Abstract
In neuronal and hormonal release, regulated exocytosis requires an essential set of proteins: the soluble N-ethylmaleimide sensitive-factor attachment receptor proteins (SNAREs) syntaxin 1, SNAP-25, VAMP, and their regulator, Munc18. Recently, it was found that Munc18-1 can interact with syntaxin 1 through distinct mechanisms: an inhibitory mode enveloping syntaxin (mode 1), sequestering it from SNARE protein interactions, and direct binding to an evolutionarily conserved N-terminal peptide of syntaxin (mode 2/3). The latter interaction has been proposed to control “priming” of the fusion reaction, defined using electrophysiology, but it is unknown how this interaction is regulated, and any dynamic effect at the molecular or vesicular level in cells remains undiscovered. We now show that a phosphorylation site in syntaxin 1 (Ser14) regulates the N-terminal interaction with Munc18-1. Probing syntaxin 1 association with Munc18-1, in real-time and in living cells, we found that modification of Ser14 modulated the dynamics of this interaction, specifically at the plasma membrane. Destabilization of this dynamic interaction enhanced vesicle immobilization at the plasma membrane with a resulting inhibition of exocytosis.
Introduction
Eukaryotic membrane fusion is a highly conserved process mediated by a defined set of proteins. The trafficking of cargo, in membrane bound vesicles, between intracellular compartments and to the plasma membrane is essential for cell function and survival. The fusion of vesicles is driven by the pairing of SNARE3 proteins that reside on the target SNARE and vesicular SNARE membranes (1, 2). This family of proteins has been the subject of intense study over the last two decades, and it is now clear that they constitute the minimal machinery for membrane fusion (3, 4). In neurons and neuroendocrine cells, the regulated fusion of secretory vesicles with the plasma membrane is mediated by the vesicular SNARE synaptobrevin (also known as VAMP2) and the target SNAREs syntaxin 1 and SNAP-25 (synaptosome-associated protein, 25 kDa). These three proteins interact to form a four-helical complex; one helix from each of syntaxin and synaptobrevin and two helices from SNAP-25 (5). The formation of this thermodynamically stable SNARE complex is thought to liberate the energy required to alter the local lipid environment and merge the two opposing lipid bilayers (6–8).
The process of membrane fusion is under strict temporal and spatial regulation. In neuronal transmission, exocytosis occurs at specific membrane sites and is tightly coupled to occur in response to depolarization of the nerve terminal. This regulation is, in part, mediated by accessory proteins that modulate the function of the SNARE proteins. One such family of SNARE regulators is the Sec1p/Munc18 (SM) protein family. These structurally conserved proteins, found throughout eukaryotic membrane trafficking, bind to their cognate syntaxin homologs to modulate SNARE function. However, the mechanisms underlying this regulation have been the subject of debate. Comparison of crystal structures of the mammalian SM protein, Munc18-1, and the yeast SM protein, Sly1p, both bound to their cognate syntaxins originally indicated two completely different modes of interaction (9, 10). This has since been reconciled by the observation that Munc18-1 can interact with syntaxin through two distinct binding mechanisms: an interaction with syntaxin in a so called “closed” conformation and binding to the N terminus of syntaxin (11–13). In light of these findings, it was proposed to call the closed binding conformation mode 1 and the N-terminal interaction mode 2 or mode 3 (depending on whether syntaxin was alone or in the SNARE complex, respectively) (14). As these latter two modes relate to the same binding site on syntaxin, for clarity, we will term them mode 2/3 as the principal aim of this nomenclature is the distinction between the different sites of the syntaxin/Munc18 interaction. These binding modes were shown to be distinct in vivo, both spatially and functionally, occurring in different cellular membrane compartments, with the mode 2/3 interaction directly influencing membrane fusion (12, 13). Crystal structures of the N-terminal peptide of syntaxin 4 bound to its cognate SM protein munc18c (Munc18-3) provided further weight to mode 2/3 binding being an important feature of mammalian SM proteins (15). This led to the reanalysis of the crystal structure diffraction data of Munc18-1 bound to syntaxin and the observation of a mode 2/3 interaction in concert with mode 1 binding (the electron density present but unattributed in the original data) (16). In combination with findings from other SM protein/syntaxin interactions (17–19), it is now becoming clear that a dual mode of interaction may be ubiquitous for these two evolutionarily conserved protein families.
In addition to defining how syntaxin and Munc18 interact, a great deal of interest has focused on the regulation of their interaction. From the initial biochemical observations of an inhibitory effect of the mode 1 interaction on the transition of syntaxin, bound by Munc18, into the ternary SNARE complex, several regulators have been proposed. Munc18 was found to be a substrate for protein kinase C which, through phosphorylation of Ser306 and Ser307, could prevent the mode 1 association of syntaxin and Munc18 (20). However, Munc18 could not be phosphorylated by protein kinase C in the assembled syntaxin-Munc18 complex and therefore may only serve a role in regulating the pool of unbound Munc18 (21). In contrast, polyunsaturated fatty acids, such as arachidonic acid, have been shown to act directly on the mode 1 interaction of syntaxin 1 with Munc18 in vitro, permitting the transition of syntaxin 1 from this inhibited state into the ternary SNARE complex (22, 23). In addition, other proteinaceous factors have been proposed to influence mode 1 binding, although their mechanisms remain enigmatic. It is likely that one or more factors act in concert to regulate the mode 1 interaction between syntaxin 1 and Munc18-1.
The characterization of the mode 2/3 interaction of syntaxin 1 with Munc18-1 provides a new target for possible regulation (11–13). However, to date, no regulators of the mode 2/3 interaction have been identified. Early work into the function of synaptotagmin, the proposed neuronal calcium sensor, observed that syntaxin 1 is phosphorylated in vivo by casein kinase II (CKII) (24). Using in vitro phosphorylation of truncation mutants and the use of phospho-specific antibodies in vivo, it was shown that Ser14 is a target for CKII (25, 26). While the phosphorylation of this site was observed to increase through development in parallel with synapse development and maturation, the authors were unable to provide a mechanism regulated by this modification. The constructs used lacked N-terminal amino acids, thus only mode 1 binding would have been detected. Importantly, this site is well placed to directly influence the mode 2/3 interaction of syntaxin 1 with Munc18-1.
We investigated the role of phosphorylation of Ser14 in syntaxin 1 on its interaction with Munc18. Through a series of phosphomimetic and phospho-null mutations, we show that Ser14 is a key regulator of interaction between syntaxin and Munc18 at the plasma membrane, previously shown to be the cellular location where mode 2/3 interaction is predominant (12). Using advanced microscopy, we were able to probe, at high frequency, changes in dynamic protein interactions at the plasma membrane to investigate the influence of Ser14 on the syntaxin/Munc18-1 interaction. Downstream of modulating this protein/protein interaction, modification of Ser14 had a direct influence on the mobility of secretory vesicles and their concomitant fusion competence.
EXPERIMENTAL PROCEDURES
Vectors and Cell Culture
Plasmid encoding glutathione S-transferase (GST) fusion protein with syntaxin 1a (amino acids 1–261, cytoplasmic domain), was described previously (27). Mammalian expression plasmids encoding mCer-syntaxin 1a (amino acids 1–288), EYFP-Munc18-1 and untagged Munc18-1 (both amino acids 1–594) were described previously (12). Dark EYFP-Munc18-1 (dkEYFP-Munc18-1) was generated by replacement of EYFP with dkEYFP (28). mCherry-NPY was generated by replacement of EGFP in EGFP-NPY (29) with mCherry. Generation of casein kinase II phosphomimetic and phospho-null mutant forms of syntaxin 1a was performed by site-directed mutagenesis using a QuikChange II XL kit (Stratagene) and verified by sequencing. Neuroblastoma 2A cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 10 mm l-glutamine, 50 units of penicillin, and 50 μg/ml streptomycin and maintained at 37 °C in 5% (v/v) CO2 and 95% (v/v) air. Pheochromocytoma (PC-12) cells were grown in RPMI medium supplemented with 10% horse serum, 5% fetal bovine serum, 10 mm GlutaMAX (Invitrogen), and 50 μg/ml gentamicin and maintained at 37 °C in 7.5% (v/v) CO2 and 92.5% (v/v) air. Transfections were performed using ExGen 500 (Fermentas) or Lipofectamine 2000 (Invitrogen).
Protein Biochemistry
Recombinant GST fusion proteins were expressed and purified as described previously (22). For in vitro phosphorylation reactions, 2 μg of GST-syntaxin 1a was incubated with 500 units of casein kinase II (New England Biolabs) in a total volume of 20 μl of 20 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, and 200 μm ATP for 1 h at 30 °C. The reaction was stopped by the addition of SDS-containing sample buffer followed by SDS-PAGE. Phosphorylated proteins were visualized by in-gel staining using the Diamond ProQ phosphoprotein stain (Invitrogen) according to the manufacturer's instructions and recorded using a Typhoon scanner (GE Healthcare). Peppermint stick markers (Invitrogen) were used as an internal control for staining efficacy. Total protein was observed by Coomassie staining. For in vivo protein phosphorylation, transfected cells were harvested, and equalized samples were analyzed by SDS-PAGE and Western immunoblotting using anti-syntaxin (HPC-1, Sigma) and anti-syntaxin (phospho-Ser14; Abcam). Protein structural alignment and presentation were performed using PyMOL (30).
Confocal Laser Scanning Microscopy and Live Cell Maintenance
Cells with the lowest detectable expression levels (∼1–10-fold overexpression) were selected for analysis, and levels were similar between experiments. All time-correlated single photon counting (TCSPC) experiments were performed using a Zeiss LSM 510 Axiovert confocal laser scanning microscope, equipped with a pulsed excitation source (MIRA 900 titanium:sapphire femto-second pulsed laser with a coupled VERDI 10-watt pump laser (Coherent). Data were acquired using a 1024 × 1024 pixel image size, using a Zeiss Plan NeoFLUAR 1.4 NA ×63 oil immersion lens or a Zeiss C-Apochromat 1.2 NA ×63 water-corrected immersion objective lens. For vesicle tracking and fusion experiments, cells were imaged under total internal reflection fluorescence microscopy (TIRFM) illumination using an Olympus CellR widefield TIRFM microscope equipped with a 561 nm diode laser. Data were acquired using a Hamamatsu ImageEM EMCCD using an Olympus PLAN APO 1.45 NA ×60 oil immersion objective. All imaging was performed using living cells, maintained at 37 °C in 5% (v/v) CO2 and 95% (v/v) air in a POC chamber (LaCon).
TCSPC-FLIM Acquisition and Analysis
TCSPC measurements were made under 800–820 nm two-photon excitation, which efficiently excited cerulean, without any detectable direct excitation or emission from EYFP, using a fast photomultiplier tube (H7422; Hamamatsu Photonics) coupled directly to the rear port of the Axiovert microscope. Full frame TCSPC recordings were acquired for between 30 and 60 s, with mean photon counts were between 105–106 counts per second. Images were recorded at 256 × 256 pixels from a 1024 × 1024 image scan with 256 time bins over a 12 ns period (31). Off-line FLIM data analysis used pixel-based fitting software (SPCImage, Becker & Hickl). The optimization of the fit parameters was performed by using the Levenberg-Marquardt algorithm, minimizing the weighted chi-square quantity. As controls for nonspecific Förster resonance energy transfer (FRET), or FRET between GFPs that may form dimers spontaneously when overexpressed in cells, we determined the fluorescence lifetimes of cerulean Syx1–288 alone, cerulean alone, or cerulean Syx1–288 cotransfected with EYFP (data not shown). No FRET was detected in any of these experiments. For point-scanning TCSPC, the “first-in, first-out” recording feature of the TCSPC card was used. First-in, first-out data describing the fluorescence decay curves were fit as before using SPCImage (with no binning), using a single component decay, and the resulting numerical data was exported. The matrix of fluorescence lifetime values was imported to ImageJ, and a look up table was applied. The fluorescence lifetime data were statistically segmented (binarized), and single molecule statistics were applied using WinEDR software (Electrophysiology Data Recorder, John Dempster, University of Strathclyde).
Vesicle Tracking
TIRFM data of PC-12 cells expressing NPY-mCherry, the corresponding syntaxin and Munc18-1, maintained at 37 °C in 5% (v/v) CO2 and 95% (v/v) air were acquired with a pixel size of 166 nm at 20 Hz. Single vesicles were identified and tracked using Imaris 5 (Bitplane). All track lengths shorter than 10 frames were discarded from the quantification. Where required, cells were stimulated by the addition of ATP to a final concentration of 300 μm.
RESULTS
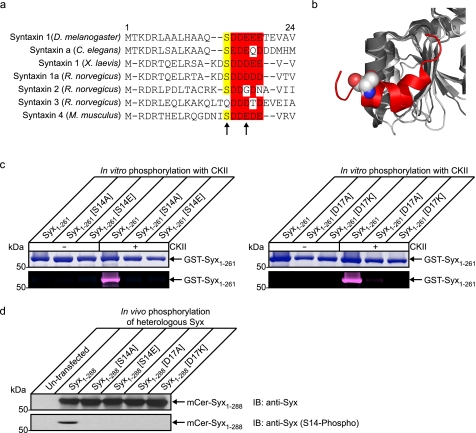
Previous work found that syntaxin 1 is phosphorylated by CKII on Ser14 (24–26). However, we were unable to identify any biological function for this modification. In the four syntaxin 1 orthologs from model organisms, the CKII phosphorylation consensus site is highly conserved (Fig. 1a). In contrast, this phosphorylation site is less conserved in mammalian syntaxin paralogs. Syntaxins 1a and 4 exhibit a high degree of similarity with only conservative changes in the acidic region of the CKII consensus site. In contrast, syntaxins 2 and 3 have only a partially conserved acidic region (essential for CKII phosphorylation) and syntaxin 3 does not possess a suitable serine residue although a threonine residue (Thr19) could support some phosphorylation (32). This loss of the CKII consensus site is correlated by previous in vitro phosphorylation experiments, which showed that syntaxins 1a and 4 were more readily phosphorylated than syntaxins 2 and 3 (26) suggesting a patterning of regulation to provide differential modulation of downstream function. A structural alignment of Munc18-1 and Munc18-3 demonstrates the highly conserved structural features surrounding the syntaxin N-terminal binding site (Fig. 1b).
FIGURE 1.
Serine 14 is phosphorylated by CKII and is a key regulator of interaction with Munc18-1 at the plasma membrane. a, sequence alignment of syntaxin 1 orthologs and paralogs. The casein kinase II consensus site is highly conserved throughout evolution but is less conserved between mammalian paralogs. This may be indicative of a divergence of function between syntaxin isoforms. b, structural alignment of Munc18-1 (dark gray; Protein Data Bank code 3C98) (16) and Munc18-3 (light gray; Protein Data Bank code 2PJX) (15) of the region involved in mode 2/3 interaction with syntaxin. This region exhibits a very high degree of structural conservation indicative of an important functional role. Shown in red is the N-terminal peptide of syntaxin 4 bound to Munc18-3 with the serine residue highlighted in a space fill representation. c, to confirm that Ser14 is a casein kinase II phosphorylation site in syntaxin 1, multiple syntaxins were expressed and purified from bacteria (GST-Syx1–261). These were subjected to in vitro phosphorylation with detection using a fluorescent in-gel phosphostain. Total protein was observed by Coomassie staining. The cytoplasmic domain of syntaxin was readily phosphorylated by CKII in vitro, and this was blocked by mutation of Ser14 or the acidic consensus site at Asp17. d, to test these mutations in a cellular context, phosphorylation of heterologously expressed proteins was detected by immunoblotting (IB) using a Ser14 phospho-specific antibody. This demonstrated that heterologous syntaxin was phosphorylated on this site in vivo and was perturbed by modification of Ser14 or the CKII consensus site.
This phosphorylation site is well placed to potentially influence the interaction of the N terminus of syntaxin 1 with Munc18-1. We thus investigated the role of Ser14 in syntaxin 1, analyzing directly its effect on the interaction with Munc18-1 and the functional consequences of this on vesicle dynamics and fusion in living neuroendocrine cells. We verified that Ser14 is a target for CKII phosphorylation and that this action could be influenced, through targeted mutagenesis, using a series of mutations generated in the CKII consensus site (Fig. 1c). The Asp17 substitution used in these experiments allowed preservation of the serine but disrupted the CKII acidic consensus sequence at the most critical position (32). Purified, bacterially expressed proteins harboring Ser14 or Asp17 substitutions were subjected to in vitro phosphorylation using pure CKII, with detection of phosphate incorporation using a fluorescent in-gel stain. This confirmed that syntaxin 1 Ser14 is the sole CKII phosphorylation site in this protein and that substitution at Asp17 completely abolishes this activity. We next confirmed that these phospho-mimetic and phospho-null mutations can perturb the phosphorylation pattern of syntaxin in vivo. Heterologous, wild-type syntaxin was efficiently phosphorylated on serine 14; this was abrogated when either Ser14 was modified, or the CKII consensus site was disrupted (Fig. 1d).
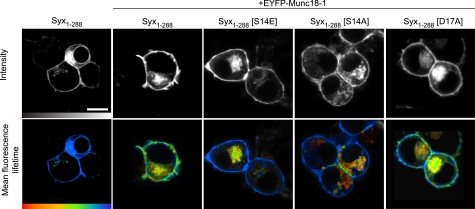
To investigate the impact of these phosphomimetic and phospho-null mutations on mode 2/3 interactions, we used mCerulean (mCer)-labeled syntaxin and EYFP-fused Munc18-1, combined with TCSPC. These functional proteins constitute an excellent FRET pair (33), which we have previously used in living cells to examine the spatial organization of the different modes of syntaxin/Munc18 interaction (12). mCer-syntaxin and EYFP-Munc18-1 were expressed in neuroendocrine cells, and image data from live cells were acquired using 2-photon excitation (Fig. 2). These data showed a principally plasma membrane localization for syntaxin/Munc18-1 with some intracellular compartments also labeled, as observed previously (34). FLIM showed a statistically significant quenching of the mean fluorescence lifetime from 2,288 ± 40 ps (mean ± S.E., n = 18; Ref. 12) in the absence of a FRET acceptor, to 1492 ± 191 ps (mean ± S.E., n = 3; t test, p < 0.001) in the presence of EYFP-Munc18-1. This compares well with our previous studies (12) and indicates interaction of syntaxin and Munc18 on the plasma membrane. The difference in donor fluorescence lifetime at the plasma membrane and on intracellular compartments is a result of the different modes of interaction which predominate at each location (12).
FIGURE 2.
Phosphorylation of syntaxin regulates its interaction with Munc18-1, specifically at the plasma membrane. TCSPC-FLIM reports FRET between mCerulean Syx1–288 (the donor) and EYFP-Munc18-1 (the acceptor) in live cells. Intensity images (upper row; gray scale) show wild-type and mutant mCerulean Syx1–288 localized to the plasma membrane of cells coexpressing EYFP-Munc18-1 (not shown in these images). TCSPC-FLIM data (bottom row) illustrate the excited state fluorescence lifetime of the donor. The mCerulean Syx1–288 exhibited a single fluorescence decay with a time constant of 2,288 ± 40 ps (mean ± S.E., n = 18) in the absence of an acceptor (EYFP and unfused Munc18-1). When coexpressed with EYFP-Munc18-1, this lifetime became significantly quenched (1,872 ± 333 ps, weighted mean of two components ± S.E., n = 5), indicative of FRET and protein interaction. The phosphomimetic form of syntaxin (Syx1–288(S14E)) returned the mean fluorescence lifetime to values not significantly different from that observed in the absence of an acceptor (2,035 ± 252 ps, weighted mean of two components ± S.E., n = 5). Notably, this change was restricted to the plasma membrane. The phospho-null syntaxin (Syx1–288(S14A)) resulted in a fluorescence lifetime not dissimilar to that observed for Syx1–288(S14E). The phospho-null syntaxin (Syx1–288(D17A)) had no detectable effect on the interaction. Scale bar, 5 μm. Color scale bar, 1,250–2,250 ps.
The same approach was used for syntaxin harboring the phosphomimetic mutation S14E. Although syntaxin localization was unaltered compared with wild-type, importantly, decreased interaction was detected between the phosphomimetic syntaxin and Munc18-1 on the plasma membrane. In comparison, there was no detectable change to the interaction in the intracellular compartments where mode 1 binding predominates (12). The D17A phospho-null mutant also exhibited efficient trafficking to the plasma membrane, with representative mean fluorescence lifetimes in agreement with wild-type mCer-syntaxin at the plasma membrane (1,968 ± 172 ps for D17A compared with 1,831 ± 178 ps in the wild-type) and at intracellular membranes (1,627 ± 94 ps for D17A compared with 1,667 ± 189 ps for wild-type). When syntaxin harboring S14A was used in this assay, the reported mean fluorescence lifetimes (plasma membrane, 1,919 ± 297 ps; intracellular membranes, 1,536 ± 86 ps) were comparable to that observed for the S14E mutation (plasma membrane 2,284 ± 214 ps; intracellular membranes 1,640 ± 139 ps). Taken together with our previous findings (12), these data indicate that Ser14 is a key residue in mode 2/3 interactions at the plasma membrane. As S14E and S14A mutations in syntaxin resulted in similar responses, as reported by fluorescence lifetime measurements, we hypothesize that specific interactions between syntaxin and Munc18 (for example part of a hydrogen bonding network), involving this amino acid, are essential for binding. Unfortunately, the position of Ser14 and its interactions are not resolved in the currently available crystal structure of syntaxin 1 and Munc18-1 (16). Nevertheless, phosphorylation of this site by CKII (24–26) would disrupt the specific interactions of Ser14 in a similar manner to that observed for the phosphomimetic mutation. As a result of these findings, we utilized D17A as a phospho-null mutation in subsequent experiments to preserve the functionality of Ser14.
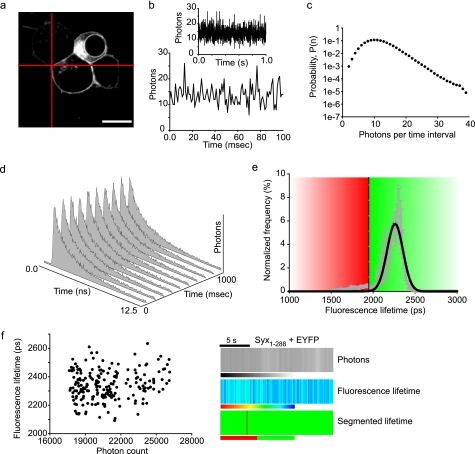
FLIM data are collated over a long acquisition time (typically tens of seconds) and therefore report the average interaction at each cellular location during this period. This is necessary to acquire sufficient photons for a robust mathematical appraisal of the data but is too slow to probe the dynamics of small ensembles of proteins, which we considered essential to gain further understanding of SM protein biology in living cells. Our approach to overcome this limitation was two-fold. First, we utilized dark EYFP as a FRET acceptor, which has greatly reduced emission (28, 35), thus permitting a broader donor filter regime. Second, we streamed data from diffraction-limited regions selectable from anywhere in the three-dimensional volume of the cell. In this way, a single excitation volume is treated as a tiny cuvette (0.1 sub-femtoliter), in a live cell, within which the dynamics of a small number of molecules (fewer than 10; Ref. 34) can be probed in real time. The recording volume was targeted to the plasma membrane of the cell, to where the effect of syntaxin N-terminal interaction with Munc18-1 was restricted, and the photons emitted from the donor mCer-syntaxin were recorded over time (Fig. 3, a and b). The generation of a probability distribution of the number of photons arriving in a time interval (known as a photon counting histogram; Fig. 3c) (36) demonstrated characteristic, non-random signal fluctuations that were not attributable to noise alone. This was performed for mCer-syntaxin in the presence of unfused dkEYFP (noninteracting control) with a complete fluorescence decay curve acquired every 100 ms (Fig. 3d). The measured fluorescence lifetime from each sampling interval and from multiple experiments were combined and fit by a Gaussian distribution (Fig. 3e). Next, we determined the 99.9% confidence interval of this distribution. Using this as a threshold, we were able to identify, in subsequent experiments, those sampling time intervals where interactions occurred. There was no correlation between donor fluorescence lifetime and intensity (Fig. 3f, left panel), demonstrating that protein concentration and fluorescence lifetime are independent. We generated separate two-dimensional kymographs of the number of photons and fluorescence lifetimes, where each vertical stripe corresponds to a single sampling window (Fig. 3f, right panel). The complex information contained in the fluorescence lifetime kymograph is simplified by applying the segmentation (derived in Fig. 3e) to show in a binary format kinetic changes in protein interaction. Importantly, using this spectroscopic approach, it is possible to observe protein interaction dynamics in living cells.
FIGURE 3.
Dynamic syntaxin/Munc18 interactions at the plasma membrane (16). Targeting the laser excitation beam on the plasma membrane (a) allows the fast streaming of TCSPC data from a single point. b, photon fluctuations are shown, with an expanded time scale in the lower graph. c, the photon fluctuations contain a non-random signal as evidenced by the non-Poisson distribution. d, the TCSPC data can be fit by an exponential decay for each time sample, and the time constant of the fluorescence decays can be calculated: in this case, decay curves were acquired every 100 ms. e, to allow segmentation of the lifetimes into intervals where interacting (red) or non-interacting (green) conditions predominate, TCSPC-FLIM data from a cell expressing mCer-syntaxin and unfused dkEYFP (as a control for the non-FRET condition) was plotted on a histogram. The distribution of lifetimes was fit by a single Gaussian and the lower 99.9% confidence interval of the distribution used to segment the data (threshold at 1,950 ps). f, the measured fluorescence lifetime exhibited no correlation with the fluorescence intensity (photon count, left panel), highlighting the independence of these two properties of the fluorophores. Presentation of fluorescence intensity, fluorescence lifetime, and segmented lifetime (using the threshold defined in e) in a human understandable format (right panel). Scales are: intensity, grayscale from 0 to 30,000 photons; fluorescence lifetime, color scale from 1,000 ps (red) to 2,500 ps (blue); segmented lifetime, binary scale (red, interacting; green, not interacting). Each vertical bar represents a single time interval of 100 ms.
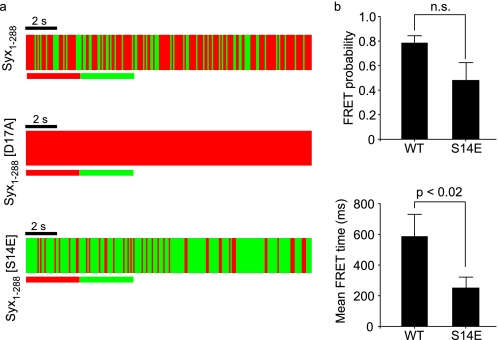
We thus examined the effect of phosphomimetic and phospho-null syntaxin on the dynamics of mode 2/3 interaction with high temporal resolution. This revealed that at the plasma membrane, interaction stability is enhanced in the phospho-null substitution (D17A) and reduced in the phospho-mimetic form of syntaxin (S14E); Fig. 4a). The absence of detectable fluctuations in the interaction state of syntaxin (D17A) precluded further analysis. Most likely, this is a result of the fluctuations in interactions falling below the sensitivity of our system. Importantly, however, statistical quantification of the interaction status and dwell time of the proteins, akin to channel conductance analyses, revealed that both the mean interaction time and the interaction probabilities are significantly reduced for the phosphomimetic mutant, compared with wild-type syntaxin (Fig. 4b).
FIGURE 4.
Modification of serine 14 modulates the dynamic syntaxin/Munc18 interaction. a, fast point TCSPC measurements from regions on the plasma membrane of living cells demonstrate that wild-type Syx1–288 interacts with Munc18-1 on the cell membrane. Similar measurements, using Syx1–288(S14E) significantly altered the dynamics of this interaction, changing the observed lifetime distribution to a “flicker,” where more time was spent in a non-interacting state (green). Syx1–288(D17A) stabilized the interaction with Munc18-1 to a level beyond the sensitivity of the system. The data from independent experiments was combined and quantified. b, these analyses confirmed that Syx1–288(S14E) destabilized the interaction with Munc18-1 at the plasma membrane compared with wild-type syntaxin, whereas Syx1–288(D17A) stabilized the interaction with Munc18-1. Error bars are S.E. (Mann-Whitney U test, n = 5). WT, wild-type; n.s., not significant.
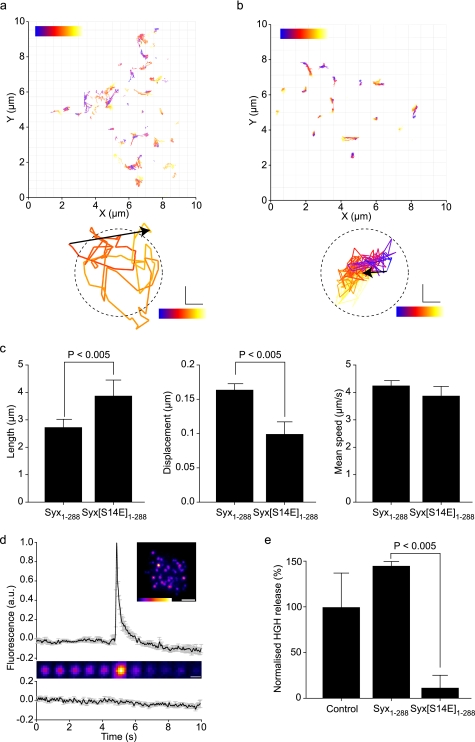
Our data demonstrate a destabilization of the dynamic interaction of the phosphomimetic syntaxin N terminus with Munc18-1 in situ and at the molecular level. As electrophysiological data suggested a role for mode 2/3 binding in priming exocytosis (37), what is the consequence of syntaxin N-terminal interaction with Munc18-1, in situ, at the single vesicle level? We therefore applied our cell biological and bio-physical approaches to quantify the effects of syntaxin Ser14 phosphorylation on both exocytosis and the underlying vesicle dynamics. To achieve this aim, we employed TIRFM. Under the appropriate conditions, this approach is at least as good as amperometry at determining single vesicle fusion events, with the added advantage of delivering quantitative data describing the spatio-temporal behavior of vesicles in the moments immediately preceding the fusion event (38–40). Employing our mutant proteins, alongside fluorescently labeled LDCVs, we were able to track secretory vesicles and measure multiple parameters under physiological temperatures (Fig. 5, a and b and supplemental movies S1 and S2). This revealed that the S14E phosphomimetic mutant acted in a dominant manner, resulting in significantly decreased LDCV displacement, but longer track lengths compared with cells expressing wild-type syntaxin 1 at similar levels (Fig. 5c). We then analyzed single vesicle fusion events employing pH-sensitive EGFP-NPY, as well as using a bulk exocytosis assay (Fig. 5, d and e). In the single vesicle assay, fusion is observed as a transient increase in fluorescence intensity due to a change in the micro-environmental pH upon fusion (supplemental movie S3). Surprisingly, phosphomimetic syntaxin reduced exocytosis to below detectable levels in the single vesicle fusion assay and significantly reduced exocytosis using a bulk exocytosis assay, compared with wild-type syntaxin in identical tests.
FIGURE 5.
Vesicle behavior and fusion are controlled by the stability of the mode 2/3 interaction of syntaxin with Munc18. Fluorescent vesicles were tracked by TIRFM in cells expressing wild-type (a) or phosphomimetic (Syx1–288(S14E)) (b) syntaxin. Individual vesicle trajectories are shown as tracks with color corresponding to time during acquisition (blue, start; red, end). Examples most closely matching the mean track length and displacement for each condition are shown below. The dashed circle corresponds to mean vesicle diameter. Scale bar, 100 nm. c, individual tracks were measured for length, displacement, and mean speed (>250 tracks per cell; n = 4 cells). The phosphomimetic mutation resulted in enhanced track length but with a shorter displacement indicative of a tighter tethering. Mean speed was unaltered. Error bars are S.E. (t test, n = 4). d, cells expressing exogenous syntaxin 1 and Munc18-1 can perform exocytosis. The fusion of single secretory vesicles upon stimulation is shown as a fluorescence intensity trace and montage (upper panel; supplemental movie S3). Non-stimulated cells exhibited no detectable fusion events (lower trace). Error bars are S.E. (n = 15). e, exocytosis is severely depleted by disrupting the mode 2/3 syntaxin interaction with Munc18. The phosphomimetic form of syntaxin 1 (Syx1–288(S14E)) exhibited a pronounced inhibition of exocytosis. Error bars are S.E. (t test, n = 3).
DISCUSSION
The role of SM proteins in regulating membrane trafficking steps throughout the Eukaryotic kingdom has for many years been shrouded in controversy (41). This confusion has resulted from conflicting information regarding Munc18-1 function and its mechanism of interaction with syntaxin. In vitro Munc18-1 potently prevents ternary SNARE complex formation, which would be predicted to inhibit exocytosis (22, 42, 43). In contrast, Munc18-1 appears to have a positive role in exocytosis, with a dramatic reduction in secretion following genetic ablation (44). Binding modes within the SM protein family also show differences with binding to their cognate syntaxin occurring in either the closed form or to the N terminus (9, 10, 15, 16). The simplest way to reconcile these differences is a model whereby Munc18-1 exhibits two independent modes of interaction with syntaxin 1 and has two (or more) important roles in the regulation of SNARE function. Evidence for dual modes of binding has been provided in a series of studies looking at the syntaxin/Munc18 interaction both in vitro and in vivo (11–13). The consensus arising from these papers is that Munc18 interacts with two independent sites on syntaxin; closed form binding (mode 1) and N-terminal binding (mode 2/3). In addition, it now appears likely that Munc18 also has at least two important roles related to SNARE function. Munc18 has a chaperone-like role to inhibit the interaction of syntaxin with other SNARE proteins while it travels through intracellular compartments (34, 45). The resulting decrease in syntaxin levels at the plasma membrane can account for the unexpected phenotype observed following genetic ablation of Munc18 (46). Munc18 has also been reported to play a positive role in membrane fusion, acting to accelerate in vitro reconstituted fusion assays (13). These differential binding modes and multiple roles in controlling SNARE function can unify previous, apparently conflicting, findings.
Previous findings have concentrated on the regulation of the mode 1 mechanism of binding (12, 20, 21). We now present a mechanism for the regulation of the mode 2/3 mechanism of interaction. The N terminus of syntaxin 1 contains a highly conserved casein kinase II consensus site, which was previously observed to be a target for phosphorylation (25, 26). We now show, using a series of phosphomimetic and phospho-null mutants that serine 14 is the sole casein kinase II target in syntaxin and that this site is a critical modulator of mode 2/3 interaction with Munc18-1. Using TCSPC-FLIM, we show that mode 2/3 interaction between syntaxin and Munc18-1, the predominate form at the plasma membrane (12), is disrupted by the phosphomimetic mutation. To overcome the inherently low temporal resolution of TCSPC-FLIM, we utilized a modification of this technique by directly streaming TCSPC-FLIM data from a fixed excitation volume. By removing the slow sampling rate inherent to scanning microscopy, the rate of acquisition could be increased to 10 Hz. This allowed us to look at rapidly changing dynamics in the interaction states of the small number of molecules in the excitation volume. This showed that the phosphomimetic mutation destabilizes the interaction between syntaxin and Munc18, but transient interactions persist.
What are the downstream effects of the mode 2/3 interaction between syntaxin and Munc18-1? In vitro, this interaction has been reported to enhance the efficacy of the fusion apparatus in a reconstituted system (13). Electron microscopy studies of Munc18-1 null models reported a decrease in the number of secretory vesicles residing at the plasma membrane (44). Furthermore, electrophysiology data reported a role of Munc18 in enhancing the initial burst phase of secretion (defined as increased priming) (37). However, at present we do not understand how the concepts of docking, priming, and fusion are underpinned by molecular interactions to give rise to the observed electrophysiological phenomena. We therefore decided to examine directly the impact of mode 2/3 binding on the behavior of secretory vesicles in living cells. The outcome of the destabilized interaction between the N terminus of syntaxin 1 and Munc18-1 at the plasma membrane, shown here, is a highly restricted vesicle motion which could not support membrane fusion. Indeed, it is known that vesicle motion increases on the molecular scale immediately prior to fusion (40). Furthermore the large majority of highly immobilized vesicles docked at the plasma membrane exhibit a low degree of fusion competence (39). Hence, transit of a vesicle from an immobilized state to a more mobile form (on the nano-scale) may be an essential step in the pathway to fusion.
Supplementary Material
Acknowledgments
We thank F. Wouters (University of Göttingen, Germany) for providing the dark EYFP vector and G. Rutter (Imperial College, UK) for providing a vector encoding neuropeptide Y. We also thank the IMPACT imaging facility (University of Edinburgh, Scotland, United Kingdom) for access to equipment and software.
This work was supported by a project grant from the Wellcome Trust (to R. R. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies S1–S3.
- SNARE
- soluble N-ethylmaleimide sensitive-factor attachment receptor protein
- SM
- Sec1p/Munc18
- VAMP
- vesicle-associated membrane protein
- CKII
- casein kinase II
- GST
- glutathione S-transferase
- TCSPC
- time-correlated single photon counting
- EGFP
- enhanced green fluorescent protein
- TIRFM
- total internal reflection fluorescence microscopy
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- Förster resonance energy transfer
- Syx
- syntaxin
- EYFP
- enhanced yellow fluorescent protein
- NPY
- neuropeptide Y
- mCer
- monomeric Cerulean.
REFERENCES
- 1.Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 2.Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E. (1993) Cell 75, 409–418 [DOI] [PubMed] [Google Scholar]
- 3.Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. (1998) Cell 92, 759–772 [DOI] [PubMed] [Google Scholar]
- 4.Rizo J., Südhof T. C. (2002) Nat. Rev. Neurosci. 3, 641–653 [DOI] [PubMed] [Google Scholar]
- 5.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 6.McNew J. A., Weber T., Parlati F., Johnston R. J., Melia T. J., Söllner T. H., Rothman J. E. (2000) J. Cell Biol. 150, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam A. D., Tryoen-Toth P., Tsai B., Vitale N., Stuenkel E. L. (2008) Mol. Biol. Cell 19, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F., Pincet F., Perez E., Eng W. S., Melia T. J., Rothman J. E., Tareste D. (2007) Nat. Struct. Mol. Biol. 14, 890–896 [DOI] [PubMed] [Google Scholar]
- 9.Bracher A., Weissenhorn W. (2002) EMBO J. 21, 6114–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misura K. M., Scheller R. H., Weis W. I. (2000) Nature 404, 355–362 [DOI] [PubMed] [Google Scholar]
- 11.Dulubova I., Khvotchev M., Liu S., Huryeva I., Südhof T. C., Rizo J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickman C., Medine C. N., Bergmann A., Duncan R. R. (2007) J. Biol. Chem. 282, 12097–12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen J., Tareste D. C., Paumet F., Rothman J. E., Melia T. J. (2007) Cell 128, 183–195 [DOI] [PubMed] [Google Scholar]
- 14.Burgoyne R. D., Morgan A. (2007) Curr. Biol. 17, R255–258 [DOI] [PubMed] [Google Scholar]
- 15.Hu S. H., Latham C. F., Gee C. L., James D. E., Martin J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8773–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. (2008) EMBO J. 27, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpp L. N., Ciufo L. F., Shanks S. G., Boyd A., Bryant N. J. (2006) J. Cell Biol. 173, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latham C. F., Lopez J. A., Hu S. H., Gee C. L., Westbury E., Blair D. H., Armishaw C. J., Alewood P. F., Bryant N. J., James D. E., Martin J. L. (2006) Traffic 7, 1408–1419 [DOI] [PubMed] [Google Scholar]
- 19.Togneri J., Cheng Y. S., Munson M., Hughson F. M., Carr C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17730–17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita Y., Sasaki T., Fukui K., Kotani H., Kimura T., Hata Y., Südhof T. C., Scheller R. H., Takai Y. (1996) J. Biol. Chem. 271, 7265–7268 [DOI] [PubMed] [Google Scholar]
- 21.Barclay J. W., Craig T. J., Fisher R. J., Ciufo L. F., Evans G. J., Morgan A., Burgoyne R. D. (2003) J. Biol. Chem. 278, 10538–10545 [DOI] [PubMed] [Google Scholar]
- 22.Rickman C., Davletov B. (2005) Chem Biol 12, 545–553 [DOI] [PubMed] [Google Scholar]
- 23.Connell E., Darios F., Broersen K., Gatsby N., Peak-Chew S. Y., Rickman C., Davletov B. (2007) EMBO Rep. 8, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett M. K., Miller K. G., Scheller R. H. (1993) J. Neurosci. 13, 1701–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foletti D. L., Lin R., Finley M. A., Scheller R. H. (2000) J. Neurosci. 20, 4535–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risinger C., Bennett M. K. (1999) J. Neurochem. 72, 614–624 [DOI] [PubMed] [Google Scholar]
- 27.Hu K., Carroll J., Fedorovich S., Rickman C., Sukhodub A., Davletov B. (2002) Nature 415, 646–650 [DOI] [PubMed] [Google Scholar]
- 28.Ganesan S., Ameer-Beg S. M., Ng T. T., Vojnovic B., Wouters F. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4089–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuboi T., Rutter G. A. (2003) Curr. Biol. 13, 563–567 [DOI] [PubMed] [Google Scholar]
- 30.DeLano W. L. (2002) The Pymol Molecular Graphics System. In [Google Scholar]
- 31.Duncan R. R., Bergmann A., Cousin M. A., Apps D. K., Shipston M. J. (2004) J. Microsc. 215, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marin O., Meggio F., Marchiori F., Borin G., Pinna L. A. (1986) Eur. J. Biochem. 160, 239–244 [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Ernst S. A., Gladycheva S. E., Lee Y. Y., Lentz S. I., Ho C. S., Li Q., Stuenkel E. L. (2004) J. Biol. Chem. 279, 55924–55936 [DOI] [PubMed] [Google Scholar]
- 34.Medine C. N., Rickman C., Chamberlain L. H., Duncan R. R. (2007) J. Cell Sci. 120, 4407–4415 [DOI] [PubMed] [Google Scholar]
- 35.Lee S. J., Escobedo-Lozoya Y., Szatmari E. M., Yasuda R. (2009) Nature 458, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian H., Elson E. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5479–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deák F., Xu Y., Chang W. P., Dulubova I., Khvotchev M., Liu X., Südhof T. C., Rizo J. (2009) J. Cell Biol. 184, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nofal S., Becherer U., Hof D., Matti U., Rettig J. (2007) J. Neurosci. 27, 1386–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becherer U., Pasche M., Nofal S., Hof D., Matti U., Rettig J. (2007) PLoS ONE 2, e505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degtyar V. E., Allersma M. W., Axelrod D., Holz R. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15929–15934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toonen R. F., Verhage M. (2003) Trends Cell Biol. 13, 177–186 [DOI] [PubMed] [Google Scholar]
- 42.Pevsner J., Hsu S. C., Braun J. E., Calakos N., Ting A. E., Bennett M. K., Scheller R. H. (1994) Neuron 13, 353–361 [DOI] [PubMed] [Google Scholar]
- 43.Yang B., Steegmaier M., Gonzalez L. C., Jr., Scheller R. H. (2000) J. Cell Biol. 148, 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voets T., Toonen R. F., Brian E. C., de Wit H., Moser T., Rettig J., Südhof T. C., Neher E., Verhage M. (2001) Neuron 31, 581–591 [DOI] [PubMed] [Google Scholar]
- 45.Arunachalam L., Han L., Tassew N. G., He Y., Wang L., Xie L., Fujita Y., Kwan E., Davletov B., Monnier P. P., Gaisano H. Y., Sugita S. (2008) Mol. Biol. Cell 19, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Wit H., Cornelisse L. N., Toonen R. F., Verhage M. (2006) PLoS ONE 1, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.