown of hnRNP A0 was accomplished using a mixture of four duplex siRNAs targeting human hnRNP A0 mRNA (GAGGAUAUCUACUCCGGUG, GAUUCGGCUUCGUGUAUUU, CCGAGAUUAUUGCCGACAA, and GUGGGAGCGACUACGGUAA) in a total final concentration of 40 nm or nontargeting control siRNA (UGGUUUACAUGUCGACUAA), both from Dharmacon (Lafayette, CO). The siRNA targeting both α and β p38MAPK isoforms (CGGCAGGAGCUGAACAAGAUU) was from Dharmacon and was employed at 100 nm. Transfection of siRNAs into HaCaT cells was performed using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. 40 h after transfection, cell culture media were changed and cells held in serum-free media for another 24 h prior to further treatment or analysis.
Reverse Transcription (RT)-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) or the Nucleo Spin RNA/Protein kit (Macherey-Nagel, Düren, Germany) for simultaneous protein isolation according to the manufacturer's instructions. Reverse transcription of RNA (1 μg) was performed using the Omniscript RT kit (Qiagen) according to the manufacturer's instructions, oligodeoxythymidine primers (10 μm), and 1 unit of RNaseOUT RNase inhibitor (Invitrogen). For PCR, 1:10 of the synthesized cDNA was added to 1 unit of Taq DNA polymerase (Qiagen), 200 μm dNTPs (Amersham Biosciences), and 1 μm (for GAPDH) or 1.5 μm (for COX-2) of the respective primer pairs (Invitrogen, GTCAGTGGTGGACCTGACCT and AGGGGTCTACATGGCAACTG for GAPDH, GCAGTTGTTCCAGACAAGCA and CAGGATACAGCTCCACAGCA for COX-2). PCR products were visualized via agarose gel electrophoresis and ethidium bromide staining.
Determination of COX-2 mRNA Stability
HaCaT cells were depleted of serum for 24 h prior to exposure to UVA or UVB. Following irradiation, cells were held in cell culture medium for 2 h at 37 °C and then washed twice with PBS and incubated in the presence of 1 μg/ml actinomycin D (Sigma), an inhibitor of transcription, in serum-free RPMI 1640 medium. At several time points after addition of actinomycin D, cells were washed with PBS and cell lysis and RNA isolation performed using TRIzol reagent (Invitrogen). COX-2 as well as GAPDH mRNA levels were quantitated via RT-PCR, agarose gel electrophoresis, and densitometric analysis of agarose gels. The obtained values for COX-2 PCR product levels were normalized according to the GAPDH controls.
PGE2 Measurement
Cells were grown to ∼90% confluence and held in serum-free media for 24 h prior to irradiation. Detection of PGE2 in cell culture supernatants post-irradiation was performed via competitive ELISA (Cayman Chemical, Ann Arbor, MI). For analysis, supernatants were diluted 1:10 in serum-free Dulbecco's modified Eagle's medium. PGE2 levels detected were normalized to cellular protein concentrations.
Western Blotting and Immunocytochemistry
For Western analyses, cells were lysed on ice by scraping and collecting lysates in cold 0.5% (w/v) SDS. Protein concentrations were determined using a detergent-compatible protein assay (Bio-Rad). 4× SDS-PAGE buffer (250 mm Tris/HCl, 5% (w/v) SDS, 20% glycerol, 100 mm dithiothreitol, and 0.01% (w/v) bromphenol blue, pH 8) was added to samples, which were then subjected to heat denaturation at 95 °C for 5 min and loaded onto SDS-polyacrylamide gels of 10% (w/v) acrylamide, followed by electrophoresis and Western blotting onto polyvinylidene difluoride membranes. For detection of COX-2 and HuR, membranes were incubated with mouse monoclonal anti-COX-2 (Cayman Chemical) and anti-HuR (Santa Cruz Biotechnology, Lake Placid, NY) antibodies, respectively, and diluted in blocking buffer (1–5% (w/v) skim milk powder in 50 mm Tris/Cl, 150 mm NaCl, 0.1% (v/v) Tween 20, pH 7.4). Further antibodies used in this study include mouse monoclonal anti-α-tubulin (Sigma), mouse monoclonal anti-GAPDH (Chemicon, Temecula, CA), mouse monoclonal anti-β-actin (Abcam, Cambridge, UK), and rabbit polyclonal anti-phospho-p38 (Cell Signaling Technology, Beverly, MA) as well as rabbit polyclonal anti-HDAC1 (Santa Cruz Biotechnology) and goat polyclonal anti-hnRNP A0 antibodies (Santa Cruz Biotechnology). Horseradish peroxidase-coupled secondary antibodies were from Amersham Biosciences and Santa Cruz Biotechnology for detection of primary antibodies from mouse, rabbit, and goat. Detection of secondary antibodies bound to polyvinylidene difluoride membranes was by chemiluminescence employing reagents from Pierce. Subcellular fractionation was performed using the ProteoExtract S-PEK kit from Calbiochem according to the manufacturer's instructions. For immunocytochemical analyses, cells were grown to ∼90% confluence on coverslips and held in serum-free cell culture medium for 24 h. Following exposure to UVB, cells were incubated in serum-free media for 2, 4, 6, or 8 h at 37 °C, washed twice with PBS, fixed with 4% (w/v) formaldehyde in PBS for 30 min at room temperature, and blocked for 1 h at room temperature in PBS containing 3% (w/v) goat normal serum and 0.3% (v/v) Triton X-100. Cells were then incubated overnight at 4 °C with an anti-HuR antibody (Santa Cruz Biotechnology; 1:250 in PBS, 1% goat normal serum), washed with PBS three times, and subsequently incubated in the presence of an Alexa Fluor 488-conjugated secondary anti-mouse antibody (Invitrogen; 1:500 in PBS) for 1 h at 37 °C. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole.
RESULTS
Induction of COX-2 Expression by UVB but Not UVA
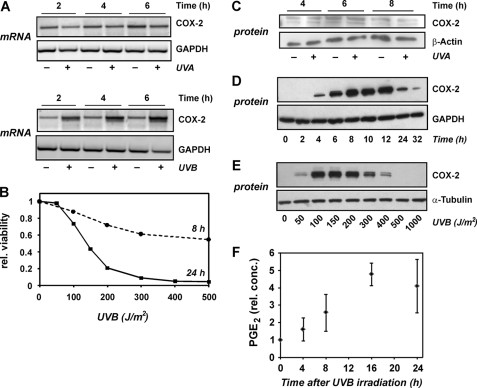
HaCaT human keratinocytes were exposed to UVA (300 kJ/m2) or UVB (100 J/m2), followed by analysis of COX-2 mRNA levels at various time points after irradiation. Although a significant elevation of COX-2 mRNA levels was already detected 2 h after exposure to UVB, no such effect was seen with UVA (Fig. 1A). The irradiation doses chosen were subcytotoxic (Fig. 1B; UVA: data not shown) and were equivalent in terms of inducing activation of a stress kinase, p38MAPK, to a similar extent at 30 min post-irradiation (see below). Although no induction of COX-2 was elicited both at mRNA (Fig. 1A) and protein levels (Fig. 1C), UVB-induced enhancement of COX-2 mRNA levels was accompanied by an even more significant stimulation of COX-2 protein levels starting at 4 h post-irradiation, with a maximum induction at about 8 h following exposure of cells to UVB (Fig. 1D). Maximum induction of COX-2 was achieved with a dose of 100–150 J/m2, whereas induction was less pronounced at higher doses (Fig. 1E), which is probably due to the cytotoxic effects of doses beyond 100 J/m2 (Fig. 1B). In line with the induced COX-2 being enzymatically active, prostaglandin E2 levels were elevated in culture supernatants of HaCaT cells exposed to UVB, with the highest accumulation of PGE2 detected at 16 h post-irradiation (Fig. 1F).
FIGURE 1.
Induction of COX-2 expression in HaCaT cells by UVB but not UVA. HaCaT cells were held under serum-free conditions for 24 h prior to exposure to UV. A, cells were exposed to UVA (300 kJ/m2) and UVB (100 J/m2) or were sham-irradiated through PBS, followed by lysis after the given periods of time. Detection of COX-2 and GAPDH mRNA levels was by RT-PCR. Data are representative of at least three independent experiments. B, viabilities of HaCaT cells following exposure to UVB at the given dose were determined in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide assay 8 and 24 h post-irradiation, respectively, and normalized for viabilities of sham-irradiated cells. Data shown are means of at least two independent experiments. C and D, cells were exposed to UVA (300 kJ/m2) or UVB (100 J/m2), followed by post-incubation in serum-free medium and lysis after the given periods of time. Detection of COX-2 protein levels was by Western blotting; GAPDH or actin served as loading controls. Data are representative of at least two (UVB) or three (UVA) independent experiments. E, cells were exposed to UVB at the given doses through PBS, followed by lysis after 8 h. Detection of COX-2 protein levels was by Western blotting; tubulin served as loading control. Data are representative of three independent experiments. F, cells were treated as described under C. Cell culture supernatants were collected at the indicated time points and analyzed for PGE2 by ELISA. The values shown are means ± S.D. (n = 3) normalized to total protein amounts in the corresponding cultures. Normalized PGE2 concentrations at 0 h post-irradiation were set to 1.
Enhanced COX-2 mRNA Stability following Exposure to UVB but Not UVA
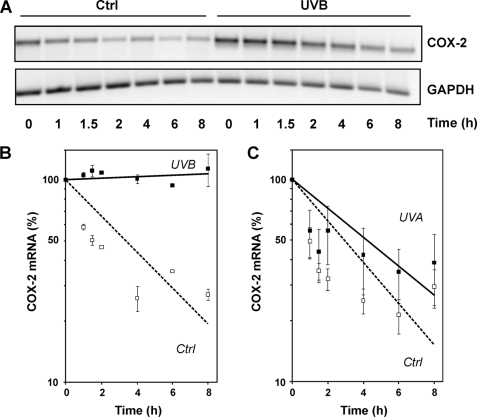
To test whether COX-2 mRNA stabilization may account for the observed induction of COX-2 mRNA and protein levels, HaCaT cells were exposed to UVB, followed by addition of the transcriptional inhibitor actinomycin D 2 h later and the analysis of remaining COX-2 mRNA at several time points thereafter. In line with previous reports (12), mRNA stability was drastically enhanced by exposure of cells to UVB (Fig. 2, A and B), whereas no significant stabilization of COX-2 mRNA was detectable in cells exposed to UVA (Fig. 2C), which is in accordance to UVA not stimulating COX-2 expression under these conditions (Fig. 1).
FIGURE 2.
Modulation of COX-2 mRNA stability by UVB but not UVA. HaCaT cells were held under serum-free conditions for 24 h, exposed to UVB (100 J/m2) and UVA (300 kJ/m2) or sham-irradiated (ctrl), followed by post-incubation in serum-free medium and the addition of actinomycin D (1 μg/ml) after 2 h. Cells were lysed at the indicated time points after addition of actinomycin D and analyzed via RT-PCR for the mRNA levels of COX-2 and GAPDH (loading control). A representative agarose gel is shown in A. Data were analyzed densitometrically, as shown in B and C. COX-2 mRNA levels were normalized over GAPDH levels; data are expressed as percent of mRNA remaining after addition of actinomycin D. The values shown are means ± S.D. (n = 3).
Crucial Role of p38MAPK in COX-2 Induction and COX-2 mRNA Stabilization by UVB
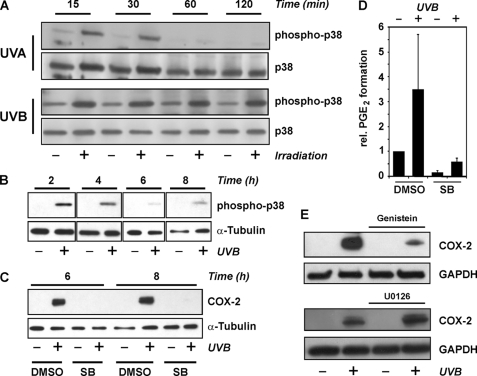
Exposure of HaCaT cells to UVB (100 J/m2) resulted in stimulation of p38MAPK (Fig. 3A). Activation of p38MAPK was detected by Western analysis of phosphorylation at Thr-180 and Tyr-182, i.e. by demonstrating that the enzymatically active form of the protein is present. Phosphorylation of p38MAPK was already detected 15 min post-irradiation both with UVB and UVA (Fig. 3, A and B), but whereas both UVA (300 kJ/m2) and UVB (100 J/m2) stimulated p38MAPK activation to a similar extent, UVB-induced stimulation was persistent and detected even 8 h post-irradiation, whereas UVA-induced p38MAPK activation was transient and no longer significant after 2 h (Fig. 3A). Inhibition of p38MAPK employing a specific inhibitor, SB202190, abrogated the induction of COX-2 expression by UVB, both at mRNA (data not shown) and protein levels (Fig. 3C), and of UVB-induced elevation of PGE2 levels in cell culture supernatants (Fig. 3D).
FIGURE 3.
p38MAPK is essential for COX-2 induction by UVB. HaCaT cells were held under serum-free conditions for 24 h prior to exposure to UVA (300 kJ/m2) or UVB (100 J/m2) through PBS, followed by post-incubation in serum-free media for the indicated periods of time. A and B, cells were lysed at the indicated time after irradiation and tested for p38MAPK phosphorylation by Western blotting employing a phospho-specific antibody. Detection of total p38 or tubulin served as loading control. C, 30 min prior to irradiation, the p38MAPK inhibitor SB202190 (SB) (5 μm) or DMSO (vehicle) was added to the culture media, followed by irradiation of cells covered in PBS and post-incubation of cells for 6 or 8 h in serum-free media containing SB202190 (5 μm) or DMSO. Cells were lysed and tested for COX-2 levels by Western blotting. Tubulin served as loading control. D, cells were treated as in B. Cell culture supernatants were collected 16 h post-irradiation and analyzed for PGE2 by ELISA. The values shown are means ± S.D. (n = 4) normalized to total protein amounts in the corresponding cultures. Normalized PGE2 concentrations determined in supernatants of sham-irradiated and vehicle-treated cells were set to 1. E, 30 min prior to UVB irradiation (100 J/m2), genistein (50 μm) or the MKK1/2 inhibitor U0126 (10 μm) or DMSO (vehicle) was added to the culture media, followed by irradiation of cells covered in PBS and post-incubation of cells for 8 h in serum-free media containing the respective inhibitor or DMSO. Cells were lysed and tested for COX-2 levels by Western blotting. Data are representative for three independent experiments.
In line with p38MAPK mediating COX-2 induction, a general tyrosine kinase inhibitor, genistein, also attenuated COX-2 induction by UVB; in contrast, no comparable inhibition of COX-2 induction was elicited by U0126, an inhibitor of the classical MKK1/2-ERK1/2MAPK pathway (Fig. 3E), pointing to the specificity of SB202190.
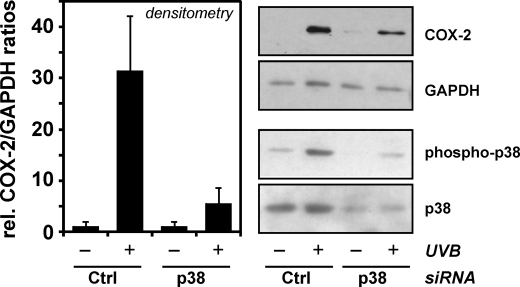
The role of p38MAPK was further investigated by depleting p38MAPK using an siRNA approach. Although cells were not fully depleted of p38MAPK, with residual p38 phosphorylation detectable after exposure to UVB (see Western blots in Fig. 4), COX-2 induction upon UVB irradiation was strongly attenuated in these cells (Fig. 4).
FIGURE 4.
Depletion of p38MAPK attenuates COX-2 induction by UVB. HaCaT cells were transfected with p38MAPK-specific or control (Ctrl) siRNA. 32 h post-transfection, culture media were changed, and cells were held in serum-free media for another ∼16 h, followed by exposure to UVB (100 J/m2) through PBS or by sham irradiation. After 8 h of post-incubation in serum-free media, cells were lysed and tested for COX-2 and p38MAPK levels and the extent of p38 phosphorylation by Western blotting. GAPDH served as loading control. For densitometric analysis of COX-2 expression, COX-2 levels as determined by densitometry were related to GAPDH levels and COX-2/GAPDH ratios calculated. Ratios of the respective control treatments (sham irradiation) were then set equal to 1. Data are means of three independent experiments ± S.E.
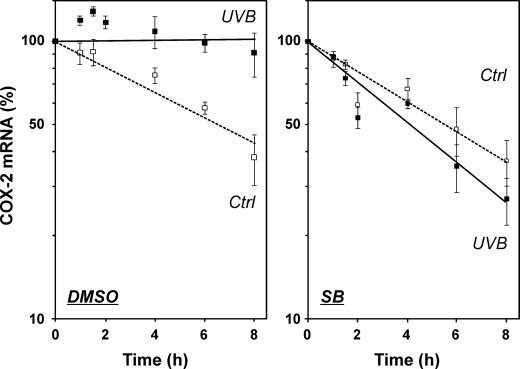
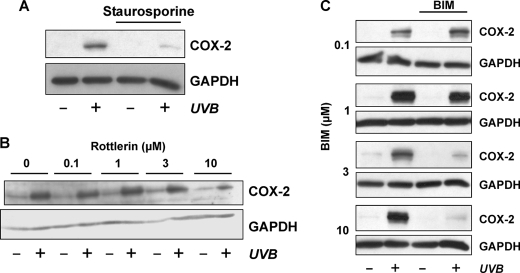
We then tested for an effect of p38MAPK inhibition on UVB-induced COX-2 mRNA stabilization. In fact, stabilization of COX-2 mRNA by UVB was completely abrogated in the presence of SB202190, whereas UVB-induced stabilization was not affected in cells treated with vehicle, i.e. DMSO (Fig. 5). In addition to inhibition of p38MAPK, it appears that protein kinase C inhibitors, staurosporine, rottlerin, or bisindolylmaleimide, attenuate, but not fully abrogate, COX-2 induction by UVB (Fig. 6).
FIGURE 5.
Stabilization of COX-2 mRNA by UVB is mediated by p38MAPK. HaCaT cells were held under serum-free conditions for 24 h. 30 min prior to exposure to UVB (100 J/m2), SB202190 (SB) (2 μm) or DMSO (vehicle) was added to the culture media, followed by irradiation of cells covered in PBS and post-incubation of cells in serum-free media and the continued presence of SB202190 or DMSO. Actinomycin D was added after 2 h (=time point 0) to a final concentration of 1 μg/ml. Cells were lysed at the indicated time points after addition of actinomycin D and analyzed by RT-PCR for COX-2 and GAPDH mRNA levels. PCR products were analyzed by agarose gel electrophoresis, followed by densitometric evaluation of gels. COX-2 mRNA levels were normalized over GAPDH levels; data are expressed as percent of mRNA remaining after addition of actinomycin D. The values shown are means ± S.D. (n = 3). Ctrl, control.
FIGURE 6.
Protein kinase C inhibitors attenuate UVB-induced elevation of COX-2 expression. HaCaT cells were held under serum-free conditions for 24 h. 30 min prior to exposure to UVB (100 J/m2), staurosporine (100 nm, A), rottlerin (B), bisindolylmaleimide I (BIM, C), or DMSO (vehicle) was added to the culture media, followed by irradiation of cells covered in PBS and post-incubation of cells in serum-free media and the continued presence of the inhibitors for 8 h. Cells were lysed and tested for COX-2 levels by Western blotting. GAPDH served as loading control. Data are representative for four (A), three (B), and two (C) independent experiments.
HuR Controls COX-2 Induction by UVB
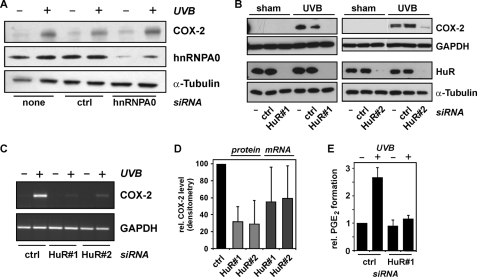
As the stabilization of COX-2 mRNA and p38MAPK appear to be involved in UVB-induced COX-2 expression, we further investigated whether selected RNA-binding proteins known to be affected by p38MAPK modulate COX-2 expression. The p38MAPK inhibitor, SB203580, was previously shown to prevent lipopolysaccharide-induced interaction of the hnRNP A0 with mRNA of COX-2 in murine macrophages (19). To test for a role of hnRNP A0 in UVB-induced COX-2 expression, hnRNP A0 was depleted from HaCaT cells by means of specific siRNA (Fig. 7A). However, the considerable reduction of hnRNP A0 levels did not impair induction of COX-2 expression by UVB (Fig. 7A). Thus, hnRNP A0 is unlikely to be involved in UVB-induced and p38MAPK-mediated induction of COX-2 expression.
FIGURE 7.
HuR controls COX-2 induction by UVB. A, HaCaT cells were transfected with siRNAs targeting hnRNP A0, with control (ctrl) siRNA, or treated with transfection agent only. Forty hours post-transfection, culture media were changed and cells held in serum-free media for another 24 h, followed by exposure to UVB (100 J/m2) through PBS or by sham irradiation. After 8 h of post-incubation in serum-free media (i.e. 72 h post-transfection), cells were lysed and tested for COX-2 and hnRNP A0 levels by Western blotting. α-Tubulin served as loading control. Data shown are representative of three independent experiments with identical results. B, cells were treated as in A but transfected with two different siRNAs targeting HuR (HuR#1 or HuR#2) or with nondepleting control siRNA (ctrl), each at 150 nm. Lysis of cells was 8 h after irradiation (i.e. 72 h post-transfection), followed by Western analysis of COX-2 and HuR levels. Data shown are representative of at least three independent experiments. C, cells were transfected and irradiated as described under B, but lysed at 6 h post-irradiation, followed by RT-PCR analysis of COX-2 and GAPDH mRNA levels. Data shown are representative of four independent experiments. D, extents of increase in COX-2 protein or mRNA levels by UVB: densitometric analysis of COX-2/GAPDH ratios of blots and agarose gels in B and C, respectively. Data are given as means ± S.D. and are normalized to the COX-2/GAPDH ratios (protein or mRNA) in UVB-irradiated cells transfected with control siRNA (ctrl). E, cells were transfected and irradiated as in B. Cell culture supernatants were collected 16 h post-irradiation and analyzed for PGE2 by ELISA. The values shown are means ± S.D. (n = 3) and normalized to total protein amounts and the PGE2 concentration found in cultures of untreated cells.
We then tested for an involvement of HuR in UVB induction of COX-2 expression. HuR is an mRNA-stabilizing protein the cytosolic localization of which is affected by p38MAPK (15). Furthermore, COX-2 mRNA was previously found to coprecipitate with endogenous HuR, suggesting that HuR interacts with COX-2 mRNA; also, overexpression of an HuR-GFP construct caused stabilization of COX-2 mRNA in unstimulated HaCaT cells (12).
In this study, knockdown of HuR in an siRNA approach resulted in a strong depletion of HuR and in significant attenuation of COX-2 induction, both at the level of protein (Fig. 7B) and mRNA (Fig. 7C). Two different siRNAs were employed, targeting the HuR coding region (HuR1) and the 3′-untranslated region (HuR2), respectively. Both siRNAs were similarly efficient in reducing HaCaT responsiveness to UVB in terms of COX-2 induction (Fig. 7D); densitometric evaluation revealed that only about 30% of the UVB-induced elevation of COX-2 levels was seen in HaCaT cells depleted of HuR, whereas slightly less efficient attenuation of induction was achieved at the level of COX-2 mRNA. Most interestingly, UVB-induced elevation of PGE2 levels in cell culture supernatants was completely abrogated in cells depleted of HuR (Fig. 7E), implying that HuR is crucial for UVB-induced enhancement of PGE2 production by HaCaT keratinocytes.
Exposure of cells to UVB (100 J/m2) did not cause changes in HuR protein levels (Fig. 7B). It is therefore unlikely that an enhanced COX-2 mRNA stability due to HuR induction is the reason for the observed COX-2 induction.
UVB Causes Elevation of Cytoplasmic HuR Levels
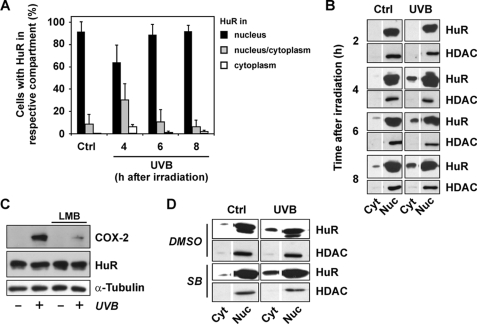
Although HuR is located predominantly in the nucleus in unstimulated cells (20), stressful stimuli such as UVC and oxidizing or alkylating agents cause its transport to the cytosol, which coincides with an enhanced mRNA stabilizing activity of HuR (14). Therefore, we tested whether exposure of HaCaT cells to UVB is capable of increasing cytosolic localization of HuR. Following exposure to UVB, cells were analyzed immunocytochemically for the predominant localization of HuR. Cells were then put in one of three categories as follows: (i) cells with predominantly nuclear HuR, (ii) cells with some HuR in the cytoplasm, and (iii) cells with predominantly cytoplasmic HuR. In nonirradiated cultures, all cells had HuR predominantly nuclear, whereas 8% displayed some cytosolic HuR (Fig. 8A). Following exposure to UVB, the percentage of cells with some or predominantly cytoplasmic HuR increased transiently, with a peak at about 4 h post-irradiation, when 6% of cells were found to carry HuR predominantly in their cytoplasm and 30% of cells with at least some detectable cytoplasmic HuR. After 8 h, numbers were back to control levels, except for the number of cells with predominantly cytoplasmic HuR, which were still slightly elevated (Fig. 8A). To confirm this UVB-induced cytoplasmic localization of HuR, we analyzed subcellular fractions of HaCaT cells exposed to UVB and found that although HuR is by far predominant in the nuclear compartment in both control and exposed cells, there are significant levels of HuR detected in the cytoplasmic fractions (Fig. 8B). Again, the maximum change of cytoplasmic levels in UVB-exposed cells versus control cells was detected at 4 h post-irradiation, and in line with the immunocytochemical analysis, some nuclear HuR remains detectable after 8 h. Cytoplasmic fractions were demonstrated to be devoid of nuclear impurities by excluding the presence of detectable levels of nuclear histone deacetylase.
FIGURE 8.
UVB-induced translocation of HuR to the cytoplasm. A, HaCaT cells were held under serum-free conditions for 24 h, exposed to UVB (100 J/m2) or sham-irradiated (ctrl), followed by post-incubation in serum-free medium and fixation in formaldehyde after the indicated periods of time. Cells were analyzed for subcellular HuR localization immunocytochemically and classified according to the cellular localization of HuR. Three categories were established as follows: (i) consisting of cells with HuR predominantly nuclear; (ii) consisting of cells with HuR predominantly nuclear but detectable cytosolic HuR; and (iii) consisting of cells with predominantly cytosolic HuR. For each time point, ∼130 cells were evaluated in each independent experiment. Data are given as means of at least three independent experiments ± S.D. B, cells were treated as described in A, followed by subcellular fractionation at the indicated time points after exposure to UVB and analysis of HuR and histone deacetylase levels in cytosolic (cyt) and nuclear (nuc) fractions by Western blotting. Data are representative of three independent experiments. C, HaCaT cells were held under serum-free conditions for 24 h. 150 min prior to exposure to UVB (100 J/m2), and leptomycin B (LMB) was added to a final concentration of 20 ng/ml, followed by irradiation of cells covered in PBS and post-incubation of cells in serum-free media and the continued presence of leptomycin B for 8 h. Leptomycin B was dissolved in methanol (70%), which was added to media in vehicle control experiments. Cells were lysed and tested for COX-2 and HuR levels by Western blotting, α-tubulin serving as loading control. Results are representative of three independent experiments. D, cells were held under serum-free conditions for 24 h, followed by incubation in the presence of the p38MAPK inhibitor, SB202190 (5 μm), for 30 min and exposure to UVB (100 J/m2) or sham-irradiation (ctrl). After 4 h of post-incubation in serum-free medium, cells were fractionated and analyzed for HuR and histone deacetylase (HDAC) content as in B. Data are representative of two independent experiments.
HuR export to the cytoplasm was previously reported to depend on the transport receptor CRM1 (chromosomal region maintenance protein 1) (21). Inhibiting CRM1-dependent nucleocytoplasmic shuttling with leptomycin B resulted in an inhibition of UVB-induced COX-2 expression (Fig. 8C), suggesting that HuR is precluded from the UVB-induced nucleocytoplasmic transport via CRM1 and thus cannot contribute to stabilization of COX-2 mRNA. UVB-induced HuR translocation was not affected by SB202190, as demonstrated both immunocytochemically and by subcellular fractionation (Fig. 8D), implying that it is independent of p38MAPK.
DISCUSSION
There is ample evidence linking COX-2 and PGE2 to the development of UV-induced skin cancer (6). In this study, we have assessed the molecular mechanisms contributing to COX-2 induction by UV radiation, employing HaCaT human keratinocytes as a model. These cells have been employed previously to investigate UV-induced modulation of COX-2 expression; UVB was demonstrated to stimulate both COX-2 expression and PGE2 production via p38MAPK (7) and to enhance COX-2 mRNA stability in HaCaT cells exposed to 250 J/m2 (12). Similarly, UVA was shown to elicit significant COX-2 mRNA stabilization and induction of COX-2 expression via p38MAPK (4).
However, our previous analyses of the relative contributions of these UV ranges to the effects of solar light on COX-2 levels in artificial human epidermis demonstrated that UVB is a far more efficient inducer of COX-2 expression than UVA; UVB and UVA-2 (320–350 nm) but not UVA-1 (350–400 nm) contributed to COX-2 induction by simulated solar light (5). Our present study was conducted for the following reasons: (i) to compare UVA and UVB at environmentally relevant doses with respect to their inducing COX-2 expression and mRNA stabilization in HaCaT cells in one study, and (ii) to identify potential approaches that would allow for a pharmacological intervention with the induction of COX-2 expression by UVB, in particular by analyzing contributions of stress kinases and RNA-binding proteins in this process.
Induction of COX-2 Expression by UVB but Not UVA
Both UVB and UVA are known stimulators of p38MAPK, which was demonstrated in this work (Figs. 3 and 4) and elsewhere (5, 7, 22) to mediate COX-2 induction by UV radiation. Nevertheless, UVA was not capable of eliciting significant elevation of COX-2 mRNA or protein levels in exposed HaCaT cells (Fig. 1) or of COX-2 mRNA stability (Fig. 2), although p38MAPK was stimulated at the doses employed. Although both UVA and UVB may affect cellular signaling cascades via the formation of reactive oxygen species (23–25), these wavelength regions differ with respect to their primary cellular target molecules and thus with respect to the following: (i) the mode of formation and the identity of generated reactive oxygen species, and (ii) the battery of molecular targets the irradiation of which may lead to the formation of active signaling molecules other than reactive oxygen species. For example, the amino acid tryptophan is a target of both UVA and UVB. However, whereas the latter is directly absorbed by Trp, causing the generation of a photoproduct actively stimulating signaling cascades that may culminate in the enhanced expression of COX-2 (9), the former only indirectly, via photoactivation of riboflavin, interacts with Trp, resulting in generation of hydrogen peroxide (26), which then acts as the active signaling molecule (27). Thus, the reason for differences in COX-2 expression in response to UVA and UVB is likely to reside in differences in patterns of molecular changes elicited in exposed cells. A major difference found in this work was that, although both stimulated p38MAPK activation, UVB-induced p38MAPK activity was more persistent than that induced by UVA (see “Results” and Fig. 3). We propose that this is a crucial factor in modulation of COX-2 expression and that these differences are due to molecular changes induced by UVB not only differing from those of UVA but also being more permanent. Nevertheless, the exact molecular changes underlying p38MAPK activation by UVB have yet to be identified.
HuR Is a Crucial Regulator of UVB-induced COX-2 Expression
The stress kinase p38MAPK was demonstrated in this study to be involved in the regulation of COX-2 expression under the influence of UVB (Figs. 3 and 4). The pharmacological inhibitors employed allow for a further specification because of their isoform specificities, with SB202190 selectively inhibiting p38MAPK α or β (28, 29), rottlerin being a fairly selective inhibitor of protein kinase Cδ isoforms (30), and bisindolylmaleimide I (also known as Gö 6850) being a rather selective inhibitor of the α, β1, δ, and ϵ isoforms of protein kinase C (31), although staurosporine is a protein kinase inhibitor with a rather broad target spectrum. The concentrations of protein kinase C inhibitors required for substantial inhibition of COX-2 induction by UVB were generally rather high, whereas the drastic effects of preincubation with SB202190 were achieved with concentrations that are generally accepted as within the safe range for inhibitor specificity (2–5 μm). We therefore believe that, although protein kinase C may be a modulator of the effects of UVB on COX-2 expression, p38MAPK is a crucial regulator of COX-2 mRNA stability (Fig. 5) and COX-2 expression (Figs. 3 and 4). This is further supported by experiments based on siRNA depletion of p38MAPK employing an siRNA targeting the α and β isoforms (Fig. 4).
We have further demonstrated in this study that induction of COX-2 expression by UVB depends on the mRNA-binding protein HuR (Fig. 7) and that HuR accumulates in the cytoplasm upon exposure of HaCaT cells to UVB (Fig. 8). Both mRNA binding activity and nucleocytoplasmic shuttling of HuR are known to be controlled by several protein kinases, including p38MAPK and protein kinase C. Although phosphorylation of HuR in its hinge region that governs nucleocytoplasmic shuttling (for review see Ref. 13) by the cell cycle regulating kinase Cdk1 (Ser-202) (32) or a yet unknown kinase (Ser-242) (33) has been demonstrated to result in a predominantly nuclear localization, phosphorylation of Ser-221 by protein kinase C isoforms coincided with its translocation to the cytoplasm (16, 17). Furthermore, checkpoint kinase Chk2-dependent phosphorylation in the RNA binding regions of HuR has been reported to cause a loss of interaction between HuR and SIRT1 mRNA (34), although in contrast p38MAPK was demonstrated to enhance HuR-dependent mRNA stabilization (15, 35). In fact, it was recently demonstrated that HuR is a direct substrate of p38MAPK and that p38-dependent phosphorylation of HuR enhances its mRNA stabilizing activity (36).
As p38MAPK was not required for UVB-induced alteration of cytoplasmic accumulation of HuR in our hands (Fig. 8D), and as HuR levels were not simply increased by UVB in our system (Figs. 7B and 8C), we propose that p38MAPK affects COX-2 mRNA stability in one of two ways. First, it may enhance HuR binding to COX-2 mRNA by phosphorylating HuR, either indirectly or directly in a not yet identified manner. Second, p38MAPK may change the affinities of mRNA-binding proteins associated with COX-2 mRNA in a way that renders COX-2 mRNA more accessible toward HuR. In fact, mRNA-binding proteins that are p38MAPK targets and that also destabilize mRNAs exist, such as “KH domain-containing RNA-binding protein” (KSRP) and tristetraproline; we hypothesize that such a destabilizing RNA-binding protein may compete with HuR for AU-rich elements in the 3′-untranslated region of COX-2 mRNA and that interaction of these destabilizing proteins with COX-2 mRNA is weakened in a p38MAPK-dependent fashion, rendering COX-2 mRNA accessible for the stabilizing RNA-binding protein HuR. The binding of HuR to COX-2 mRNA and the enhanced stability of COX-2 mRNA in cells overexpressing an HuR-GFP fusion protein has recently been demonstrated (12).
Conclusions
In this study, COX-2 expression in HaCaT keratinocytes was found to be induced in cells exposed to UVB but not UVA. The reported UVB effects strongly depended on p38MAPK, which mediated UVB-induced stabilization of COX-2 mRNA and an increase in COX-2 mRNA and protein levels, as demonstrated employing a specific inhibitor of p38MAPK, SB202190, and p38-specific siRNA. Furthermore, depletion of the mRNA-stabilizing protein HuR by means of an siRNA approach employing one of two different specific siRNAs emphasized the crucial role of HuR in COX-2 induction upon exposure to UVB. The involvement of p38MAPK and HuR in UVB-induced COX-2 expression was highlighted also at the level of PGE2 production.
These data not only provide insight into molecular cascades mediating UVB-induced COX-2 expression but also point at two different approaches potentially interfering with COX-2 induction by UVB, which is of interest with respect to the prevention of UVB-induced skin carcinogenesis; both approaches employed in this study, i.e. low molecular mass inhibitors, such as SB202190, and siRNA-based strategies, equally and efficiently interfered with UVB-induced COX-2 expression and PGE2 production. Furthermore, and more specifically, two potential target molecules for approaches aiming at interfering with UVB-induced COX-2 expression were established, p38MAPK and HuR.
Acknowledgments
We thank Dr. Peter Schroeder, Leibniz-Institut für Umweltmedizinische Forschung, for helpful discussion.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB728 and by Bundesministerium für Bildung und Forschung Grant 03NUK003C.
- PG
- prostaglandin
- RT
- reverse transcription
- siRNA
- small interfering RNA
- MAPK
- mitogen-activated protein kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1.Smith W. L., Garavito R. M., DeWitt D. L. (1996) J. Biol. Chem. 271, 33157–33160 [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekharan N. V., Dai H., Roos K. L., Evanson N. K., Tomsik J., Elton T. S., Simmons D. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13926–13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckman S. Y., Gresham A., Hale P., Hruza G., Anast J., Masferrer J., Pentland A. P. (1998) Carcinogenesis 19, 723–729 [DOI] [PubMed] [Google Scholar]
- 4.Bachelor M. A., Silvers A. L., Bowden G. T. (2002) Oncogene 21, 7092–7099 [DOI] [PubMed] [Google Scholar]
- 5.Mahns A., Wolber R., Stäb F., Klotz L. O., Sies H. (2004) Photochem. Photobiol. Sci. 3, 257–262 [DOI] [PubMed] [Google Scholar]
- 6.Rundhaug J. E., Fischer S. M. (2008) Photochem. Photobiol. 84, 322–329 [DOI] [PubMed] [Google Scholar]
- 7.Chen W., Tang Q., Gonzales M. S., Bowden G. T. (2001) Oncogene 20, 3921–3926 [DOI] [PubMed] [Google Scholar]
- 8.Tang Q., Chen W., Gonzales M. S., Finch J., Inoue H., Bowden G. T. (2001) Oncogene 20, 5164–5172 [DOI] [PubMed] [Google Scholar]
- 9.Fritsche E., Schäfer C., Calles C., Bernsmann T., Bernshausen T., Wurm M., Hübenthal U., Cline J. E., Hajimiragha H., Schroeder P., Klotz L. O., Rannug A., Fürst P., Hanenberg H., Abel J., Krutmann J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8851–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollig F., Winzen R., Kracht M., Ghebremedhin B., Ritter B., Wilhelm A., Resch K., Holtmann H. (2002) Eur. J. Biochem. 269, 5830–5839 [DOI] [PubMed] [Google Scholar]
- 11.Gowrishankar G., Winzen R., Bollig F., Ghebremedhin B., Redich N., Ritter B., Resch K., Kracht M., Holtmann H. (2005) Biol. Chem. 386, 1287–1293 [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Bowden G. T. (2008) Mol. Carcinog. 47, 974–983 [DOI] [PubMed] [Google Scholar]
- 13.Hinman M. N., Lou H. (2008) Cell. Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) Mol. Cell. Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. (2008) Biol. Chem. 389, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doller A., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2007) Mol. Biol. Cell 18, 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doller A., Akool el-S, Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2008) Mol. Cell. Biol. 28, 2608–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rousseau S., Morrice N., Peggie M., Campbell D. G., Gaestel M., Cohen P. (2002) EMBO J. 21, 6505–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan C. M., Steitz J. A. (2001) Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallouzi I. E., Brennan C. M., Steitz J. A. (2001) RNA 7, 1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachelor M. A., Cooper S. J., Sikorski E. T., Bowden G. T. (2005) Mol. Cancer Res. 3, 90–99 [DOI] [PubMed] [Google Scholar]
- 23.Brenneisen P., Wenk J., Klotz L. O., Wlaschek M., Briviba K., Krieg T., Sies H., Scharffetter-Kochanek K. (1998) J. Biol. Chem. 273, 5279–5287 [DOI] [PubMed] [Google Scholar]
- 24.Klotz L. O., Pellieux C., Briviba K., Pierlot C., Aubry J. M., Sies H. (1999) Eur. J. Biochem. 260, 917–922 [DOI] [PubMed] [Google Scholar]
- 25.Klotz L. O. (2002) Biol. Chem. 383, 443–456 [DOI] [PubMed] [Google Scholar]
- 26.Mahns A., Melchheier I., Suschek C. V., Sies H., Klotz L. O. (2003) Free Radic. Res. 37, 391–397 [DOI] [PubMed] [Google Scholar]
- 27.von Montfort C., Fernau N. S., Beier J. I., Sies H., Klotz L. O. (2006) Free Radic. Biol. Med. 41, 1478–1487 [DOI] [PubMed] [Google Scholar]
- 28.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gschwendt M., Müller H. J., Kielbassa K., Zang R., Kittstein W., Rincke G., Marks F. (1994) Biochem. Biophys. Res. Commun. 199, 93–98 [DOI] [PubMed] [Google Scholar]
- 31.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. (1993) J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 32.Kim H. H., Abdelmohsen K., Lal A., Pullmann R., Jr., Yang X., Galban S., Srikantan S., Martindale J. L., Blethrow J., Shokat K. M., Gorospe M. (2008) Genes Dev. 22, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H. H., Yang X., Kuwano Y., Gorospe M. (2008) Cell Cycle 7, 3371–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subbaramaiah K., Marmo T. P., Dixon D. A., Dannenberg A. J. (2003) J. Biol. Chem. 278, 37637–37647 [DOI] [PubMed] [Google Scholar]
- 36.Lafarga V., Cuadrado A., Lopez de Silanes I., Bengoechea R., Fernandez-Capetillo O., Nebreda A. R. (2009) Mol. Cell. Biol. 29, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]