Abstract
Transcription of the yeast mitochondrial genome is carried out by an RNA polymerase (Rpo41p) that is related to single subunit bacteriophage RNA polymerases but requires an additional factor (Mtf1p) for initiation. In this work we show that Mtf1p is involved in multiple roles during initiation including discrimination of upstream base pairs in the promoter, initial melting of three to four base pairs around the site of transcript initiation, and suppression of nonspecific initiation. It, thus, appears that Mtf1p is functionally analogous to initiation factors of multisubunit RNA polymerases, such as σ. Photocross-linking experiments reveal close proximity between Mtf1p and the promoter DNA and show that the C-terminal domain makes contacts with the template strand in the vicinity of the start site. Interestingly, Mtf1p is related to a class of RNA methyltransferases, suggesting an early evolutionary link between RNA synthesis and processing.
Keywords: RNA/Polymerase, Transcription, Transcription/Initiation Factors, Mtf1p, Mitochondrial RNA Polymerase, Photocross-linking
Introduction
Energy production in most eukaryotic cells depends largely upon the function of mitochondria, and a number of human diseases have been attributed to disruption of the activity of these organelles. Whereas most proteins that are essential for mitochondrial processes are encoded by nuclear genes and imported into the organelle, a subset of critical proteins is encoded by the mitochondrial genome (1). Expression of the nuclear and mitochondrial genes must, therefore, be coordinately regulated to ensure proper mitochondrial function. However, little is known about how this regulation occurs.
Mitochondrial genomes are transcribed by RNA polymerases (RNAPs)3 that are related to the single subunit RNAPs encoded by T7-like phages (2–4). Unlike T7 RNAP, however, mitochondrial RNAPs require additional factors for transcription initiation. In the yeast Saccharomyces cerevisiae the catalytic subunit of mitochondrial RNAP (Rpo41p) requires a 40-kDa protein, Mtf1p, for efficient initiation. Earlier work suggested that Mtf1p might be a functional analog of the bacterial initiation factor σ (5–7). For example, as is the case for σ, Mtf1p is required for promoter binding (8) and the formation of an open complex (9) but is lost from the complex shortly after initiation (10). However, subsequent results revealed that Mtf1p is structurally unrelated to σ factors (11) and that Rpo41p can initiate transcription on a pre-melted promoter in the absence of Mtf1p (12). The latter observation led to a revised view that promoter specificity determinants are intrinsic to Rpo41p and that the role of Mtf1p is limited to melting of the promoter and/or to stabilization of an open promoter complex (12).
The manner in which the Rpo41p·Mtf1p complex recognizes the promoter and initiates transcription has not yet been determined. Because of conservation among phage and mitochondrial RNAPs (see Fig. 1), it might be anticipated that a similar mechanism of promoter recognition and melting would be employed during the formation of an open promoter complex by mitochondrial RNAP. Indeed, it has recently been shown that, like T7 RNAP, yeast mitochondrial RNAP melts a region of the promoter from −4 to +2 in the open complex (9). In T7 RNAP, promoter recognition and formation of the open complex involve three elements that interact with the upstream region of the promoter (13). One of these elements (the specificity loop, which establishes major groove base-specific interactions in the region from −7 to −11), is readily identifiable in Rpo41p based on sequence homology with the flanking regions in T7 RNAP and appears to function in a similar manner (14). However, the organization of promoters is different in the T7 and mitochondrial systems, and the distance from the start site to the groups of residues that are required for promoter recognition is not the same (Fig. 1). These considerations indicate that a different geometry of interactions is involved in recognition and initiation in the T7 system versus the mitochondrial system and suggest the possibility that alternative or additional mechanisms may be involved.
FIGURE 1.
Comparison of Rpo41p and T7 RNAP. Regions of homology between the phage and mitochondrial RNAPs are represented as boxes labeled A–L (4); substantial insertions in the sequences are represented by loops. The consensus sequences of the phage T7 and S. cerevisiae (S.c.) mitochondrial promoters are aligned relative to their start sites at +1 (bent arrows). Positions that determine specificity are underlined; single underline indicates positions at which substitutions reduce promoter strength by at least a factor of 3; the double underline indicates positions at which substitutions lead to at least a 10-fold reduction (18, 19, 29, 30). Interactions of the three elements in T7 RNAP that are primarily responsible for promoter recognition and melting are shown by arrows (solid arrow, specificity loop; dashed arrow, intercalating loop; dotted arrow, AT-rich recognition loop) (13). Asterisks below the Rpo41p scheme indicate the location of residues in Rpo41p that have been implicated in interactions with Mtf1p (31, 32).
The manner in which Mtf1p participates in promoter binding and initiation is not clear. In particular, it was not known whether Mtf1p acts through direct interactions with the promoter DNA or by modulating Rpo41p activity in an indirect manner. In this work we used DNA-protein cross-linking methods to identify residues in the promoter that contact Rpo41p and Mtf1p and mapped a region in Mtf1p that approaches the promoter DNA in the vicinity of the transcription start site. These results present the first demonstration of close contacts of Mtf1p with the promoter DNA and reveal a more direct role in transcription initiation than had previously been appreciated. In addition, we found that Mtf1p is involved in discriminating upstream elements of the promoter and may affect selection of the correct start site. Taken together, the results indicate striking functional similarities between Mtf1p and bacterial transcription initiation factors, such as σ, and present an intriguing evolutionary overlap in the initiation strategies used by single- and multisubunit RNAPs.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
N-terminal histidine-tagged Rpo41p and Mtf1p were expressed and purified as described elsewhere (15).
Mutagenesis
Mtf1p mutants were generated using the QuikChange® site-directed mutagenesis kit (Stratagene). The sequences of oligonucleotides used as mutagenesis primers (IDT) and all other oligonucleotides are listed in supplemental Table 1. Expression and purification of Mtf1p mutants was performed as previously described (15). All Mtf1p mutants used in mapping of photocross-linking sites were fully active in transcription initiation assays.
Transcription Assays
A synthetic DNA template containing the yeast mitochondrial consensus promoter was prepared by annealing together oligonucleotides MS75 (template (T) strand) and MS63 (nontemplate (NT) strand). Transcription reactions were performed in 10 μl of transcription buffer (20 mm Tris·HCl, pH 7.9, 50 mm KCl, 10 mm MgCl2, 0.1% Tween 20, 5 mm Tris(2-carboxyethyl)phosphine). The reaction mixtures contained Rpo41p, Mtf1p, and DNA template at 50 nm each. The reactions were initiated by simultaneous addition of substrate NTPs as indicated (300 μm each) together with 0.3 μCi of [α-32P]ATP, incubated for 10 min at 30 °C, and stopped by the addition of 10 μl of stop-buffer (0.05 m EDTA, 90% formamide, 0.02% bromphenol blue, and 0.02% xylene cyanol). The products of the reactions were resolved by electrophoresis in 20% (19:1, acrylamide:bisacrylamide) gels in the presence of 6 m urea, visualized by a Typhoon 9200 PhosphorImager (GE Healthcare), and quantified using ImageQuant 5.2 software (GE Healthcare).
Photocross-linking
Templates that incorporated 4-thio-dTMP at specific positions were prepared by extension of unmodified DNA primers using the Klenow fragment of DNA polymerase I as described in Ma et al. (16) and Temiakov et al. (17). Details are given in the supplemental data.
The templates were incubated with Rpo41p and Mtf1p (50 nm each) in 20 μl of transcription buffer (20 mm Tris·HCl, pH 7.9, 50 mm KCl, 10 mm MgCl2, 0.1% Tween 20, 5 mm Tris(2-carboxyethyl)phosphine) for 5 min at 30 °C, and the solutions were irradiated with a 6-watt model 9815 UV lamp equipped with a 312-nm filter (Cole-Parmer) from a distance of ∼30 mm for 10 min at 30 °C. The products of the reactions were resolved by 4–12% Bis-Tris PAGE using MES running buffer (Invitrogen) and visualized by autoradiography.
Mapping of Photocross-linking Site
The template used in cross-linking mapping experiments, which included a radioactive label adjacent to the photoreactive probe, were prepared by a procedure similar to that described in Ma et al. (16) (see the supplemental data for details). Cross-linking reactions were carried out as described above, except that the concentrations of photoreactive DNA template, Rpo41p, and Mtf1p were 1 μm each, and the reaction volume was 50 μl. After UV irradiation, 20 units of DNase I (Roche Applied Science) were added, and the mixture was incubated for 1 h at 37 °C. The Mtf1p-DNA photoadduct was resolved by 10% Tris-glycine PAGE and extracted from the gel by 0.25 ml of extraction buffer (10 mm Tris·HCl, pH 7.4, 200 mm NaCl, 6 m urea, 0.1% SDS, 5 mm β-mercaptoethanol) overnight at 4 °C. Cleavage and analysis of the cross-linked products was carried out as described in the supplemental data.
RESULTS
Rpo41p Can Initiate Transcription from a Minimal 3–4-nt Bubble
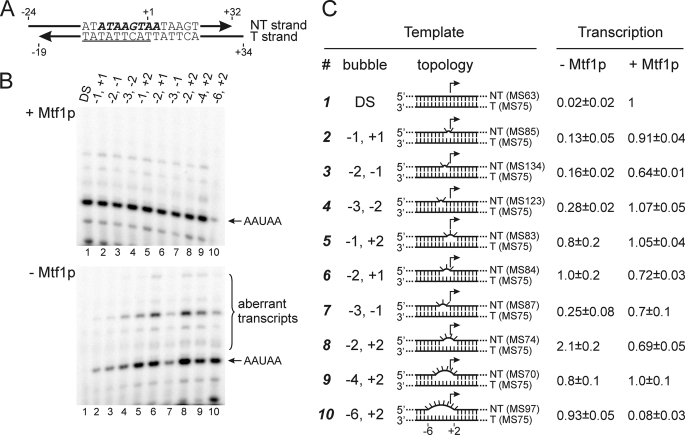
Earlier studies showed that Rpo41p can initiate transcription accurately in vitro without Mtf1p if the promoter is “premelted”; i.e. if the non-template (NT) strand of the promoter is substituted with a sequence that is not complementary to the template (T) strand in the region from position −4 to +2 (a “bubble” template) (12). However, it was not known whether the entire region from −4 to +2 must be melted for Rpo41p to initiate transcription on its own or whether a more limited melted region might suffice. To explore this, we prepared a series of artificially melted templates in which the bubble varied in size from two to eight base pairs and its boundaries ranged from −6 to +2 (Fig. 2).
FIGURE 2.
Initiation on partially melted promoters by Rpo41p in the presence and absence of Mtf1p. A, a T strand having the consensus promoter sequence (underlined) was annealed with NT strands that were complementary (DS) or mismatched within the interval from −6 to +2 (bold italics) as noted in panel C (numbers indicate the first and the last mismatched bases relative to the start site at +1). B, transcription was carried out in the presence of GTP and α-[32P]ATP in the presence (upper panel) or absence (lower panel) of Mtf1p, and the products were resolved by electrophoresis; the positions of the expected 5-nt initiation product (AAUAA) and aberrant products are indicated. C, the number of the template, the boundaries of the bubble, and a schematic diagram of the topology of the DNA templates are provided. Mismatched regions are represented by flipped-out bases. The efficiency of transcription relative to control (DS template in the presence of Mtf1p) is indicated in the right columns. Average intensities from two (templates 3 and 4) or three (all other templates) are presented along with S.D. The identities of the T and NT strand oligomers used to assemble the templates are given in parentheses (see supplemental Table 1).
In the presence of Mtf1p all templates allowed efficient initiation, with the exception of a template in which the bubble extended from −6 to +2, in which case the presence of Mtf1p resulted in a decrease in synthesis of the 5-nt initiation product (Fig. 2B, lane 10). As will be discussed below, this apparent inhibitory effect is due to the choice of a different start site induced by Mtf1p on this template, which does not support transcription with the subset of NTPs used in this experiment.
In the absence of Mtf1p, introduction of as few as two mismatches in the NT strand led to some increase in transcription levels relative to that observed with the full-duplex promoter (Fig. 2B, lower image). However, efficient initiation that was comparable to the control lane (DS template in the presence of Mtf1p; lane 1) was observed only when the size of the bubble was at least 3 nt and the boundaries extended from either −2 to +1 or −1 to +2. The most efficient initiation occurred on a template in which the bubble extended from −2 to +2 (lane 8), in which case transcription at a greater level than the control was observed. This result indicates that the interval from −2 to +2 is likely to be the region that Mtf1p helps to melt and/or stabilize in the open form during transcription initiation. This is consistent with photocross-linking data (below), which reveal contacts between Mtf1p and promoter DNA in the region around the start site.
Interestingly, the bubble templates that allowed efficient initiation in the absence of Mtf1p also directed the formation of substantial amounts of aberrant transcripts (Fig. 2B). These could originate either from misincorporation or from initiation at alternative start sites (or both). In the presence of Mtf1p, the formation of the aberrant transcripts relative to the correct message (AAUAA) was greatly reduced, indicating a role for Mtf1p in the fidelity of initiation.
Potential Role for Mtf1p in Start-site Selection
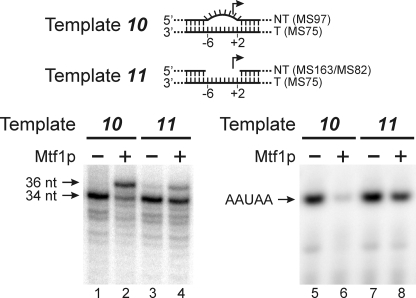
We noted in Fig. 2 that the presence of Mtf1p led to an inhibition of initiation from template 10, which includes a bubble that extends from −6 to +2. To examine this in more detail, we carried out reactions under different conditions that allowed detection of full-length run-off transcripts (Fig. 3). Although Rpo41p correctly initiated on the bubble template (template 10, lane 1), the addition of Mtf1p led to a predominant false start at position −2 on this template (lane 2), resulting in a longer run-off product (36 versus 34 nt). To a lesser extent, this effect was also seen with a gap template (template 11; lanes 3 and 4) in which the NT strand was deleted in the region from −6 to +2. This effect led to an apparent inhibition of initiation by Mtf1p on these templates (lanes 6 and 8) because initiation at −2 would require the presence of GTP, which is missing in the reactions used to detect initiation products. This result explains the apparent inhibitory effect of Mtf1p in the initiation assays shown in Fig. 2B, lane 10.
FIGURE 3.
Mtf1p affects the start site of initiation. Templates were assembled by annealing together the oligonucleotides indicated (parentheses). In the bubble template (10) the NT strand was mismatched from −6 to +2, and in the “gap” template (11) the NT strand was deleted from position −6 to +2. The templates were incubated with Rpo41p and nucleotide triphosphates for 10 min at 30 °C in the presence or absence of Mtf1p as indicated, and the products were resolved by gel electrophoresis. The positions of the expected run-off product (34 nt) and the product of a false start at position −2 (36 nt) synthesized in the presence of [α-32P]ATP, -CTP, -GTP, and -UTP are indicated in the left panel. Short initiation products synthesized in the presence of only [α-32P]ATP and UTP are shown in the right panel. The position of the correct product, initiated at +1 (AAUAA), is indicated.
Mtf1p Plays a Role in Recognition of the Upstream Region of the Promoter
Previous work has shown that certain base pairs in the consensus sequence are critical for promoter utilization on a double-stranded template (18, 19). As summarized in Fig. 1, these include the base pairs at positions −2, −4, −6, and −7. To explore whether these base pairs remain important for initiation after the open complex is formed, we used templates in which NT strand was non-complementary from position −4 to +2, and the sequence in the region from −5 to −8 of these promoters was either consensus or scrambled (Fig. 4A). As expected from previous reports, substitutions in the upstream region resulted in an ∼20-fold decrease in transcription (Fig. 4B, lane 8 versus lane 2). Surprisingly, however, the same substitutions allowed synthesis of the correct initiation product with ∼50% efficiency on the bubble template (template 13, lane 5). From this we conclude that once the promoter has been melted, Rpo41p does not require the correct base pairs at positions −6 and −7 for efficient initiation. Strikingly, initiation from the scrambled bubble template (template 13) was drastically reduced in the presence of Mtf1p (lane 6), indicating that Mtf1p helps to discriminate against promoters that are altered in the upstream region. This could occur through direct interactions of Mtf1p with the promoter or indirectly by enabling Rpo41p to reject incorrect base pairs in that region. As Mtf1p is required for both promoter binding and melting, these observations raise the possibility that Mtf1p may contribute to identification of base pairs at −6 and/or −7 even before promoter melting has begun.
FIGURE 4.
Potential role for Mtf1p in discriminating base pairs in the upstream region of the promoter. A, templates for transcription were either double-stranded (1 and 14) or non-complementary in the region from −2 to +4 (12 and 13) and carried either consensus (1 and 12) or modified (13 and 14) promoter sequences in the region from −5 to −8. Non-complementary nucleotides in the NT strand are italicized; the consensus sequence in the T strand is underlined. B, products synthesized during a 10-min incubation with [α-32P]ATP and UTP and Rpo41p and Mtf1p (as indicated) were resolved by gel electrophoresis; the position of the expected 5-nt initiation product (AAUAA) is indicated. The accumulation of products generated in the absence (white) or presence (gray) of Mtf1p was quantified by PhosphorImager analysis and is expressed relative to the level observed in the presence of Mtf1p on template 1 (lane 2); average intensities from five separate experiments are shown along with S.D.
Mtf1p Makes Contacts with the Promoter DNA
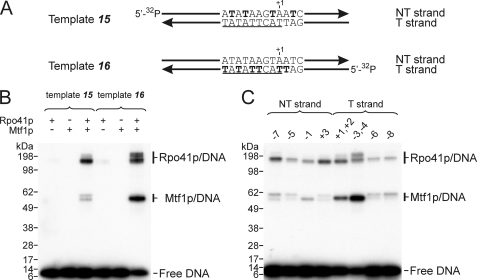
In principal, Mtf1p may support melting of the promoter and other steps of initiation by direct interactions with the DNA or indirectly if it enables Rpo41p to carry out these functions. Previously, it was not known whether Mtf1p establishes any contacts with the DNA in the open promoter complexes or en route to its formation. To reveal potential sites of interaction between Rpo41p and Mtf1p with the promoter, we used DNA-protein photocross-linking methods in which a photoreactive probe, 4-thio-dTMP, was incorporated into the DNA (Fig. 5). Initially, all dTMP residues in the promoter were substituted with 4-thio-dTMP, either in the NT strand at positions +3, −1, −5, and −7 (template 17) or in the T strand at positions+2, +1, −3, −4, −6, and −8 (template 18). These photoreactive promoter templates were incubated with Rpo41p and Mtf1p, and the complexes were subjected to UV-irradiation to induce the formation of covalent linkages. The products were resolved by SDS-PAGE, and the species that were radioactively labeled via the attached DNA were visualized by autoradiography (Fig. 5B). Both Rpo41p and Mtf1p formed cross-linked products with promoter DNA. Whereas intense cross-linking to Rpo41p was observed with both strands of the promoter, Mtf1p reacted primarily with photoprobes in the T strand and to a lesser extent in the NT strand. Importantly, efficient cross-linking required the presence of both proteins, attesting to the specificity of the reaction.
FIGURE 5.
Photocross-linking reveals contacts of both Rpo41p and Mtf1p with promoter DNA. A, double-stranded templates having a 5′ 32P-labeled T strand (template 15) or NT strand (template 16) and 4-thio-dTMP at the positions indicated in bold were prepared as described under “Experimental Procedures.” The promoter consensus sequence is underlined. B, templates were incubated with Rpo41p and Mtf1p (separately or together) for 5 min and subjected to UV irradiation for 10 min, and the products were resolved by gel electrophoresis in 4–12% Bis-Tris SDS gel. The slight variability in mobility of the photoproducts is likely due to the presence of DNA attached to different regions of the proteins. C, the templates were similar to those shown in panel A, but the photoactive probes were incorporated specifically at the positions indicated above each lane. Rpo41p and Mtf1p were both present in the cross-linking reactions.
To examine this in more detail, we utilized a set of templates in which the photoprobe was incorporated at specific positions in the promoter (Fig. 5C). The most efficient cross-linking to Mtf1p was observed when the photoprobe was positioned at −3 and −4 in the T strand. Rpo41p exhibited a broader spectrum of contacts, and efficient cross-linking was observed at positions +1, +2, −3, and −4 in the T strand and at positions −7 and +3 in the NT strand.
Contacts of Mtf1p with the Template Strand Map to the C-terminal Domain
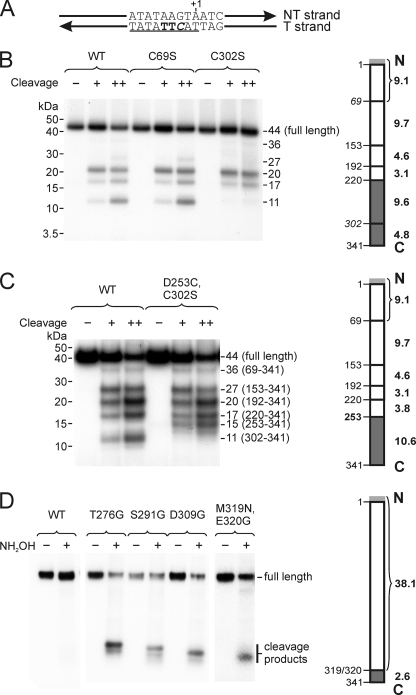
The structure of Mtf1p has been reported (11), and it was, therefore, of interest to determine which regions of the protein are involved in promoter contacts. In this study we focused on mapping the site in Mtf1p that cross-links to positions −3/−4 of the T strand, as photoprobes at these positions provided the highest efficiency of cross-linking. We, therefore, constructed a template that carried 4-thio-dTMP at positions −3 and −4 in the T strand and a radioactive nucleotide at the adjacent position (Fig. 6A). Promoter complexes formed on this template were UV-irradiated and treated with DNase to reduce the influence of DNA on protein mobility during electrophoresis, and the product corresponding to the T strand promoter DNA fragment cross-linked to Mtf1p was resolved by gel electrophoresis and isolated from the gel.
FIGURE 6.
The C terminus of Mtf1p contacts the template strand near positions −3 and −4. A, the DNA template contained 4-thio-dTMP residues at positions −3 and −4 (bold) and [32P]dCMP at position −2 (italics). The consensus sequence is underlined. B, NTCB-induced cleavage of Mtf1p-DNA photocross-linking products. The template shown in panel A was incubated with Rpo41p and Mtf1p (WT or mutant as indicated), and the resulting complexes were irradiated with UV. The cleavage patterns generated in the presence of either 150 (+) or 300 (++) mm NaOH were resolved by gel electrophoresis; the apparent molecular weights of the fragments (including attached DNA) are indicated. The diagram to the right of the autoradiogram indicates the positions of NTCB cleavage sites in WT Mtf1p (left side of the bar) and the molecular masses of the expected protein fragments (without contribution of the attached DNA; in kDa, right side). The Mtf1p protein used in these experiments contained an N-terminal His tag (MAHHHHHH), indicated in light gray. The results of this experiment indicate that the region that is involved in cross-linking extends from residues 220 to 341 (dark gray). C, this panel is the same as panel B except using Mtf1p mutant D253C/C302S, in which the NTCB cleavage site at position 302 was eliminated, and a new site was introduced at position 253. The region that is involved in cross-linking is indicated in dark gray. D, hydroxylamine-induced cleavage of Mtf1p-DNA photocross-linking products. Mtf1p mutants having hydroxylamine cleavage sites (NG) at the positions indicated were constructed and used to prepare complexes as above. The cross-linking products were subjected to hydroxylamine-induced cleavage (NH2OH) and resolved by gel electrophoresis. As the NG sites were moved incrementally toward the C terminus of the protein, the sizes of the labeled products became correspondingly smaller. The results indicate that the site of cross-linking lies in the interval from 320 to 341 (dark gray).
To map the site of the photocross-linking reaction we subjected the photoproduct to treatment with NTCB, which cleaves N-terminal to cysteine residues (Fig. 6B). Cleavage of wild type (WT) Mtf1p revealed a pattern that was difficult to interpret due to the contribution of the remaining DNA to the mobility of the cross-linked product. To circumvent this, we constructed Mtf1p mutants that eliminated certain NTCB cleavage sites and examined the effects of these changes on the cleavage pattern. The C69S mutant exhibited a set of cleavage products that was identical to that of the WT Mtf1p, which excluded the involvement of regions 1–68 and 69–152 as targets of cross-linking. However, in the C302S cleavage pattern, the smallest product was missing, indicating that the cross-linking site lies between residues 220–301 or 302–341 (shaded dark gray in Fig. 6B).
Mtf1p has a two-domain structure in which a large N-terminal domain (residues 1–250) is connected to a smaller four-helix C-terminal domain (residues 257–324) through a linker region (residues 251–256) (11). The region that was identified in these experiments as the site of cross-linking to the T strand bases at −3/−4 includes a fraction of the N-terminal domain of Mtf1p, the flexible interdomain linker, and the entire C-terminal domain. To refine the site of promoter contact, we constructed an additional Mtf1p mutant, D253C/C302S, in which the NTCB cleavage site at 302 was eliminated and a new site was generated at position 253. Promoter complexes assembled using this mutant were UV-irradiated, and the resulting Mtf1p-DNA photoproduct was analyzed by NTCB-induced cleavage as described above (Fig. 6C). Movement of the NTCB cleavage site resulted in a change in the mobility of the minimal labeled cleavage product from 11 kDa in WT Mtf1p to 15 kDa in the mutant, indicating that the site of cross-linking lies between residues 253–341 (shaded dark gray in Fig. 6C).
Finally, we introduced cleavage sites for hydroxylamine, which recognizes NG pairs (which are not present in WT Mtf1p). We constructed a series of mutants that had one NG site each at positions 237/238, 276/277, 290/291, 308/309, and 319/320, taking advantage of native asparagines occurring at most of these positions. Based on the formation of a series of low molecular weight cleavage fragments that progressively decreased in size as the hydroxylamine cleavage site was moved toward the C terminus of the protein (Fig. 6D), we conclude that the region of Mtf1p that is involved in contacts with the template strand at positions −3/−4 lies between amino acids 320 and 341.
DISCUSSION
The question as to whether Mtf1p is a functional analog of bacterial initiation factors, such as σ, has been a subject of discussion for many years (20). Similar to σ, Mtf1p participates in promoter melting (9), suppresses nonspecific initiation by the catalytic core RNAP (Ref. 21 and this work), and dissociates from the complex upon transition into the elongation phase (10). On the other hand, Mtf1p is structurally unrelated to σ factors and instead resembles rRNA methyltransferases such as Bacillus subtilis ErmC′ (11) and Escherichia coli KsgA (22). Furthermore, it was shown that Rpo41p can initiate transcription in the absence of Mtf1p on a template in which the initiation region is melted from −4 to +2 (12). The latter observation led to the current view that Rpo41p carries specificity elements necessary for promoter recognition and initiation and that the role of Mtf1p is limited to promoter melting and/or stabilization of the open complex (12). Nevertheless, the role of Mtf1p in initiation was unclear, and in particular, it was not known whether Mtf1p contacts the promoter directly or exerts its effect on Rpo41p in an indirect manner.
Here, we demonstrate that Mtf1p makes direct contacts with the promoter DNA. In addition, we find that Mtf1p supports a number of σ-like functions that include melting of an initial 3–4-bp interval around the start site, discrimination of base pairs in the upstream region of the promoter, and suppression of nonspecific initiation. Thus, even though Mtf1p exhibits no sequence or structural homology to bacterial σ factors, it appears to be involved in many of the same functions. The results suggest intriguing overlaps in the initiation strategies used by single- and multisubunit RNAPs that have developed during evolution.
To characterize the interactions of Mtf1p and Rpo41p with the DNA, we carried out cross-linking experiments using DNA templates in which the photoreactive probe 4-thio-dTMP had been introduced into the consensus region of the promoter and detected contacts between both of these proteins and the promoter. The 4-thio-dTMP probe was chosen because it is the least disturbing photoreactive nucleotide analog and because cross-linking with this probe reveals intimate contacts with the base (within chemical bond length) (23). In related work, Paratkar and Patel (34) used a phenylazide photoreactive probe that is attached to the DNA backbone and has a wider radius of action (∼10 Å). Despite differences in the probes, the results of the two studies are in good agreement, and both demonstrate close approaches of Mtf1p to the promoter (34).
Previous results demonstrated that Rpo41p can initiate transcription on a template in which the initiation region is melted from −4 to +2. In this work we found that the minimal region that must be melted to allow efficient initiation by Rpo41p in the absence of Mtf1p is limited to a 3–4-bp interval that surrounds the start site, suggesting that a primary role for Mtf1p is to nucleate or stabilize the initial melting of this region. In addition, we found that the presence of Mtf1p suppresses nonspecific initiation and may induce false starts on some templates, suggesting a role for Mtf1p in start site selection. Consistent with a direct role for Mtf1p in promoter melting and initiation, we observed cross-links between Mtf1p and bases in the T and NT strands in the initiation region.
Results from transcription initiation on various bubble templates indicate that Mtf1p may also play a role in recognition of upstream promoter elements (Fig. 4). We observed that whereas some base pairs in the upstream region of the promoter are critical for initiation before melting (on a double-stranded promoter), they are not essential afterward, as Rpo41p tolerates substitutions in this region on a bubble template. Strikingly, the presence of Mtf1p resulted in discrimination against these substitutions on the bubble template. This effect does not seem to be related to the mechanism of inhibition of initiation from template 10 by Mtf1 (as shown in Fig. 3), as template 13 directed the formation of correct run-off products (34 nt) in the presence of Mtf1 (not shown). As Mtf1p is associated with Rpo41p during both promoter binding and melting, we cannot distinguish the role of the initiation factor during these stages using the methods described here. The observation that the presence of Mtf1p results in discrimination against substitutions in the upstream region even when promoter melting has already occurred (on a bubble template) indicates that Mtf1p may be involved in recognition of upstream base pairs during the initial stages of promoter recognition before melting. These findings are consistent with the cross-linking results presented here and obtained independently by Paratkar and Patel (34), which reveal contacts between Mtf1p and upstream promoter elements.
In the absence of structural information about the Rpo41p·Mtf1p complex, it is not known how the two proteins bind to one another or to the promoter. As an initial approach toward this question, we mapped the site of cross-linking between Mtf1p and the bases at −3/−4 in the T strand (which lie at the upstream edge of the melted region in the open complex (9)). We found that the contacts involve a C-terminal region of Mtf1p from residue 320 to 341 (Fig. 6). Previous data concerning the importance of the C-terminal portion of Mtf1p are somewhat controversial. Shadel and Clayton (20) had indicated that a deletion in the C-terminal region that extended from residues 292 to 341 (Δ292–341) behaved normally in vivo at permissive temperatures but exhibited a petite phenotype at 37 °C. However, Cliften et al. (6) reported severe defects in binding to Rpo41p even when substantially smaller regions (Δ297–341 and Δ316–341) were deleted. More biochemical and genetic studies are needed to elucidate the function of the C-terminal domain.
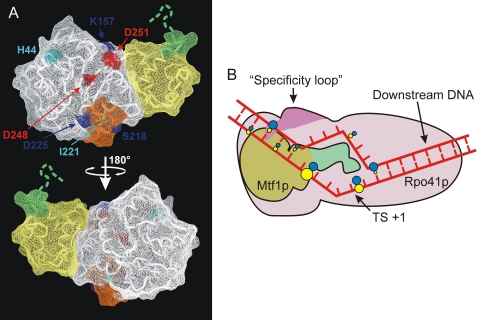
Genetic studies concerning interactions between Rpo41p and Mtf1p identified a number of mutants of Mtf1p that are deficient in either binding to Rpo41p or stimulating its ability to initiate (6, 20). Most of the identified amino acids were later found to be buried inside the protein (11), diminishing the significance of these observations for prediction of a binding surface. However, mapping the positions of only the surface-exposed substitutions that affect binding suggests a face of Mtf1p that is likely to be involved in interaction with Rpo41p (Fig. 7A). A schematic summarizing the interactions of Mtf1p and Rpo41p with the promoter is presented in Fig. 7B.
FIGURE 7.
Interactions of Rpo41p and Mtf1p with the promoter. A, potential interface of Mtf1p involved in interaction with Rpo41p. The positions of surface-exposed amino acids that were demonstrated to be critical (dark blue) or important (light blue) for binding to Rpo41p (6) are superimposed on the structure of Mtf1p (PDB code 1I4W). Two residues in this potential interface, Asp-248 and -251, that were shown to be important for transcription activity in vitro are shown in red (20). A two-helix loop element that has been previously implicated as the Rpo41p binding determinant (11) is shown in orange. The N-terminal and C-terminal domains are highlighted in white and yellow, respectively, and the C-terminal fragment involved in contacts with the −3/−4 region of the promoter is shown in green. B, a diagram is shown of interactions of Rpo41p and Mtf1p with the promoter. Rpo41p is indicated in pink; the interaction of the specificity loop (purple) with the −7 bp is indicated (14). Mtf1p is indicated in green and brown; green denotes the C-terminal region that is involved in cross-linking to the bases at −3 and −4 in the T strand. The figure is not to scale. The promoter DNA is drawn to depict the melted region in the open complex from −4 to +2 (9) and a bent conformation consistent with previous biochemical data (33). The colored dots indicate positions where 4-thio-dTMP residues cross-link with the corresponding protein; blue indicates contacts with Rpo41p, and yellow indicates contacts with Mtf1p. The sizes of the dots indicate the relative intensities of the cross-links.
We attempted to dock Rpo41p and Mtf1p with the promoter using the known structure of the T7 RNAP initiation complex (IC), taking into account the genetic data on Rpo41p/Mtf1p interactions noted above as well as the proximity of the C-terminal region of Mtf1p with the −3/−4 region of the promoter (this work). However, these modeling attempts resulted in major steric clashes between Mtf1p and elements in the N-terminal domain of T7 RNAP, including the intercalating loop and possibly the AT-rich recognition loop (data not shown). We propose that if these elements are present in Rpo41p, their orientation and function in promoter recognition may be different than in the T7 RNAP IC. Such a situation has recently been observed in promoter recognition by the T7-like N4 virion RNAP (24). There is also the possibility that additional mechanisms are involved in promoter recognition and/or that these elements are missing from Rpo41p and that their functions are carried out by Mtf1p. Further cross-linking and mapping studies will be required to resolve this issue.
The results presented here indicate significant functional similarities between Mtf1p and bacterial transcription initiation factors and present an intriguing overlap in the initiation strategies used by single- and multisubunit RNAPs. It is thought that mitochondria evolved from an ancient bacterial endosymbiont and that over time many of the genes involved in mitochondrial function were transferred to the cell nucleus, leaving a small genome that encodes proteins with specialized functions. Mtf1p is structurally related to a class of bacterial rRNA methyltransferases such as B. subtilis ErmC′ (11) and E. coli KsgA (22). Other mitochondrial RNAPs also use proteins that are related to methyltransferases as initiation factors. For example, initiation by the human mitochondrial RNAP requires either mtTFB1 or mtTFB2, both of which are related to KsgA (25, 26). Similar to the results reported here, it has recently been found that the human mtTFB2 also cross-links to the mitochondrial promoter in an in vitro system (35), yet surprisingly, it is the N terminus of mtTFB2 that cross-links to the promoter versus the C terminus of Mtf1p (this work). Thus, even though a methyltransferase-like protein has been recruited as an initiation factor in both systems, the nature of the protein and its interactions with the promoter are different. The use of this class of proteins as initiation factors may reflect a need to couple RNA synthesis and processing. Proteins with dual functions are not unknown in mitochondrial transcription systems and may reflect pressure to reduce the size of the genome during evolution. For example, it has been shown that the human mitochondrial RNAP is associated with mitochondrial ribosomal protein MRPL12 and that this protein modulates the activity of the RNAP in vitro, suggesting a coupling of transcription and translation (27). And it has recently been found that the yeast mitochondrial DEAD box helicase Mss116, which is involved in RNA processing and splicing, is associated with the yeast transcription complex and modulates the activity of Rpo41p in vitro (15).
T7 and mitochondrial RNAPs are members of a structurally related family of nucleotide polymerases that includes DNA polymerase I and human immunodeficiency virus reverse transcriptase. Members of the family share a conserved catalytic core domain and mechanism of nucleotide incorporation yet have distinct properties with regard to template specificity. Although T7 RNAP is considered to be the prototype of the RNAPs that are members of this family, it is clear that there are multiple strategies for transcript initiation within this group, some of which involve the use of auxiliary factors, as observed in the unrelated multisubunit RNAPs. The capture and use of other proteins to perform auxiliary functions is also observed in other members of the family; for example, T7 DNA polymerase (which is related to polymerase I) uses the host protein thioredoxin to enhance processivity (28). The evolution of this family of enzymes, therefore, represents an intriguing example of adaptation and differentiation at the molecular level.
Supplementary Material
Acknowledgments
We are grateful to Alexander Kukarin for help in cloning experiments, Dmitry Litonin for help in optimization of protein purification, and Dmitriy Markov, Smita Patel, Dmitry Vassylyev, Steve Emanuel, and Vadim Molodtsov for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM38147 (to W. T. M.). This work was also supported by grants from the Research Foundation of University of Medicine and Dentistry of New Jersey (to M. A., D. T., and W. T. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- RNAP
- RNA polymerase
- WT
- wild type
- IC
- initiation complex
- T
- template
- NT
- nontemplate
- Bis-Tris
- 2-[bis(2-hydroxyethyl)imino]-2-(hydroxymethyl)propane-1,3-diol
- MES
- 2-(4-morpholino)ethanesulfonic acid
- nt
- nucleotide(s)
- NTCB
- 2-nitro-5-thiocyanobenzoic acid.
REFERENCES
- 1.Perocchi F., Jensen L. J., Gagneur J., Ahting U., von Mering C., Bork P., Prokisch H., Steinmetz L. M. (2006) PLoS Genet. 2, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters B. S., Stohl L. L., Clayton D. A. (1987) Cell 51, 89–99 [DOI] [PubMed] [Google Scholar]
- 3.Cermakian N., Ikeda T. M., Cedergren R., Gray M. W. (1996) Nucleic Acids Res. 24, 648–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cermakian N., Ikeda T. M., Miramontes P., Lang B. F., Gray M. W., Cedergren R. (1997) J. Mol. Evol. 45, 671–681 [DOI] [PubMed] [Google Scholar]
- 5.Jang S. H., Jaehning J. A. (1991) J. Biol. Chem. 266, 22671–22677 [PubMed] [Google Scholar]
- 6.Cliften P. F., Park J. Y., Davis B. P., Jang S. H., Jaehning J. A. (1997) Genes Dev. 11, 2897–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlok M. A., Jang S. H., Jaehning J. A. (2002) J. Biol. Chem. 277, 28143–28149 [DOI] [PubMed] [Google Scholar]
- 8.Schinkel A. H., Groot Koerkamp M. J., Tabak H. F. (1988) EMBO J. 7, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang G. Q., Paratkar S., Patel S. S. (2009) J. Biol. Chem. 284, 5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangus D. A., Jang S. H., Jaehning J. A. (1994) J. Biol. Chem. 269, 26568–26574 [PubMed] [Google Scholar]
- 11.Schubot F. D., Chen C. J., Rose J. P., Dailey T. A., Dailey H. A., Wang B. C. (2001) Protein Sci. 10, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsunaga M., Jaehning J. A. (2004) J. Biol. Chem. 279, 44239–44242 [DOI] [PubMed] [Google Scholar]
- 13.Cheetham G. M., Jeruzalmi D., Steitz T. A. (1999) Nature 399, 80–83 [DOI] [PubMed] [Google Scholar]
- 14.Nayak D., Guo Q., Sousa R. (2009) J. Biol. Chem. 284, 13641–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markov D. A., Savkina M., Anikin M., Del Campo M., Ecker K., Lambowitz A. M., De Gnore J. P., McAllister W. T. (2009) Yeast 26, 423–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma K., Temiakov D., Jiang M., Anikin M., McAllister W. T. (2002) J. Biol. Chem. 277, 43206–43215 [DOI] [PubMed] [Google Scholar]
- 17.Temiakov D., Anikin M., Ma K., Jiang M., McAllister W. T. (2003) Methods Enzymol. 371, 133–143 [DOI] [PubMed] [Google Scholar]
- 18.Biswas T. K., Getz G. S. (1986) J. Biol. Chem. 261, 3927–3930 [PubMed] [Google Scholar]
- 19.Biswas T. K., Ticho B., Getz G. S. (1987) J. Biol. Chem. 262, 13690–13696 [PubMed] [Google Scholar]
- 20.Shadel G. S., Clayton D. A. (1995) Mol. Cell. Biol. 15, 2101–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schinkel A. H., Groot Koerkamp M. J., Van der Horst G. T., Touw E. P., Osinga K. A., Van der Bliek A. M., Veeneman G. H., Van Boom J. H., Tabak H. F. (1986) EMBO J. 5, 1041–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Farrell H. C., Scarsdale J. N., Rife J. P. (2004) J. Mol. Biol. 339, 337–353 [DOI] [PubMed] [Google Scholar]
- 23.Favre A., Saintomé C., Fourrey J. L., Clivio P., Laugâa P. (1998) J. Photochem. Photobiol. B. 42, 109–124 [DOI] [PubMed] [Google Scholar]
- 24.Gleghorn M. L., Davydova E. K., Rothman-Denes L. B., Murakami K. S. (2008) Mol. Cell 32, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidel-Rogol B. L., McCulloch V., Shadel G. S. (2003) Nat. Genet. 33, 23–24 [DOI] [PubMed] [Google Scholar]
- 26.Cotney J., Shadel G. S. (2006) J. Mol. Evol. 63, 707–717 [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Cotney J., Shadel G. S. (2007) J. Biol. Chem. 282, 12610–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabor S., Huber H. E., Richardson C. C. (1987) J. Biol. Chem. 262, 16212–16223 [PubMed] [Google Scholar]
- 29.Diaz G. A., Raskin C. A., McAllister W. T. (1993) J. Mol. Biol. 229, 805–811 [DOI] [PubMed] [Google Scholar]
- 30.Imburgio D., Rong M., Ma K., McAllister W. T. (2000) Biochemistry 39, 10419–10430 [DOI] [PubMed] [Google Scholar]
- 31.Cliften P. F., Jang S. H., Jaehning J. A. (2000) Mol. Cell. Biol. 20, 7013–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsunaga M., Jaehning J. A. (2004) J. Biol. Chem. 279, 2012–2019 [DOI] [PubMed] [Google Scholar]
- 33.Schinkel A. H., Groot Koerkamp M. J., Teunissen A. W., Tabak H. F. (1988) Nucleic Acids Res. 16, 9147–9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paratkar S., Patel S. S. (2010) J. Biol. Chem. 285, 3949–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. (2009) Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.