Abstract
In mammals, NRF-2 (nuclear respiratory factor 2), also named GA-binding protein, is an Ets family transcription factor that controls many genes involved in cell cycle progression and protein synthesis as well as in mitochondrial biogenesis. In this paper, we analyzed the role of NRF-2 in the regulation of human genes involved in mitochondrial DNA transcription and replication. By a combination of bioinformatic and biochemical approaches, we found that the factor binds in vitro and in vivo to the proximal promoter region of the genes coding for the transcription termination factor mTERF, the RNA polymerase POLRMT, the B subunit of the DNA polymerase-γ, the DNA helicase TWINKLE, and the single-stranded DNA-binding protein mtSSB. The role of NRF-2 in modulating the expression of those genes was further established by RNA interference and overexpression strategies. On the contrary, we found that NRF-2 does not control the genes for the subunit A of DNA polymerase-γ and for the transcription repressor MTERF3; we suggest that these genes are under regulatory mechanisms that do not involve NRF proteins. Since NRFs are known to positively control the expression of transcription-activating proteins, the novelty emerging from our data is that proteins playing antithetical roles in mitochondrial DNA transcription, namely activators and repressors, are under different regulatory pathways. Finally, we developed a more stringent consensus with respect to the general consensus of NRF-2/GA-binding protein when searching for NRF-2 binding sites in the promoter of mitochondrial proteins.
Keywords: DNA polymerase gamma, MTERF3, Mitochondrial biogenesis, NRF-2, POLRMT, TWINKLE helicase, mTERF, mtSSB
Introduction
The basal components of mammalian mtDNA2 transcription and replication machineries have been extensively characterized (1). mtDNA transcription is carried out by the bacteriophage-related POLRMT (mitochondrial RNA polymerase), which, in the initiation step, is assisted by TFAM (mitochondrial transcription factor A) and TFB2M (mitochondrial transcription factor B2). Transcription termination is promoted by mTERF (mitochondrial termination factor), which binds simultaneously the termination site within the tRNALeu(UUR) gene and a site placed in the heavy strand promoter region. This favors a POLRMT recycling mechanism that accounts for the high rate of rDNA transcription and for the stimulatory effect exerted by mTERF on transcription initiation (2). mtDNA expression tuning requires not only activation factors but also the transcription repressor MTERF3 (3), which, together with mTERF, belongs to the MTERF protein family (4); it exerts its function by binding the promoter region of mtDNA. mtDNA synthesis is carried out by the DNA polymerase-γ (Pol-γ) (5). It consists of two polypeptides, the catalytic subunit Pol-γA and the accessory subunit Pol-γB. The latter was shown to increase the enzyme polymerase and exonuclease activities as well as processivity. mtDNA replication basic machinery includes also the bacteriophage-like DNA helicase TWINKLE and the single-stranded DNA-binding protein (mtSSB) (1).
Because the coding capacity of mammalian mtDNA is limited to 13 oxidative phosphorylation polypeptides, 22 tRNAs, and 2 rRNAs, the vast majority of mitochondrial proteins, including those involved in mtDNA replication and expression, are nucleus-encoded. Biogenesis and function of mitochondria require, therefore, the tightly coordinated expression of nuclear and mitochondrial genomes. Promoter structural and functional analyses have shown that various combinations of nuclear transcription factors, such as NRF-1 (nuclear respiratory factor 1) and NRF-2, Sp1 (specificity protein 1), YY1 (ying-yang protein 1), ERRα (estrogen-related receptor α), and others, regulate the expression of many nuclear genes encoding mitochondrial proteins (6). Moreover, it is well established that three co-activators of the PGC-1 family (PGC-1α (PPARγ coactivator 1α), PGC-1β (PPARγ coactivator 1β), and PRC (PGC-1-related coactivator)) mediate the response of mitochondrial biogenesis to environmental stimuli, such as energy deprivation, cold, and fasting (6, 7).
NRF-2, also known as GA-binding protein (GABP), is one of the about 30 mammalian factors belonging to the Ets (E26 transformation-specific) factor family (8). Ets factors share an evolutionarily conserved DNA binding domain that preferentially recognizes sequences containing the GGAA core motif. NRF-2/GABP is involved in the control of basic cellular processes, such as cell cycle progression, protein synthesis, and mitochondrial biogenesis (8, 9). NRF-2/GABP is the only Ets factor showing a multimeric composition. It consists of the 51-kDa subunit α, containing the Ets DNA binding domain, and of the four subunits β1, β2, γ1, and γ2, each of them being able to form heterodimeric complexes with subunit α (10). The β and γ polypeptides are splicing variants of the same gene and contain a transcription activation domain. Subunits β (41–42 kDa) display a homodimerization domain in the C-terminal region, whereas the shorter subunits γ (37–38 kDa) lack this domain and are not able to interact with each other. NRF-2/GABP α/β or α/γ heterodimers form spontaneously in solution; however, only α/β dimers can associate and form, in the presence of two tandemly arranged binding sites, an α2/β2 tetramer that binds DNA cooperatively.
NRF-2 has been shown to be a master coordinator of the expression of all 10 nucleus-encoded complex IV subunits (11). However, the transcriptional activation is not restricted to oxidative phosphorylation polypeptides because it was shown to control also the expression of a variety of proteins, including enzymes (12), TOM complex receptors (13), complex IV assembly factors (14, 15), and peroxiredoxin V (16).
Regarding NRF-2 control on mtDNA transcription and replication proteins, the only available information indicates that it activates the expression of the transcription initiation factors TFAM (17) and TFB2M (18). Under NRF-2 control is also TFB1M (mitochondrial transcription factor B1), the TFB2M paralogue, that was recently shown to be mainly involved in mitochondrial protein synthesis (18, 19). In the current study, we investigated on the dependence on NRF-2/GABP of the genes for the other proteins of the minimal transcription and replication machineries. By a combination of in vitro and in vivo experiments, we demonstrate that NRF-2 positively regulates the expression of POLRMT, mTERF, Pol-γB, TWINKLE, and mtSSB; on the contrary, no control by NRF-2 was observed for MTERF3 and Pol-γA genes.
EXPERIMENTAL PROCEDURES
Bioinformatics
Promoter sequences of the analyzed human genes were retrieved by the Ensembl Genome Browser release 53 (available on the World Wide Web). Transcription factor binding sites were predicted by using the MAPPER search engine (20), applying factor-derived models from the TRANSFAC and JASPAR data bases; all of the predicted sites were confirmed by the Genomatix MatInspector tool (21). Multiple alignments were performed at the NPS@ Web server of the Pôle Bioinformatique Lyonnais using the International Union of Biochemistry (IUB) weight matrix and formatted with the ESPript 2.2 Web tool (available on the World Wide Web). Graphical representation of NRF-2 binding site consensus was obtained using the WebLogo tool (22).
Cell Culture and Nuclear Extract Preparation
HeLa cells (ECACC) were maintained in Dulbecco's modified Eagle's medium (EuroClone) supplemented with 10% fetal bovine serum in the presence of 50 units/ml penicillin and 50 μg/ml streptomycin at 37 °C in 5% CO2. Nuclear extract was prepared from monolayer cultured HeLa cells as described (23) and partially purified by heparin-Sepharose chromatography. To this purpose, total nuclear proteins (about 7 mg) in 20 mm Hepes, pH 7.9, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 25% glycerol (v/v), 0.5 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol were diluted to 100 mm NaCl with column buffer (20 mm HEPES, pH 7.3, 20% glycerol (v/v), 0.2 mm EDTA, 0.5 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride) and loaded onto a 4-ml heparin-Sepharose CL-6B (GE Healthcare) column equilibrated with column buffer containing 100 mm KCl. NRF-2 complexes were eluted with the same buffer containing 300 mm KCl.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA probes were obtained by annealing the single-stranded oligonucleotides reported in Table 1. Probes were 3′-end-labeled with [α-32P]dATP and Klenow enzyme and then purified using Micro Bio-Spin P-30 columns (Bio-Rad). Binding reaction mixtures were set up by combining 10 μg of heparin-Sepharose purified nuclear extract, 80 fmol of DNA probe, and 2 μg of poly(dI-dC)·poly(dI-dC) (GE Healthcare) in a 20-μl final volume containing 25 mm Tris-HCl, pH 7.9, 10% glycerol (v/v), 0.5 mm EDTA, 6.25 mm MgCl2, 1 mm dithiothreitol. Unlabeled competitor oligonucleotides were added to the binding reactions in a 100-fold molar excess. Samples were incubated at room temperature for 15 min. In order to produce supershifted complexes, specific anti-NRF-2α and anti-NRF-2β sera (from R. C. Scarpulla (Northwestern University Medical School, Chicago, IL)) were added 10 min after the binding reaction was started. After an additional 5 min of incubation at room temperature, samples were electrophoresed onto a 5% polyacrylamide gel in 0.5× TBE. Bands were visualized by the Typhoon 8600 phosphorimaging system (Amersham Biosciences).
TABLE 1.
EMSA probe and competitor oligonucleotides
For each probe and competitor, the complementary sequences are shown. Oligonucleotide positions are numbered with respect to the transcription initiation site. NRF-2 core recognition sequences are in boldface type, and mutated nucleotides are underlined; gt nucleotides were artificially added to each oligonucleotide.
| Promoter | Sequence | Position |
|---|---|---|
| RCOX4 | 5′-gtCTTGCTCTTCCGGTGCGGGACCCGCTCTTCCGGTCGCGA-3′ | See Ref. 18 |
| 5′-gtTCGCGACCGGAAGAGCGGGTCCCGCACCGGAAGAGCAAG-3′ | ||
| mTERF (−48/−29 probe) | 5′-gtGCGACCCGAGGCCGGAAGTTAGTCTTTTCCACTTCCGCTCCTCCTAGCC-3′ | −61/−13 |
| 5′-gtGGCTAGGAGGAGCGGAAGTGGAAAAGACTAACTTCCGGCCTCGGGTCGC-3′ | ||
| POLRMT (−61/−45 probe) | 5′-gtTCCTGGGAGTCTACTTCCGGCTGGGGTTTCCCTTCGCAGCCTCCGTCG-3′ | −75/−28 |
| 5′-gtCGACGGAGGCTGCGAAGGGAAACCCCAGCCGGAAGTAGACTCCCAGGA-3′ | ||
| POLRMT (mut-61 competitor) | 5′-gtTCCTGGGAGTCTACTAGAGGCTGGGGTTTCCCTTCGCAGCCTCCGTCG-3′ | −75/−28 |
| 5′-gtCGACGGAGGCTGCGAAGGGAAACCCCAGCCTCTAGTAGACTCCCAGGA-3′ | ||
| POLRMT (mut-45 competitor) | 5′-gtTCCTGGGAGTCTACTTCCGGCTGGGGTTTCAGATCGCAGCCTCCGTCG-3′ | −75/−28 |
| 5′-gtCGACGGAGGCTGCGATCTGAAACCCCAGCCGGAAGTAGACTCCCAGGA-3′ | ||
| TWINKLE (−45 probe) | 5′-gtACTAAACCTCGAGGCTTCCGGTTCCGGGACGACCGCTCCC-3′ | −60/−21 |
| 5′-gtGGGAGCGGTCGTCCCGGAACCGGAAGCCTCGAGGTTTAGT-3′ | ||
| mtSSB (−11 probe) | 5′-gtTTTGCGTTCCCTGTGCGCCGGAAGTGATCCCCTGCGTGGC-3′ | −30/+10 |
| 5′-gtGCCACGCAGGGGATCACTTCCGGCGCACAGGGAACGCAAA-3′ | ||
| Pol-γA (−22 probe) | 5′-gtCCTAGCTGGGTGCAGACGGGAAGTTGCGGCTGCCAGCGAA-3′ | −40/−1 |
| 5′-gtTTCGCTGGCAGCCGCAACTTCCCGTCTGCACCCAGCTAGG-3′ | ||
| Pol-γB (−61 probe) | 5′-gtCCGGAACGCAGACATGCGCTTCCGGGGTGGGGCCTGCCGC-3′ | −80/−41 |
| 5′-gtGCGGCAGGCCCCACCCCGGAAGCGCATGTCTGCGTTCCGG-3′ | ||
| Pol-γB (−255 probe) | 5′-gtTGCAAAACAGTGGCAGAAGGAAGTGACTCAAGTGGCTACA-3′ | −273/−234 |
| 5′-gtTGTAGCCACTTGAGTCACTTCCTTCTGCCACTGTTTTGCA-3′ |
Chromatin Immunoprecipitation (ChIP)
HeLa cells (106/immunoprecipitation sample) were grown in monolayer at 60% confluence in 75-cm2 flasks and cross-linked by the addition of 1% formaldehyde. Cross-linking was allowed to proceed for 10 min at room temperature and was stopped by adding 125 mm glycine. Cells were trypsinized, scraped, and collected by centrifugation at 1000 × g for 5 min and then washed with phosphate-buffered saline and recollected. Cell pellets were resuspended in 100 μl of ice-cold lysis buffer (10 mm HEPES-KOH, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiothreitol and 0.2 mm phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. Nuclei were collected by centrifugation at 1000 × g for 5 min at 4 °C, and the resulting pellets were resuspended in 200 μl of sonication buffer (10 mm EDTA, 50 mm Tris-HCl, pH 7.6, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A) at room temperature, gently mixed, and incubated on ice for 20 min. Nuclei were then sonicated with 15 pulses of 10 s (Branson Sonifier 250, 30% duty cycle) with 20-s cooling on ice between pulses, yielding DNA fragments mostly between 200 and 700 bp, as verified by gel electrophoresis on agarose gel.
Immunoprecipitation was carried out by using salmon sperm DNA/Protein A-agarose (Upstate) according to the manufacturer's instructions and rabbit polyclonal anti-NRF-2α serum or p53 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) as mock control. Immunoprecipitated DNA (one-fifteenth of the total amount) was used as template for semiquantitative PCR amplification using the promoter-specific primers reported in Table 2. The PCR products, obtained after 25 cycles within the linear range of amplification, were electrophoresed onto a 2.5% agarose gel and visualized by ethidium bromide staining.
TABLE 2.
ChIP primers
Amplicon positions are numbered with respect to the transcription initiation site; the position of the β-actin amplicon is in reference to the mRNA sequence (GenBankTM accession number NM_001101). For each primer, F and R indicate forward and reverse orientation, respectively.
| Promoter | Sequence | Amplicon position |
|---|---|---|
| TFB2M | F 5′-GGTCGGTCGCTCTCCTCAA-3′ | −114/+7 |
| R 5′-AAACACTAGAGCCTGCGCATG-3′ | ||
| mTERF | F 5′-GACCAACGACATCACCTCTGC-3′ | −84/+37 |
| R 5′-CACCCATCCACTGTAGTTCGC-3′ | ||
| POLRMT | F 5′-AAAACAGCAGGAGGAACCAATC-3′ | −136/−6 |
| R 5′-CCGGGAGTTGTGGTTTCATG-3′ | ||
| MTERF3 | F 5′-CTGTCTCCCCGCGTAACC-3′ | −100/−2 |
| R 5′-CTCCTCAGCCCGCCCTAC-3′ | ||
| TWINKLE | F 5′-GGCGGGACTAAACCTCGAG-3′ | −66/+65 |
| R 5′-GGTTACCACTTTTCTCTCTCCCAC-3′ | ||
| mtSSB | F 5′-GGCAGTATTTCCAAGGCGC-3′ | −88/+33 |
| R 5′-GACGATCTAACCCGAGCAGC-3′ | ||
| Pol-γA | F 5′-CCTCTCGGGTAGCCGCG-3′ | −60/+30 |
| R 5′-TAGCGTGTGGCCTCCACC-3′ | ||
| Pol-γB | F 5′-GAGAACCATCCGAGCCGG-3′ | −130/+21 |
| R 5′-GGAGGTGAGCGTGCTTGC-3′ | ||
| β-actin | F 5′-CCCAGCCATGTACGTTGCTA-3′ | 471/556 |
| R 5′-CGTCACCGGAGTCCATCAC-3′ |
RNA Interference (RNAi) and Overexpression
RNAi was performed using Silencer Select predesigned siRNAs (Ambion) directed against NRF-2α and NRF-2β. The sense sequences of NRF-2α and NRF-2β siRNAs were 5′-GAAUUCAGCAUGACCGAUAtt-3′ and 5′-GGUGGAACUUUUAAUCAAAtt-3′, respectively. Silencer siRNA mixture kit-RNase III (Ambion) was used to generate a mixture of siRNAs on the sequence of the bacterial gene lacZ (mock control); lacZ double-stranded RNA was synthesized as reported (24), purified, and digested with RNase III. HeLa cells (2 × 105) in 6-well plates were reverse-transfected with 30 pmol of both NRF-2α and NRF-2β siRNAs or 60 pmol of the lacZ siRNAs in the presence of 5 μl of siPORT NeoFX transfection agent (Ambion). After 48 h of growth, cells were either collected or transfected again in the same conditions and grown for an additional 48 h.
To obtain overexpression constructs pcDNA/NRF-2α and pcDNA/NRF-2β1, cDNAs of human NRF-2α and NRF-2β1 were amplified by PCR using as template pET3d/NRF-2α and pET3a/NRF-2β1 (10), respectively. To generate NRF-2α cDNA, primers were NRF2α-For (5′-CGCGGTACCATGGCTAAAAGAGAAGCAGAGG-3′), containing a KpnI site (underlined) and the ATG codon (boldface type) in the Kozak consensus context, and NRF2α-Rev (5′-CGCTCTAGATCAATTATCCTTTTCCGTTTG-3′), containing an XbaI site (underlined) and the stop codon (boldface type). To generate NRF-2β1 cDNA, primers were NRF2β-For (5′-CGCGGTACCATGGCCCTGGTAGATTTGGGA-3′), containing a KpnI site (underlined) and the ATG codon (boldface type) in the Kozak consensus context, and NRF2β-Rev (5′-CGCTCTAGATTAAACAGCTTCTTTATTAGTC-3′), containing an XbaI site (underlined) and the stop codon (boldface type). The cDNAs were inserted into the MCS of pcDNA3.1/myc-HisB vector (Invitrogen) to obtain pcDNA/NRF-2α and pcDNA/ NRF-2β1. They were transiently co-transfected into HeLa cells by cationic lipid-mediated transfection. Briefly, 2 × 105 cells were seeded in 6-well plates and transfected the next day with 0.5 μg of both pcDNA/NRF-2α and pcDNA/NRF-2β1 or 1 μg of the empty vector in the presence of 3 μl of Cellfectin reagent (Invitrogen), according to the manufacturer's instructions. Total cellular proteins and RNA were extracted from both NRF-2-depleted and -overexpressing cells using the RNeasy Midi kit (Qiagen) and subjected to Western blotting and real-time reverse transcription-PCR analyses, respectively, as described below.
Western Blotting Analysis
Total cellular proteins (40 μg) were fractionated on 12% SDS-polyacrylamide gel and electroblotted for 1 h onto polyvinylidene difluoride membrane (Hybond-P, GE Healthcare). Membranes were preincubated for 1 h with 1× Blotto (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100) containing 5% nonfat dry milk, followed by incubation for 1 h in the same solution, with rabbit polyclonal antibodies against NRF-2α, NRF-2β, or β-actin (Sigma). Filters were washed three times for 20 min with 1× Blotto, 5% milk; incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology, Inc.); and washed with 1× Blotto. Protein bands were visualized using the ECL Plus Western blotting reagents (GE Healthcare).
Real-time Reverse Transcription-PCR Assay
Total RNA (300 ng) was reverse-transcribed in a final volume of 20 μl using the enhanced avian myeloblastosis virus reverse transcriptase kit (Sigma), according to the manufacturer's instructions. Real-time PCR was performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems) and the ABI PRISM 7000 sequence detection system (Applied Biosystems). Primers were designed using Primer Express 2.0 software (Applied Biosystems); the resulting sequences are reported in Table 3. Each reaction was run in triplicate and contained 1 μl of reverse transcription reaction along with 200 nm primers in a final reaction volume of 20 μl. Amplification conditions were as follows: 95 °C for 10 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. To ensure that only a single product was amplified, the melting curve analysis was performed using the Dissociation Curves software (Applied Biosystems). All PCR products were run on a 2.5% agarose gel to confirm specificity.
TABLE 3.
Real-time PCR primers
Amplicon position is numbered according to the respective mRNA sequence; accession numbers are relative to the GenBankTM data base. For each primer, F and R indicate forward and reverse orientation, respectively.
| Transcript | Sequence | Accession no. | Amplicon position |
|---|---|---|---|
| mTERF | F 5′-GGCTTTTTGGTGTGAAGTGTCA-3′ | NM_006980 | 243/363 |
| R 5′-ACTCCAGGCTGTCGTTTCCTT-3′ | |||
| POLRMT | F 5′-AGCAGAAGAACGGCTTCCC-3′ | NM_005035 | 3397/3507 |
| R 5′-CGTGCACAGAGACGAAGGTC-3′ | |||
| MTERF3 | F 5′-CCTTCCATGAATGAACAGTCACA-3′ | NM_015942 | 343/453 |
| R 5′-AATTGGCTGCATTGGAGACAA-3′ | |||
| TWINKLE | F 5′-AACCCCAAACGATGCTTCTTG-3′ | AF292005 | 1161/1261 |
| R 5′-GCGGTACGAAGAATACGAGAAAG-3′ | |||
| mtSSB | F 5′-CAGTCAAAAGACAACATGGCACA-3′ | NM_003143 | 234/324 |
| R 5′-AATTCGAGACCCCTTTTTCACA-3′ | |||
| Pol-γA | F 5′-AAACGTATCAGCTCCCAGATGGT-3′ | NM_002693 | 2698/2798 |
| R 5′-GCCCCATAGAGGCCTTCCT-3′ | |||
| Pol-γB | F 5′-AAGGTTGCTTTGGATGTAGGAAGA-3′ | NM_007215 | 1227/1327 |
| R 5′-GGCCACACAGAAATCCCATT-3′ | |||
| β-Actin | F 5′-CCCAGCCATGTACGTTGCTA-3′ | NM_001101 | 471/556 |
| R 5′-CGTCACCGGAGTCCATCAC-3′ |
RESULTS
Identification of Transcription Factor Binding Sites in the Promoter of Human Genes Encoding Proteins for mtDNA Replication and Transcription
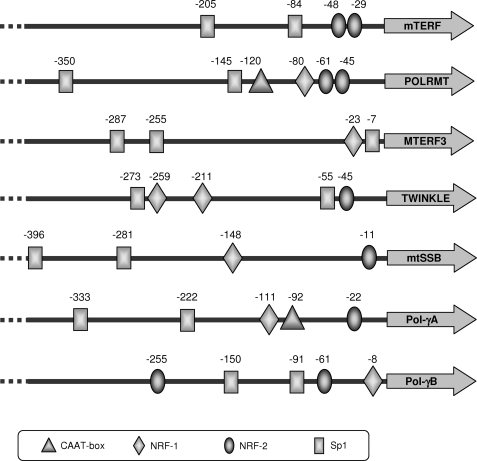
To identify putative sites for NRF-2 in the promoter proximal regions of human genes encoding proteins of the basal mtDNA replication and transcription machineries, we performed a careful in silico search using bioinformatic tools such as MAPPER and MatInspector. As illustrated in Fig. 1, NRF-2 recognition sites with different scores were identified in all the analyzed promoters except that of MTERF3. Sites showing higher probability (MAPPER score 4.3–3.5) were identified at positions −48 and −29 for mTERF, −61 for POLRMT, −45 for TWINKLE, −11 for mtSSB, and −61 for Pol-γB. Lower score sites (1.4) were identified at positions −22 for Pol-γA and −255 for Pol-γB. Moreover, sequence visual inspection identified an NRF-2 core motif in position −45 of the POLRMT promoter, 16 nt from that at −61, a distance suggesting a cooperative binding of an NRF-2 heterotetrameric complex to the tandemly arranged sites. Fig. 1 also reports the location of putative binding sites for the nuclear factors NRF-1 and Sp1. Binding sites for Sp1 were predicted in all the analyzed promoters; no prediction of NRF-1 sites was obtained for mTERF. Finally, the in silico analysis of the promoters indicated the absence of canonical TATA and CAAT boxes, a feature shared by many respiratory genes; the only exceptions are the POLRMT and Pol-γA genes, which exhibit high scoring CAAT boxes at positions −120 and −92, respectively (Fig. 1).
FIGURE 1.
Location of predicted CAAT boxes and NRF-1, NRF-2, and Sp1 binding sites in the promoter proximal region of human genes coding for mtDNA transcription and replication proteins. Position numbers refer to the transcription start sites of each gene as deduced from mRNA sequence extracted by the Ensembl search engine. The −45 NRF-2 site in the POLRMT promoter was detected by visual inspection of the sequence.
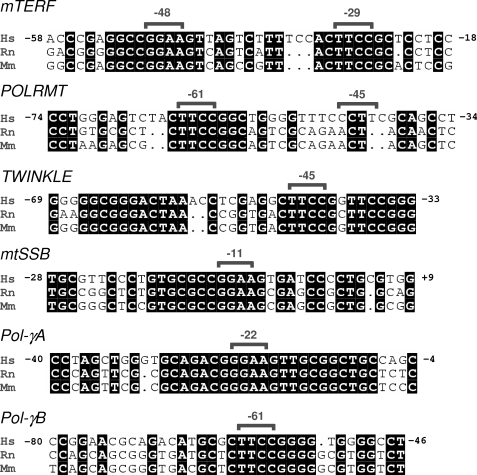
We analyzed the conservation of all the putative NRF-2 sites by comparing the human promoter sequences with the corresponding sequences of mice and rats. As reported in Fig. 2, all of the predicted sites are conserved, at least in the core sequence, in all of the three analyzed species, with the only exception being the low score Pol-γB −255 site, which falls in an extended region of the human promoter that is not conserved in mice and rats. No conservation was also displayed by the NRF-2 core motif identified at −45 in the POLRMT promoter.
FIGURE 2.
Conservation in different species of NRF-2 binding sites in the analyzed promoters. Shown are alignments of corresponding human (Hs), rat (Rn), and mouse (Mm) promoter sequences containing the predicted NRF-2 binding sites. Nucleotides conserved in all sequences are shaded in black; the numbers on both sides of the human sequences indicate nucleotide positions in reference to the transcription start site. Predicted NRF-2 site core sequences are indicated by brackets together with their respective positions.
In Vitro NRF-2 Binding to the Putative Sites
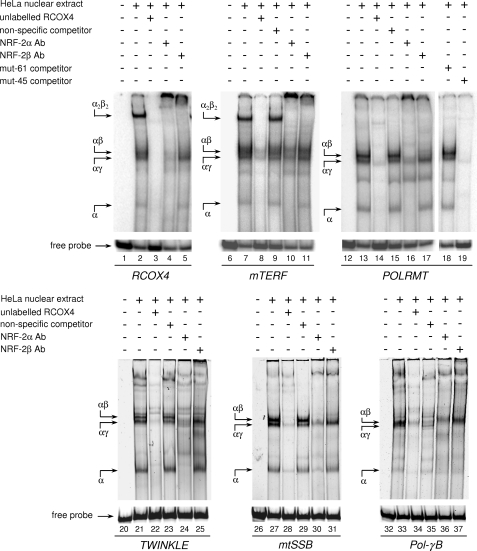
To test NRF-2 binding to the putative recognition sequences in the analyzed promoters, we carried out EMSAs using a HeLa cell partially purified nuclear extract and the seven 32P-labeled double-stranded oligonucleotide probes listed in Table 1. Each probe was centered on one binding site with the exception of mTERF and POLRMT probes that contained two tandemly arranged sites. A probe containing the two 20-nt distant sites of the rat COX4 (RCOX4) promoter, which are known to cooperatively bind two α/β NRF-2 dimers, was used as positive control. Electrophoretic analysis of the binding products shows (Fig. 3, lane 2) a pattern consistent with that reported by Gleyzer et al. (18). It consists of a faster migrating faint DNA-protein complex containing the DNA-binding subunit α, two close intermediate migrating complexes containing the α/γ and α/β heterodimers, respectively, and a slower migrating abundant complex containing the α2/β2 heterotetramer. All of the complexes were competed by an excess of unlabeled homologous competitor (Fig. 3, lane 3); moreover, the NRF-2α antiserum supershifted all of the complexes, whereas the NRF-2β antiserum specifically supershifted those complexes containing NRF-2 subunit β (Fig. 3, lanes 4 and 5). These results confirmed that the protein fraction used in the assay contained all of the NRF-2 subunits in their active form.
FIGURE 3.
In vitro binding of NRF-2 to the predicted sites in the promoter of the selected genes. NRF-2 binding was evaluated by EMSA using a heparin-Sepharose-purified nuclear extract from HeLa cells and radiolabeled double-stranded oligonucleotide probes containing one or two NRF-2 putative binding sites. The unlabeled specific (RCOX4), nonspecific, and mutated (mut-61 and mut-45) competitor oligonucleotides (100-fold molar excess) and NRF-2 antisera are indicated above each lane. Probes are shown below the lanes. DNA-protein complexes are indicated by arrows on the left of each panel. Ab, antibody.
A pattern similar to that obtained for RCOX4 probe was observed for the mTERF probe, which contains two NRF-2 sites in tandem; also in this case (Fig. 3, lane 7) an abundant heterotetrameric complex is present, thus showing that the −48 and −29 sites are both functional and able to bind NRF-2 cooperatively with high affinity. All of the complexes were strongly competed by an excess of an unlabeled oligonucleotide containing authentic NRF-2 sites (RCOX4 probe) and unaffected by the addition of an excess of an unlabeled nonspecific competitor (Fig. 3, lanes 8 and 9). NRF-2α and NRF-2β antisera supershifted completely the α2/β2 heterotetramer and, although at a lower extent, the other α- or β-containing complexes (Fig. 3, lanes 10 and 11).
Contrary to the mTERF probe, that of POLRMT, although containing two putative NRF-2 sites (−61 and −45), formed only complexes containing subunit α and the α/γ and α/β heterodimers the latter being the most abundant (Fig. 3, lane 13). All of the complexes were specific, as observed in the presence of competitors or antibodies (Fig. 3, lanes 14–17). The absence of the complex containing the α2/β2 heterotetramer suggested that the putative −45 site was not functional. To confirm this hypothesis, we performed further competition experiments using as unlabeled competitor two versions of the POLRMT oligonucleotide probe mutated in the −61 and −45 site, respectively (Fig. 3, lanes 18 and 19). The oligonucleotide containing the mutation at the −45 site still kept a high competition capacity, whereas the oligonucleotide mutated at the −61 site showed no capacity to compete; this clearly implies that the −61 site is the only functional NRF-2 site in the POLRMT proximal promoter.
NRF-2 also bound TWINKLE probe containing the single site at −45 (Fig. 3, lanes 20–25), mtSSB probe containing the single site at −11 (Fig. 3, lanes 26–31), and, although with a lower affinity, Pol-γB probe containing the single site at −61 (Fig. 3, lanes 32–37). No NRF-2 complexes were obtained, instead, using a probe containing the low scoring Pol-γB −255 site, not conserved in rats and mice (data not shown). The Pol-γA −22 site, although conserved in rats and mice, was unable to bind NRF-2, even in the presence of high protein concentrations (see supplemental Fig. S1) .
In Vivo NRF-2 Binding to the Proximal Promoter of the Analyzed Genes
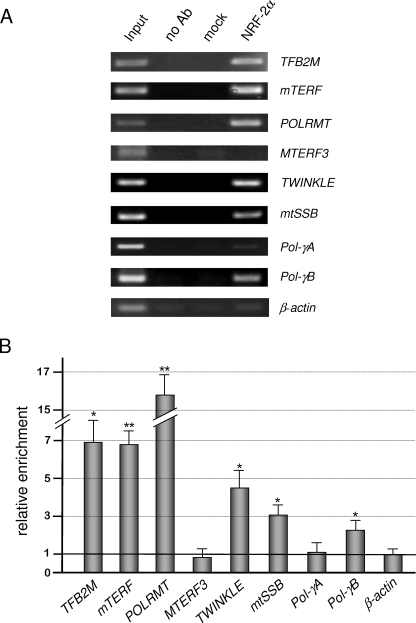
The in vivo binding of NRF-2 to all of the analyzed promoters was tested by ChIP analysis on human HeLa cultured cells. Primer pairs were selected to obtain amplicons ranging from 90 to 151 nt and containing the EMSA-positive or -negative NRF-2 sites. In the case of the proximal promoter of the MTERF3 gene, where no NRF-2 sites were identified, the amplicon consisted of about 100 bp immediately upstream of the transcription initiation site. As positive and negative controls, amplicons containing the already characterized NRF-2 sites in the TFB2M promoter (18) and part of the β-actin coding sequence, respectively, were used.
As illustrated in Fig. 4, A and B, for mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB, a significant enrichment of immunoprecipitated promoter fragments was obtained using specific NRF-2 antibodies, with respect to the mock (p53 antibodies) or no antibody control samples. The amount of amplified DNAs was roughly comparable with that obtained using as template about 0.3% of the total input chromatin (Input lane). The specificity of the enrichment obtained for the analyzed promoters was confirmed by the absence of a significant signal depending on NRF-2 antibodies when using actin primers. No significant enrichment was observed for Pol-γA amplicon (containing the EMSA-negative −22 site) and for the MTERF3 amplicon. For these genes, we extended the analyzed promoter region, selecting a further amplicon located about 500 nt upstream of the transcription initiation site. In both cases, no positive signals were observed (not shown). In conclusion, the ChIP assay confirmed the in vivo binding of NRF-2 to all of the promoters containing EMSA-positive sites and ruled out the in vivo binding of NRF-2 to Pol-γA and MTERF3 proximal promoter regions.
FIGURE 4.
ChIP assay for in vivo binding of NRF-2 to the analyzed promoters. A, nuclei obtained from formaldehyde-fixed HeLa cells were lysed, and chromatin was fragmented by sonication. Chromatin was immunoprecipitated either in the absence of antibodies (no Ab) or in the presence of p53 commercial antibodies (mock) or polyclonal NRF-2α antiserum. Immunoprecipitated DNAs were subjected to semiquantitative PCR using primers specific for each examined promoter. Template DNAs, immunoprecipitated in the different conditions, are indicated above each lane; input indicates PCRs containing as template 0.3% of the total amount of chromatin used in immunoprecipitation reactions. Analyzed promoters are indicated to the right. Reactions addressed to TFB2M and β-actin were used as positive and negative control, respectively. B, histogram showing the quantification of PCR products obtained on NRF-2α immunoprecipitated DNA, relative to input signal; the relative enrichment of β-actin DNA was fixed as 1. Data represent the average of independent quantifications (n = 6) from three ChIP experiments; values are expressed as a ratio, and results are means ± S.D. Statistical analysis was performed using paired two-tailed Student's t test (*, p < 0.05; **, p < 0.01).
Effect of NRF-2 Silencing and Overexpression on the mRNA Level of the Analyzed Genes
We finally wished to investigate the in vivo control by NRF-2 of the expression of all of the analyzed genes by measuring the abundance of the corresponding mRNAs in cells where altered levels of the factor were obtained.
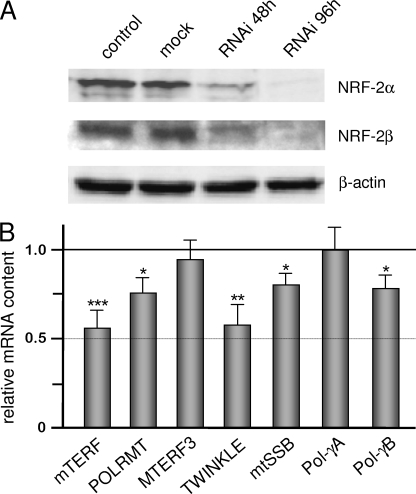
We used an RNAi procedure to obtain depletion of NRF-2 α and β subunits in HeLa cells. We treated cells for 48 and 96 h with a mixture of NRF-2α and NRF-2β siRNAs and monitored the level of both polypeptides by a Western blotting assay. We obtained (Fig. 5A) a decrease of more than 60% in NRF-2 α and β subunit level at 48 h and of more than 80% at 96 h; the effect of NRF-2 siRNAs was specific because no change was observed in cells treated with siRNA containing the sequence of the lacZ gene (mock control). We measured in 96 h-treated cells the steady-state level of mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB mRNAs whose gene promoters were demonstrated to be contacted in vitro and in vivo by NRF-2. We also measured the level of MTERF3 and Pol-γA mRNAs for which EMSA and ChIP experiments indicated no NRF-2 binding to the promoter. Results obtained from three independent RNAi experiments (Fig. 5B) indicate that in NRF-2-depleted cells the level of mRNAs for mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB decreased from 45 to 25% with respect to non-depleted cells. On the contrary, no effect of NRF-2 depletion was observed on the MTERF3 and Pol-γA mRNA level.
FIGURE 5.
Effect of NRF-2 complex knockdown on the level of the mRNA from all analyzed genes. A, NRF-2α and -β subunits were knocked down in HeLa cells by means of RNAi. Cells, either untreated (control) or treated with lacZ (mock) or NRF-2α/NRF-2β siRNAs, were harvested 48 and 96 h after transfection; total cellular proteins were subjected to Western blotting analysis with polyclonal antibodies against NRF-2α, NRF-2β, and β-actin. B, total RNA was extracted from 96-h treated and control cells, and relative quantification of mRNAs was carried out by real-time reverse transcription-PCR. Bars indicate the relative content of transcripts, normalized to β-actin mRNA (endogenous control), in treated with respect to control cells, fixed as 1. The relative quantification was performed according to the Pfaffl equation (25). Values are expressed as a ratio, and results are means ± S.D. (n = 9). Statistical analysis was performed using paired two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
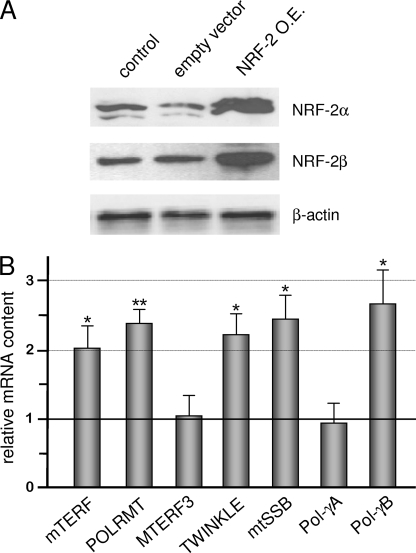
Next, we overexpressed NRF-2 α and β subunits in HeLa cells by co-transfection with two recombinant vectors, each containing the cDNA for one subunit under the control of the constitutive CMV promoter. Twenty-four hours after transfection, we measured the level of α and β subunits and detected an about 5-fold increase for both polypeptides (Fig. 6A). By measuring the content of the mRNAs of all of the analyzed genes in NRF-2-overexpressing cells (Fig. 6B), we found a significant increase (2–2.6-fold) in the level of mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB mRNAs. Also in this case, no effect of NRF-2-altered quantity was observed on the expression of MTERF3 and Pol-γA genes.
FIGURE 6.
Effect of NRF-2 complex overexpression on the level of the mRNA from all analyzed genes. A, NRF-2α and -β subunit overexpression was obtained by co-transfecting HeLa cells with DNA constructs containing the cDNA of the two subunits under the control of the constitutive CMV promoter. Cells either untreated (control) or transfected with empty pcDNA3.1 vector or co-transfected with pcDNA/NRF-2α and pcDNA/NRF-2β1 constructs (NRF-2 O.E.) were harvested 24 h after transfection; total cellular proteins were subjected to Western blotting analysis with polyclonal antibodies against NRF-2α, NRF-2β, and β-actin. B, total RNA was extracted from treated and control cells, and relative quantification of mRNAs was carried out by real-time reverse transcription-PCR as described in the legend to Fig. 5. Values are expressed as a ratio, and results are means ± S.D. (n = 5). Statistical analysis was performed using paired two-tailed Student's t test (*, p < 0.05; **, p < 0.01).
These results show that variation in mRNA levels for all of the analyzed genes following NRF-2 loss or gain of function are consistent with the data obtained from the in vitro and in vivo binding assays and demonstrate that NRF-2 is a transcription factor positively controlling the expression of mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB genes.
A Refined Model for NRF-2 Binding Site Prediction
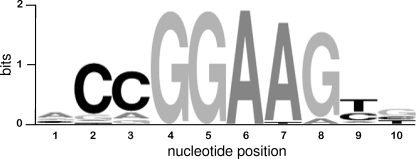
Our data add six new examples to the collection of NRF-2 sites in the promoter of genes for mitochondrial proteins whose binding by the factor was demonstrated in vitro and/or in vivo. We performed a multialignment using a sequence data set composed of 30 functional NRF-2 binding sites of mitochondrial protein genes, including those described in the present paper (see supplemental Table S1 and Fig. S2). The obtained consensus, formatted as WebLogo (Fig. 7), shows a higher representation of G in position 8 with respect to the NRF-2/GABP recognition motif obtained by ChIP-Seq data (26). Hence, the canonical NRF-2/GABP recognition core GGAA could be extended to the conserved GGAAG block in the case of sites regulating mitochondrial proteins.
FIGURE 7.
Motif analysis of functional NRF-2 binding sites in mitochondrial gene promoters. The binding site alignment is shown as WebLogo output. The information content of the motifs is expressed in bits. The relative size of the letters is a measure of the relative representation of each nucleotide in a given position; letters are sorted in descending order depending on their frequencies.
Finally, we extended the in silico promoter analysis to several other proteins involved in DNA replication and/or repair and associated with mitochondria (for a review, see Refs. 27 and 28). As shown in supplemental Table S2, 4 of 15 analyzed promoters (FEN1, MPG, NTHL1, and NUDT1) exhibit NRF-2 binding sites.
DISCUSSION
mtDNA transcription and replication basic machineries are composed of well characterized enzymatic activities and accessory protein factors. However, with the only exceptions being the transcription initiation factors TFAM and TFBM, for which the control by both NRF-1 and NRF-2 factors and consequently by PGC-1 family co-activators was well established (17, 18), still limited is the information concerning the regulation of the expression of the mitochondrial transcription and replication proteins. The only available data come from an extensive ChIP-on-chip analysis (29) that indicated NRF-1 binding to the promoters of POLRMT and Pol-γB genes.
In this paper, we focused our attention on the regulation by NRF-2 of nuclear genes coding for proteins involved in mtDNA transcription and replication. By bioinformatic approaches, we identified NRF-2 putative binding sites in the promoter proximal regions of most of them. In vitro and in vivo binding assays showed correspondence between the bioinformatic prediction and the actual NRF-2 binding for those high scoring sites located in the promoters of mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB genes. All of the NRF-2-bound sequences are conserved in mice and rats. The putative site at −22 of the Pol-γA promoter, although lying in an extended conserved sequence block, showed a lower score and was unable to interact with NRF-2. Interestingly, we found that the conserved sequence block contains a high scoring recognition element for the Maf transcription factors (MARE) (30). No NRF-2 binding was predicted and experimentally observed for the promoter proximal region of the MTERF3 gene. By using knockdown and overexpression strategies, we confirmed the positive regulation by NRF-2 on the expression of mTERF, POLRMT, TWINKLE, mtSSB, and Pol-γB genes and ruled out the control on MTERF3 and Pol-γA genes. The mRNA level profile observed in NRF-2-depleted cells suggests that NRF-2 control is stronger for mTERF and TWINKLE than for POLRMT, mtSSB, and Pol-γB, for which the mRNA decrease is less evident. For these genes, it is possible that NRF-2 plays a less determinant role in promoter activation and/or that its depletion is, at least partially, compensated by factors such as NRF-1 or Sp1. The observation that mTERF and TWINKLE mRNAs decrease similarly and yet mTERF promoter only contains two tandemly arranged sites that bind with high affinity two α/β NRF-2 monomers indicates a lack of correlation between the extent of mRNA level decrease and the number and arrangement of NRF-2 sites. It is notable that the TWINKLE promoter contains, in close proximity of the −45 NRF-2 site, a sequence recognized by Sp1, a factor that is known to physically and functionally interact with NRF-2 on several promoters (8, 31).
An interesting observation comes from the analysis of functional NRF-2 binding sequences in mitochondrial protein genes. Their alignment produces a more stringent consensus with respect to the general consensus for GABP/NRF-2 recognition sites that is shared by the other Ets-related factors (26). It follows that a model based on a more stringent consensus might be used when searching for new NRF-2 binding sites in the promoters of genes involved in mitochondrial biogenesis. Moreover, it is possible to speculate that mitochondrial protein promoters have evolved a more strict consensus to preferentially recruit NRF-2 rather than other Ets factors; such a selection might be less crucial in the case of non-mitochondrial genes.
Our findings concerning POLRMT and mTERF, together with those of Scarpulla and co-workers (17, 18) regarding TFAM and TFB2M, draw a picture in which control by NRF-2 appears to be a common feature of transcription-activating proteins. This indicates that they are all subjected to regulatory pathways triggered by events requiring an enhanced mitochondrial biogenesis and mediated by the PGC-1 family co-activators (6). Recently, Scarpulla (7) reported an up-regulation of NRF-2 as well as POLRMT, TFAM, and TFB1/2 M expression in systems characterized by active mitochondrial biogenesis, such as serum-stimulated fibroblasts, C2C12 cells overexpressing PGC-1α, and differentiated L1 adipocytes. The only mitochondrial transcription factor that seems to escape NRF-2 control is MTERF3. It is interesting to note that, although in silico analysis predicted an NRF-1 site in the MTERF3 promoter, our ChIP data3 tend to exclude in vivo binding of the factor; moreover, MTERF3 promoter was not included in the list of sequences contacted by NRF-1 deriving from ChIP-on-chip analysis (29). Therefore, it appears that MTERF3 also eludes NRF-1 control. This is somehow not surprising, because MTERF3 is a transcription repressor and therefore regulation of its expression could require pathways that are different from those controlling proteins activating transcription and are probably responsive to different environmental or metabolic signals.
Very recently, a new member of the MTERF protein family has been characterized; this factor, named MTERF2, is a mitochondrial protein that positively modulates transcription by acting at the heavy strand promoter region (32). By means of bioinformatic search, we detected two tandemly arranged sites in its proximal promoter region, suggesting that also MTERF2 is subjected to NRF-2 control, this being consistent with its effect on mitochondrial biogenesis.
Our data concerning TWINKLE, mtSSB, Pol-γA, and Pol-γB genes represent the first findings on the control by NRF-2 on proteins belonging to the mtDNA replication machinery. They indicate that TWINKLE helicase, mtSSB, and the accessory subunit of Pol-γ are activated by NRF-2, whereas the catalytic subunit of the enzyme is not. An increase in the level of Pol-γB mRNA was also described in systems characterized by enhanced mitochondrial biogenesis, where NRF-2 and the transcription proteins are up-regulated (7). The in silico analysis predicted a low scoring NRF-1 site in the Pol-γA promoter; however, our ChIP data3 tend to exclude in vivo binding of the factor, as for MTERF3. Therefore, also Pol-γA appears to elude the control by both NRF-1 and -2 transcription factors. A similar situation was described in Drosophila, where Pol-γB, but not Pol-γA, is controlled by DREF (DNA replication-related element factor) (33), a transcription factor activating the expression of several mitochondrial proteins, such as mtSSB and transcription initiation and termination factors, as well as proteins involved in cell cycle progression (24, 34). It appears, therefore, that a pattern of expression control different from that of the other replication proteins could be a peculiar feature of the catalytic moiety of the mitochondrial DNA polymerase. Moreover, the observation that NRF-2, which, similarly to DREF, is implicated in cell cycle control (9), regulates TWINKLE, mtSSB, and Pol-γB suggests that also in mammals those proteins, but not Pol-γA, contribute to link nuclear and mitochondrial DNA replication.
In addition, our findings are consistent with what was observed in muscle responding to continuous motor nerve stimulation, a potent activator of mitochondrial biogenesis (35). In this system, Pol-γA expression does not increase, in contrast with mtSSB, which is up-regulated. It is not possible, however, to exclude the possibility that during development and/or in different tissues, specific regulatory factors could differentially regulate Pol-γA gene expression. In early embryogenesis of Xenopus laevis, a correspondence between the level of Pol-γA transcript and that of mtDNA was observed (36). Interestingly, as mentioned above, the human Pol-γA promoter contains the recognition element for Maf proteins, which are transcriptional activators involved in the control of development and differentiation (28). Finally, the differential regulation of Pol-γ subunit genes can also be evaluated in the light of recent findings (37) suggesting that Pol-γB, but not Pol-γA, is involved in mitochondrial nucleoid maintenance.
In conclusion, the current study extends the collection of mitochondrial genes controlled by NRF-2 and provides novel information on the regulation of mitochondrial biogenesis in humans. In particular, it appears that proteins playing antithetical roles in mtDNA transcription are subjected to distinct regulatory pathways, that is, all of the activators are governed by NRFs, whereas the MTERF3 repressor seems to elude such a control. Moreover, the observed differential regulation of Pol-γA and Pol-γB genes might be functional in the fine tuning of mitochondrial DNA polymerase activity during development and differentiation as well as in the additional role proposed for the accessory subunit. All of these findings arouse interest in the study of the pathways responsible for the regulation of MTERF3 and Pol-γA expression.
The picture emerging from our study strengthens the role of NRF-2 in mitochondrial biogenesis as a bigenomic coordinator because it activates directly the expression of nucleus-encoded oxidative phosphorylation subunits and indirectly that of mitochondria-encoded subunits by promoting the expression of components of mitochondrial transcription and replication machineries.
Supplementary Material
Acknowledgments
We are very grateful to R. C. Scarpulla (Department of Cell and Molecular Biology, Northwestern Medical School, Chicago, IL) for the kind gift of anti-NRF-2α and -β sera and of vectors containing NRF-2α and -β cDNAs. We also thank T. De Filippis (Department of Biochemistry and Molecular Biology, University of Bari) for valuable technical assistance.
This work was supported by grants from Università di Bari, Progetto di Ricerca di Ateneo, and Telethon-Italy (Grant GGP06233).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
F. Bruni, unpublished results.
- mtDNA
- mitochondrial DNA
- GABP
- GA-binding protein
- EMSA
- electrophoretic mobility shift assay
- ChIP
- chromatin immunoprecipitation
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- nt
- nucleotide(s).
REFERENCES
- 1.Falkenberg M., Larsson N. G., Gustafsson C. M. (2007) Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 2.Martin M., Cho J., Cesare A. J., Griffith J. D., Attardi G. (2005) Cell 123, 1227–1240 [DOI] [PubMed] [Google Scholar]
- 3.Park C. B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C. M., Larsson N. G. (2007) Cell 130, 273–285 [DOI] [PubMed] [Google Scholar]
- 4.Roberti M., Polosa P. L., Bruni F., Manzari C., Deceglie S., Gadaleta M. N., Cantatore P. (2009) Biochim. Biophys. Acta 1787, 303–311 [DOI] [PubMed] [Google Scholar]
- 5.Graziewicz M. A., Longley M. J., Copeland W. C. (2006) Chem. Rev. 106, 383–405 [DOI] [PubMed] [Google Scholar]
- 6.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 7.Scarpulla R. C. (2008) Ann. N. Y. Acad. Sci. 1147, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosmarin A. G., Resendes K. K., Yang Z., McMillan J. N., Fleming S. L. (2004) Blood Cells Mol. Dis. 32, 143–154 [DOI] [PubMed] [Google Scholar]
- 9.Yang Z. F., Mott S., Rosmarin A. G. (2007) Nat. Cell Biol. 9, 339–346 [DOI] [PubMed] [Google Scholar]
- 10.Gugneja S., Virbasius J. V., Scarpulla R. C. (1995) Mol. Cell. Biol. 15, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ongwijitwat S., Liang H. L., Graboyes E. M., Wong-Riley M. T. (2006) Gene 374, 39–49 [DOI] [PubMed] [Google Scholar]
- 12.Gong Q., Brown L. J., MacDonald M. J. (2000) J. Biol. Chem. 275, 38012–38021 [DOI] [PubMed] [Google Scholar]
- 13.Blesa J. R., Prieto-Ruiz J. A., Hernández J. M., Hernández-Yago J. (2007) Gene 391, 198–208 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y., Kako K., Arai H., Ohishi T., Inada Y., Takehara A., Fukamizu A., Munekata E. (2002) Biochim. Biophys. Acta 1574, 359–364 [DOI] [PubMed] [Google Scholar]
- 15.Gaston K., Fried M. (1995) Gene 157, 257–259 [DOI] [PubMed] [Google Scholar]
- 16.Kropotov A., Usmanova N., Serikov V., Zhivotovsky B., Tomilin N. (2007) FEBS J. 274, 5804–5814 [DOI] [PubMed] [Google Scholar]
- 17.Virbasius J. V., Scarpulla R. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleyzer N., Vercauteren K., Scarpulla R. C. (2005) Mol. Cell. Biol. 25, 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotney J., McKay S. E., Shadel G. S. (2009) Hum. Mol. Genet. 18, 2670–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marinescu V. D., Kohane I. S., Riva A. (2005) BMC Bioinformatics 6, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 22.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Moreno M. A., Bruni F., Adán C., Sierra R. H., Polosa P. L., Cantatore P., Garesse R., Roberti M. (2009) Biochem. J. 418, 453–462 [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valouev A., Johnson D. S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R. M., Sidow A. (2008) Nat. Methods 5, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegde M. L., Hazra T. K., Mitra S. (2008) Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holt I. J. (2009) Trends Biochem. Sci. 34, 358–365 [DOI] [PubMed] [Google Scholar]
- 29.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., Kluger Y., Dynlacht B. D. (2004) Mol. Cell. 16, 399–411 [DOI] [PubMed] [Google Scholar]
- 30.Kataoka K. (2007) J. Biochem. 141, 775–781 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K., Hayashi N., Shimokawa T., Umehara N., Kaminogawa S., Ra C. (2008) J. Biol. Chem. 283, 15134–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenz T., Luca C., Torraco A., Moraes C. T. (2009) Cell Metab. 9, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Lefai E., Fernandez-Moreno M. A., Alahari A., Kaguni L. S., Garesse R. (2000) J. Biol. Chem. 275, 33123–33133 [DOI] [PubMed] [Google Scholar]
- 34.Matsukage A., Hirose F., Yoo M. A., Yamaguchi M. (2008) Biochim. Biophys. Acta 1779, 81–89 [DOI] [PubMed] [Google Scholar]
- 35.Schultz R. A., Swoap S. J., McDaniel L. D., Zhang B., Koon E. C., Garry D. J., Li K., Williams R. S. (1998) J. Biol. Chem. 273, 3447–3451 [DOI] [PubMed] [Google Scholar]
- 36.Ye F., Carrodeguas J. A., Bogenhagen D. F. (1996) Nucleic Acids Res. 24, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Re M., Sembongi H., He J., Reyes A., Yasukawa T., Martinsson P., Bailey L. J., Goffart S., Boyd-Kirkup J. D., Wong T. S., Fersht A. R., Spelbrink J. N., Holt I. J. (2009) Nucleic Acids Res. 37, 5701–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.