Abstract
The thioredoxin-interacting protein TXNIP is a ubiquitously expressed redox protein that promotes apoptosis. Recently, we found that TXNIP deficiency protects against type 1 and 2 diabetes by inhibiting beta cell apoptosis and maintaining pancreatic beta cell mass, indicating that TXNIP plays a key role in beta cell biology. However, very little is known about the intracellular localization and function of TXNIP, and although TXNIP has been thought to be a cytoplasmic protein, our immunohistochemistry studies in beta cells surprisingly revealed a nuclear TXNIP localization, suggesting that TXNIP may shuttle within the cell. Using immunohistochemistry/confocal imaging and cell fractionation/co-immunoprecipitation, we found that, under physiological conditions, TXNIP is localized primarily in the nucleus of pancreatic beta cells, whereas oxidative stress leads to TXNIP shuttling into the mitochondria. In mitochondria, TXNIP binds to and oxidizes Trx2, thereby reducing Trx2 binding to ASK1 and allowing for ASK1 phosphorylation/activation, resulting in induction of the mitochondrial pathway of apoptosis with cytochrome c release and caspase-3 cleavage. TXNIP overexpression and Trx2 (but not cytosolic Trx1) silencing mimic these effects. Thus, we discovered that TXNIP shuttles between subcellular compartments in response to oxidative stress and identified a novel redox-sensitive mitochondrial TXNIP-Trx2-ASK1 signaling cascade.
Keywords: Cell/Apoptosis, Cell/Fractionation, Cell/Trafficking, Diseases/Diabetes, Subcellular Organelles/Mitochondria, Tissue/Organ Systems/Pancreatic Islet, Thioredoxin
Introduction
The thioredoxin-interacting protein TXNIP, also known as VDUP1 (vitamin D3 up-regulated protein-1) (1) or TBP-2 (thioredoxin-binding protein-2), binds to and negatively regulates Trx1 and controls the cellular redox state (2–6). Trx1 is a redox protein with important anti-oxidative and cell signaling functions that undergoes reversible oxidation of its two active-site cysteine residues, the reduction of which is catalyzed by the NADPH-dependent flavoenzyme thioredoxin reductase (2–4, 7). TXNIP expression is ubiquitous and is induced by a variety of stresses, including UV light, γ-rays, heat shock, and H2O2 (3, 8), as well as glucose (9–13). TXNIP overexpression renders cells more susceptible to oxidative stress and promotes apoptosis (11, 14–16).
In pancreatic beta cells, we originally identified TXNIP as the gene that was most dramatically induced by glucose in a human islet microarray study (12) and went on to show that TXNIP is essential for glucotoxicity-induced beta cell apoptosis (17). Interestingly, TXNIP induces beta cell apoptosis through the intrinsic mitochondrial death pathway (17). Recently, we further found that TXNIP deficiency protects against type 1 and 2 diabetes by inhibiting beta cell apoptosis and maintaining pancreatic beta cell mass and function (18), indicating that TXNIP plays a key role in beta cell biology. However, very little is known about the intracellular localization and function of TXNIP, and although TXNIP has been thought to be a cytoplasmic protein (3, 10, 14), our immunohistochemistry studies in pancreatic beta cells surprisingly revealed a nuclear TXNIP localization pattern, suggesting that TXNIP may shuttle within the cell. The aim of this study was therefore to investigate the possibility of TXNIP shuttling between different compartments in the beta cell, to elucidate the factors regulating the subcellular localization of TXNIP, and to determine any implications this may have in terms of TXNIP function.
EXPERIMENTAL PROCEDURES
Tissue Culture
INS-1 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1 mm sodium pyruvate, 2 mm l-glutamine, 10 mm HEPES, and 0.05 mm 2-mercaptoethanol. INS-1 cells with constitutive TXNIP overexpression (INS-TXNIP) and control cells overexpressing LacZ (INS-LacZ) were generated and selected as described previously (11).
Immunohistochemistry
Wild-type C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) were killed, and their pancreases were harvested, fixed in 4% formaldehyde, and processed in an automated Shandon Citadel 100 machine before paraffin embedding and preparation of 10-μm sections. Beta cells were visualized by insulin staining using guinea pig anti-insulin antibody (Zymed Laboratories Inc., South San Francisco, CA) and Cy3-conjugated anti-guinea pig IgG (1:500; Jackson ImmunoResearch Laboratories, West Grove, PA). TXNIP was detected using the mouse anti-TXNIP primary antibody (1:100; JY2, MBL International Co., Woburn, MA) and the fluorescein isothiocyanate-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories). Adequate staining was confirmed by comparing INS-TXNIP and INS-LacZ cells, revealing a dramatic increase in the TXNIP-overexpressing cells (supplemental Fig. S1). VECTASHIELD with 4′,6-diamidino-2-phenylindole mounting solution (Vector Labs, Burlingame, CA) was used for visualization of nuclei.
To visualize the mitochondria, CMXRos (200 nm final concentration; MitoTracker Red, Molecular Probes, Eugene, OR) was added directly to INS-1 cells grown on coverslips in a 6-well plate for 30 min at 37 °C and 5% CO2, followed by the addition of 0.1% (v/v) paraformaldehyde and incubation on ice for 5 min. Cells were then fixed with cold methanol (−20 °C) for 5 min, permeabilized with cold acetone (−20 °C) for 5 min, rinsed with phosphate-buffered saline, and nonspecific sites were blocked with phosphate-buffered saline containing 2% bovine serum albumin, 10% goat serum (Invitrogen), and 0.1% Triton X-100 for 1 h. TXNIP and Trx2 were detected using mouse anti-TXNIP (1:200) and rabbit anti-Trx2 (1:200) antibodies (Santa Cruz Biotechnology) overnight at 4 °C and goat anti-mouse Alexa Fluor 594-conjugated or goat anti-rabbit Alexa Fluor 488-conjugated secondary antibody (Molecular Probes) at 1:1000 or 1:5000 dilution for 1 h at room temperature. Microscopy was performed on a Nikon C1 laser scanning confocal microscope.
Co-immunoprecipitation
Cytosolic, mitochondrial, and nuclear fractions were prepared as described previously (17, 19), boiled in SDS sample buffer, and stored at −80 °C until used.
Nuclear, cytosolic, and mitochondria proteins (250 μg) were incubated with 2 μg of primary antibody (anti-TXNIP/Trx2/ASK1 (apoptosis signal-regulating kinase-1)) in 200 μl (final volume) of immunoprecipitation buffer (20 mm HEPES (pH 7.9), 0.15 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture) for 4 h at 4 °C, followed by incubation for 2 h with 50 μl of washed 50% protein A- or G-Sepharose slurry (Pierce). After three 10-min washes with immunoprecipitation buffer, the pellets were resuspended in electrophoresis SDS sample buffer (Invitrogen), boiled for 5 min at 95 °C, and analyzed by 10–20% SDS-PAGE. Immunoblotting was performed using the following primary antibodies: anti-TXNIP JY2 (1:400); anti-Trx2 (1:600), anti-cytochrome c (1:500), and anti-USF2 (1:500) (Santa Cruz Biotechnology); anti-ASK1 (1:300), anti-phospho-ASK1 Ser967 (1:300), anti-cyclooxygenase IV (1:300), anti-Bad (1:500), and monoclonal anti-cleaved caspase-3 (1:200) (Cell Signaling, Boston, MA); anti-Trx1 (1:1000; Millipore, Billerica, MA); and anti-importin-α (1:300) and anti-β-actin (1:200) (Abcam, Cambridge, MA). The secondary antibodies used were as follows: anti-mouse IgG (1:5000; Amersham Biosciences) and anti-rabbit IgG (1:5000; Bio-Rad). Bands were visualized by ECL Plus detection reagent (Amersham Biosciences) and quantified by ImageQuant Version 5.1 (GE Healthcare).
Assessment of Trx2 Redox Status
INS-1 cell mitochondrial fractions were dissolved in lysis/derivatization buffer (20 mm Tris-HCl (pH 8) and 15 mm AMS2 (Molecular Probes), a thiol-reactive agent used to alkylate protein sulfhydryl groups) and incubated on a Nutator for 3 h at 4 °C and for 30 min at 37 °C. Redox Western blotting was performed as described previously (20, 21), and the differences in molecular weight of the two Trx2 redox forms after the addition of AMS were resolved on an SDS/10–20% Tricine-polyacrylamide gel in the presence of nonreducing loading buffer.
Transfection Experiments and RNA Interference
INS-1 cells were grown in 6-well plates and transfected with specific siRNA oligonucleotides for rat importin-α1 (Invitrogen 1330001), rat Trx1 (Dharmacon L-080167-01-0010), rat Trx2 (Dharmacon L091907-01-0010), and rat TXNIP (Dharmacon; siGENOME SMARTpool gene ID 117514) or with scrambled oligonucleotide (0.1 μm; Dharmacon D-001810-01-20) using DharmaFECT transfection reagent (5 μl/well; Dharmacon/Thermo Scientific, Chicago, IL). The final concentration of oligonucleotides used was 25 nm. Cells were harvested after 48 h for RNA extraction and after 72 h for subcellular fractionation and protein extraction (as described above), and samples were stored at −80 °C until further use. Effective knockdown was confirmed at the mRNA and protein levels.
Reverse Transcription and Quantitative PCR
Total RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA)) according to the manufacturer's instructions. 1 μg of RNA was reverse-transcribed to cDNA using a first-strand cDNA synthesis kit (Roche Applied Science).
Quantitative real-time PCR was performed on a PRISM 7000 sequence detection system using SYBR Green (Applied Biosystems, Foster City, CA). Primers were designed using Primer Express 3.0 (Applied Biosystems) and were as follows: rat TXNIP, CGAGTCAAAGCCGTCAGGAT (forward) and TTCATAGCGCAAGTAGTCCAAGGT (reverse); Trx1, CGTGGATGACTGCCAGGAT (forward) and GGTCGGCATGCATTTGACT (reverse); Trx2, TGTGGCCCCTGCAAGATC (forward) and CCGTGCTGTTTGGCTACCAT (reverse); and importin-α1, CAGAGTTCCGGACGAGAAAGG (forward) and GATCCTCCAGAAGTTGCATTTGT (reverse). All samples were analyzed in triplicates and corrected for the 18 S ribosomal subunit (Applied Biosystems) run as an internal standard.
Statistical Analysis
To calculate the significance of a difference between two means, we used two-tailed Student's t tests. A p value of <0.05 was considered statistically significant.
RESULTS
TXNIP Is Localized in the Nucleus in Pancreatic Beta Cells
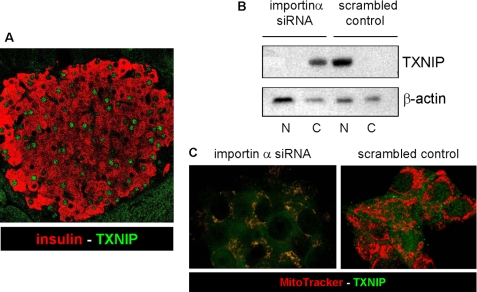
When studying the role of TXNIP in pancreatic beta cells, we surprisingly found that TXNIP is localized predominantly in the nucleus in primary mouse islets (Fig. 1A) as well as in INS-1 beta cells as shown by immunohistochemistry and confocal microscopy (Fig. 2A). To study the mechanisms contributing to this nuclear localization, we investigated the involvement of importin-α, a known major nuclear transport system (22). Transfection with importin-α1 siRNA led to a marked knockdown of importin-α1 in INS-1 cells compared with scrambled siRNA, whereas the expression of TXNIP, Trx1, and Trx2 was not reduced as confirmed by quantitative real-time reverse transcription-PCR as well as Western blotting (supplemental Fig. S2). Cell fractionation into nuclear and cytoplasmic fractions confirmed the nuclear localization of TXNIP in control INS-1 cells transfected with scrambled oligonucleotide, whereas importin-α1 siRNA knockdown resulted in a complete shift of TXNIP out of the nucleus and into the cytoplasmic fraction (Fig. 1B). Confocal imaging further revealed that TXNIP also accumulated in mitochondria (Fig. 1C). This suggests that the nuclear localization of TXNIP in pancreatic beta cells is mediated mainly by importin-α1, which is consistent with the observation that the importin-α1 subfamily of nuclear transport proteins interacts with TXNIP (23). However, because several previous reports demonstrated a cytoplasmic localization of TXNIP (3, 10, 14), these data suggest that TXNIP may be shuttling within the cell.
FIGURE 1.
Nuclear localization of TXNIP in pancreatic beta cells. A, immunofluorescent detection of endogenous TXNIP in wild-type C57BL/6 mouse pancreas sections by confocal microscopy. TXNIP was detected with the JY2 antibody and fluorescein isothiocyanate-labeled secondary antibody (green); beta cells were visualized by anti-insulin antibody and Cy3-conjugated secondary antibody (red). The pancreases of three mice were analyzed by staining of two separate 10-μm sections each; a representative pancreatic islet is shown. B, effects of importin-α1 siRNA knockdown on subcellular localization of TXNIP in INS-1 cells as assessed by cell fractionation and immunoblotting. N, nuclear fractions; C, cytoplasmic fractions. β-Actin was used as a loading control. One representative of three independent experiments is shown. C, confocal imaging of changes in subcellular localization of TXNIP in response to importin-α1 siRNA knockdown in INS-1 cells. Merged images were taken after 48 h: TXNIP (green) and MitoTracker Red (red)).
FIGURE 2.
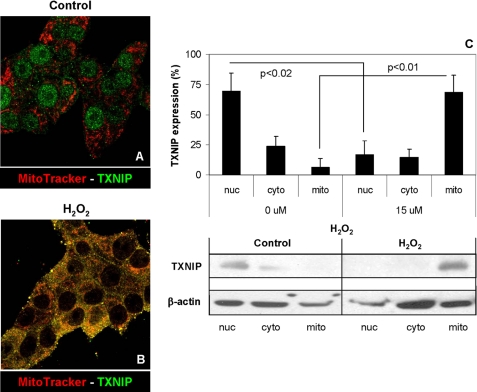
Shuttling of TXNIP into the mitochondria in response to oxidative stress. Shown is the immunofluorescent double staining of TXNIP (Alexa Fluor 488, green) and mitochondria (MitoTracker Red, red) in INS-1 beta cells treated without (A) or with (B) 15 μm H2O2 for 4 h. Merged confocal images (×200) are shown and are representative of four independent experiments. C, effects of H2O2 on the percentage of TXNIP protein localized in nuclear (nuc), cytoplasmic (cyto), and mitochondrial (mito) INS-1 cell fractions as analyzed by immunoblotting. Error bars represent means ± S.E. of five independent experiments.
TXNIP Shuttles from the Nucleus to Mitochondria in Response to Oxidative Stress
We therefore set out to study the subcellular localization of TXNIP, focusing on three compartments, the nucleus, the cytoplasm, and the mitochondria, this especially as we recently found that TXNIP induces apoptosis through the intrinsic mitochondrial death pathway (17). Because TXNIP plays an important role in the cellular redox state, we investigated the possibility that oxidative stress might regulate intracellular TXNIP shuttling. Although under normal physiological conditions TXNIP was again localized mainly in the nucleus (Fig. 2A), oxidative stress induced by hydrogen peroxide led to translocation of TXNIP from the nucleus into the mitochondria as demonstrated by co-localization with the mitochondrion-specific probe MitoTracker Red (Fig. 2B) as well as by cell fractionation and Western blotting (Fig. 2C). Appropriate fractionation into the three subcellular fractions was confirmed by subcellular markers under control and oxidative stress conditions (supplemental Fig. S3, A and B, respectively), and very similar confocal microscopy findings were also observed with oxidative stress induced by staurosporine (supplemental Fig. S3C).
TXNIP Interacts with Mitochondrial Trx2, and This Interaction Is Increased in Response to Oxidative Stress
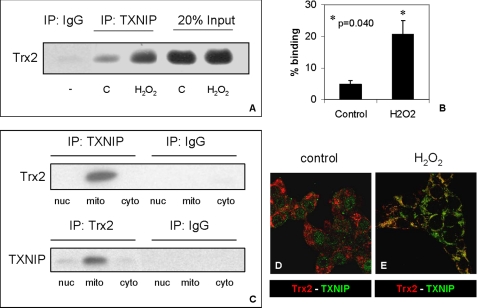
The predominant mitochondrial localization of TXNIP in response to oxidative stress prompted us to further study the role of TXNIP in mitochondria. Many studies have reported the interaction between TXNIP and thioredoxin (giving TXNIP its name), but based on the presumed cytosolic localization of TXNIP, the focus had remained on cytosolic thioredoxin (Trx1) (10). However, more recently, a mitochondrion-specific form of thioredoxin (Trx2) has been cloned that contains a mitochondrial signal sequence at the N terminus, conferring its specific localization to mitochondria (24). Trx2 has been shown to play a critical role in controlling reactive oxygen species-induced mitochondrial stress and mitochondrion-dependent apoptosis (25) whereby Trx2 helps repair reversible oxidative protein damage by reducing disulfide bonds that are formed by the oxidation of cysteines. Like other mammalian thioredoxins, Trx2 also contains the conserved thioredoxin-active site Trp-Cys-Gly-Pro-Cys, which would allow for its interaction with TXNIP (10, 24), and we therefore investigated the possibility of a TXNIP-Trx2 interaction in INS-1 beta cells. In fact, co-immunoprecipitation studies using mitochondrial fractions revealed that TXNIP interacted with Trx2 (Fig. 3A) and that this interaction was significantly increased by oxidative stress (Fig. 3B), consistent with the observed shuttling of TXNIP into the mitochondria (and confirmed by more TXNIP in the H2O2-treated mitochondrial input) (supplemental Fig. S4). Furthermore, this interaction was restricted to the mitochondria as demonstrated by a comparison of nuclear, cytoplasmic, and mitochondrial cell fractions (Fig. 3C) and immunohistochemistry and confocal microscopy (Fig. 3D), which is in alignment with the mitochondrion-specific expression pattern of Trx2.
FIGURE 3.
TXNIP interaction with mitochondrial Trx2. A, effects of oxidative stress on TXNIP-Trx2 co-immunoprecipitation. INS-1 cells were treated without (control (C)) or with H2O2 (15 μm for 4 h) prior to isolation of their mitochondrial fractions and immunoprecipitation (IP) with anti-TXNIP antibody JY2 and immunoblotting for Trx2. B, quantification of TXNIP-Trx2 binding in mitochondrial fractions of INS-1 cells treated with or without H2O2. Error bars represent means ± S.E. of three independent experiments. C, subcellular specificity of the TXNIP-Trx2 interaction. Co-immunoprecipitation experiments were performed using nuclear (nuc), mitochondrial (mito), and cytoplasmic (cyto) fractions of H2O2-treated INS-1 cells, and anti-TXNIP and anti-Trx2 antibodies were used for pulldown assays. One representative of three independent immunoblots is shown. D and E, representative confocal images of immunohistochemical localization of TXNIP (green) and Trx2 (red) in control and H2O2-treated INS-1 cells, respectively.
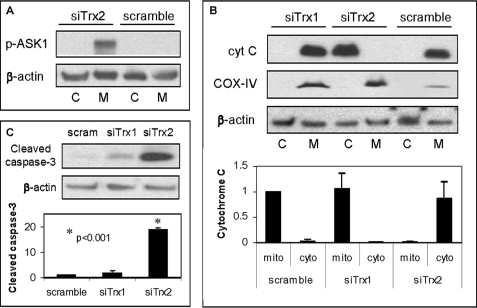
Trx2 Knockdown Results in Phosphorylation/Activation of ASK1 and Activation of the Mitochondrial Death Pathway
To study the role of mitochondrial Trx2 in beta cells, we silenced its expression or that of cytosolic Trx1 in INS-1 beta cells using isoform-specific siRNAs. Transfection with Trx2 siRNA resulted in 78.5 ± 3.2% Trx2 knockdown compared with the scrambled control while Trx1 and TXNIP expression remained unchanged, and Trx1 siRNA led to 85 ± 2.1% Trx1 knockdown while Trx2 and TXNIP expression was unaffected. Similar findings were also observed at the protein level (supplemental Fig. S5), confirming effective and highly specific silencing. Because in endothelial cells Trx2 has been shown to interact with ASK1 and to inhibit its activity (26), we first investigated the effects of Trx2 knockdown on ASK1. ASK1 is a major MAPKKK (mitogen-activated protein kinase kinase kinase) involved in mediating oxidative stress-initiated signals and regulating apoptosis (27, 28). ASK1 is expressed in the cytosol as well as in mitochondria (26), and its activity is regulated by phosphorylation/dephosphorylation whereby phosphorylation of serine 967 results in activation of ASK1 (27, 28). Interestingly, Trx2 down-regulation resulted in a dramatic increase in phosphorylated/activated ASK1 (Fig. 4A). In addition, it led to major cytochrome c release from the mitochondria into the cytosol, a classical marker for the intrinsic mitochondrial death pathway, whereas knockdown of Trx1 had no such effect (Fig. 4B and data not shown). Moreover, knockdown of Trx2 resulted in a dramatic activation of caspase-3 in the cytoplasm (Fig. 4C), whereas the effect of Trx1 siRNA was not significant, suggesting that mitochondrial Trx2 plays a unique and critical role in beta cell survival.
FIGURE 4.
Effects of Trx2 siRNA knockdown in INS-1 beta cells. INS-1 cells were transfected with specific siRNAs targeting rat Trx2 (siTrx2) or Trx1 (siTrx1) or with control scrambled (scram) siRNA 72 h prior to fractionation into cytoplasmic (C, cyto) and mitochondrial (M, mito) fractions and analysis of phosphorylated ASK1 (p-ASK1; A), cytochrome c (cyt C; B), and cytoplasmic cleaved caspase-3 (C) by immunoblotting. Three independent experiments were performed, and error bars represent means ± S.E. corrected for β-actin. Cyclooxygenase IV (COX-IV) was run as a mitochondrial marker.
Increased Mitochondrial TXNIP Expression Leads to Trx2 Oxidation, Decreased Trx2-ASK1 Binding, and Phosphorylation/Activation of ASK1
As redox proteins, thioredoxins can exist in either a reduced or oxidized form, which heavily impacts on their ability to interact with other proteins such as TXNIP and ASK1 (5, 29). In fact, when Trx1 is oxidized by reactive oxygen species, its binding to ASK1 is disrupted, and ASK1 is activated, thereby transmitting the apoptotic signal (29). Even though specific experiments showing that the same is true for Trx2 are not available, the fact that Trx2 contains the two critical cysteine residues at the active center suggested that it may act very similarly to its cytosolic counterpart. We therefore wanted to test whether oxidative stress or increased mitochondrial TXNIP expression and TXNIP-Trx2 interaction may also induce Trx2 oxidation and/or affect Trx2-ASK1 binding.
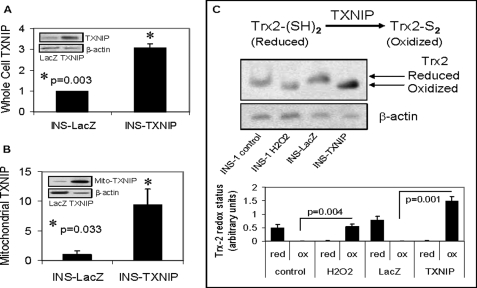
Using our stably transfected INS-TXNIP cells and cell fractionation, we confirmed not only that INS-TXNIP cells demonstrated significantly increased whole cell TXNIP expression (Fig. 5A) but that TXNIP overexpression also resulted in a dramatic almost 10-fold increase in mitochondrial TXNIP expression compared with control INS-LacZ cells (Fig. 5B and supplemental Fig. S1). Interestingly, TXNIP led to a strong induction of oxidized Trx2 in the INS-TXNIP cells that was even more pronounced than what was observed in response to H2O2 in INS-1 cells run as a comparison (Fig. 5C).
FIGURE 5.
Overexpression of mitochondrial TXNIP and Trx2 oxidation. A and B, whole cell and mitochondrial TXNIP expression, respectively, in control INS-LacZ and TXNIP-overexpressing INS-TXNIP cells as assessed and quantified by immunoblotting. Error bars represent means ± S.E. of at least three independent experiments corrected for β-actin. Insets, representative immunoblots. C, redox immunoblot and quantification of Trx2 in mitochondrial fractions of control and H2O2-treated INS-1 cells and control INS-LacZ and INS-TXNIP cells. Cell extracts were alkylated with AMS as described under “Experimental Procedures.” Upper bands represent reduced (red) Trx2, and lower bands represent oxidized (ox) Trx2. β-Actin was used as a loading control. One representative of three independent experiments is shown. Error bars represent means ± S.E.
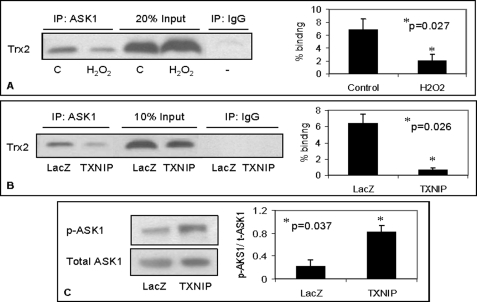
Because Trx2 has been suggested to act as an antioxidant in mitochondria and to bind and inhibit ASK1, we wanted to assess how oxidative stress or TXNIP overexpression may affect this interaction. Our finding of TXNIP-Trx2 binding (Fig. 3) further suggested that TXNIP may compete with ASK1 for binding to Trx2 and thereby release ASK1 from its inhibition by Trx2. In fact, after confirming that Trx2-ASK1 binding also occurs in beta cells, our co-immunoprecipitation data revealed that increased mitochondrial TXNIP (be it caused by H2O2 or directly by overexpression) (supplemental Fig. S4) significantly decreased Trx2-ASK1 interaction (Fig. 6, A and B). The same results were also obtained when protein complexes were immunoprecipitated with anti-Trx2 antibody (supplemental Fig. S6). Interestingly, this TXNIP-mediated reduction in binding between ASK1 and Trx2 led to phosphorylation/activation of ASK1 (Fig. 6C), thereby initiating this apoptotic signaling cascade.
FIGURE 6.
Effects of increased TXNIP expression on Trx2-ASK1 interaction and ASK1 phosphorylation/activation. Shown are the effects of increased mitochondrial TXNIP induced by H2O2 treatment of INS-1 cells (A) or by TXNIP overexpression in INS-TXNIP cells (B) on the interaction between Trx2 and ASK1 as determined by co-immunoprecipitation (IP). Protein complexes were precipitated with anti-ASK1 antibody and analyzed for Trx2 by Western blotting, and the percent binding was quantified. C, control. Shown are the TXNIP effects on ASK1 phosphorylation/activation (C). Phosphorylated ASK1 (p-ASK1) expression in mitochondrial fractions of control INS-LacZ and INS-TXNIP cells was assessed by Western blotting with phosphorylated ASK1 (Ser967)-specific antibody and normalized for total ASK1 (t-ASK1). Error bars represent means ± S.E. of three independent experiments.
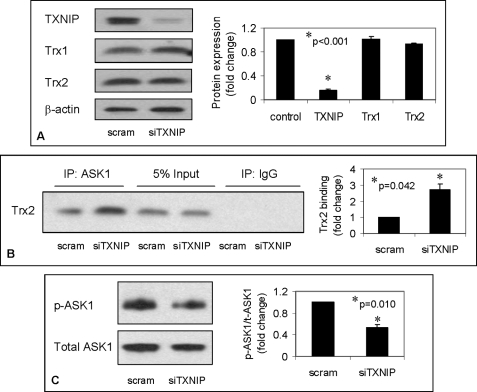
TXNIP Knockdown Promotes Trx2- ASK1 Interaction and Blocks ASK1 Phosphorylation/Activation
To further determine the role of TXNIP in these processes, we down-regulated TXNIP by siRNA, resulting in a highly significant and specific knockdown (Fig. 7A). This led to an ∼3-fold increase in Trx2-ASK1 binding as assessed again by immunoprecipitation (Fig. 7B) and blocked ASK1 phosphorylation/activation (Fig. 7C). Given the exact opposite findings in response to TXNIP overexpression, these results strongly support the critical role TXNIP plays in this newly identified signaling pathway.
FIGURE 7.
Effects of TXNIP knockdown on Trx2-ASK1 interaction and ASK1 phosphorylation/activation. A, demonstration of specific TXNIP knockdown by siRNA in INS-1 cells. Cells were transfected with TXNIP siRNA (siTXNIP) or scrambled (scram) oligonucleotides as described under “Experimental Procedures” and analyzed 48 h later by immunoblotting for TXNIP as well as for Trx1 and Trx2 protein expression. B and C, Trx2-ASK1 interaction and ASK1 phosphorylation/activation, respectively, in response to TXNIP knockdown as assessed by co-immunoprecipitation (IP). Error bars represent means ± S.E. of at least three independent experiments. p-ASK1, phosphorylated ASK1; t-ASK1, total ASK1.
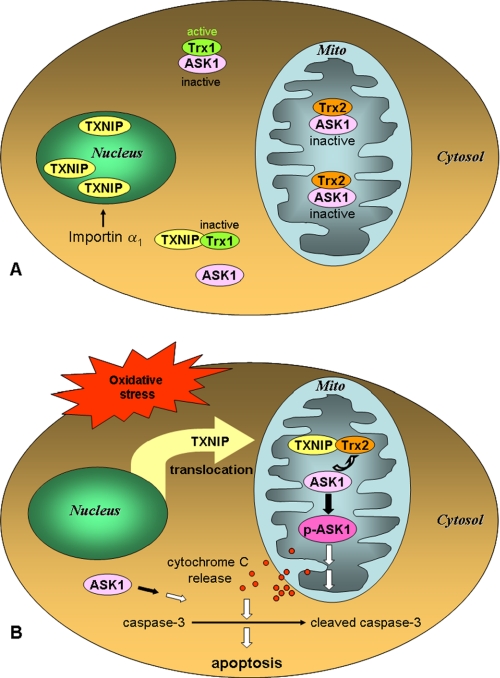
Combined, the results suggest that TXNIP is located predominantly in the nucleus under basal conditions and that ASK1 is bound to Trx2 in the mitochondria and thereby maintained inactive, assuring beta cell survival (Fig. 8A). In contrast, in response to oxidative stress, TXNIP translocates into mitochondria, where it forms a complex with Trx2, removing Trx2 from ASK1. This alleviation of Trx2-mediated inhibition results in phosphorylation of ASK1, cytochrome c release, activation of caspase-3, and apoptosis (Fig. 8B).
FIGURE 8.
Schematic representation of the effects of oxidative stress-induced subcellular TXNIP shuttling. A, in the cytosol, TXNIP has been shown to bind to Trx1, leading to its inactivation and dissociation from ASK1. However, our data now indicate that, under basal conditions, importin-α ensures that TXNIP is primarily localized in the nucleus. This also allows mitochondrial Trx2 to remain bound to ASK1, preventing its activation and promoting cell survival. B, in contrast, in response to oxidative stress, TXNIP translocates from the nucleus into the mitochondria (Mito), where it competes for Trx2 binding, leading to ASK1 phosphorylation/activation and initiation of the apoptotic cascade. p-ASK1, phosphorylated ASK1.
DISCUSSION
The results of this study demonstrate for the first time that endogenous TXNIP shuttles between cell compartments and specifically translocates from the nucleus into the mitochondria in response to oxidative stress. In addition, they reveal that, once in the mitochondria, TXNIP binds to Trx2, thereby releasing ASK1 from its inhibition and resulting in ASK1 phosphorylation/activation and initiation of mitochondrion-mediated beta cell apoptosis.
Our initial observation of endogenous TXNIP being predominantly localized in the nucleus in pancreatic beta cells under basal conditions was surprising, as previous studies had described TXNIP as a cytosolic protein (3, 10, 14). On the other had, our results are consistent with the finding that, in MCF-7 breast cancer cells, overexpression of enhanced green fluorescent protein-tagged TXNIP or induction of TXNIP expression by histone deacetylase inhibition results in nuclear localization of TXNIP (23). Interestingly, TXNIP translocation into the nucleus in MCF-7 cells is also dependent on importin-α, very similar to our observation in pancreatic beta cells (Fig. 1). An additional indication for a role of TXNIP in the nucleus comes from recent data suggesting that TXNIP acts as a transcriptional co-repressor by interacting with HDAC1 (histone deacetylase-1) (30) and that TXNIP promotes nuclear localization of Trx1 and DnaJb5 (a heat shock protein) in cardiac myocytes and thereby modulates nuclear localization of HDAC4 (31). The notion of TXNIP acting as a co-repressor is also in alignment with our observation in beta cells that TXNIP overexpression led to a decrease in the expression of most transcripts as assessed by oligonucleotide microarray analysis (32).
Interestingly, we further discovered that oxidative stress leads to TXNIP shuttling from the nucleus into the mitochondria (Fig. 2). This represents a completely novel concept, even though some TXNIP localization in nuclei and mitochondria has been described previously in rat renal proximal tubular cells (33). Because oxidative stress down-regulates the importin-α nuclear import pathway (34), this inhibition may have contributed to the observed change in TXNIP localization. Overall, the oxidative stress-induced translocation of TXNIP into the mitochondria seemed to fit well with the major role TXNIP plays as a regulator of the cellular redox state and mitochondrial death pathway (17), but its exact significance remained elusive. We therefore examined the biological role and function of this mitochondrial TXNIP accumulation further. Interestingly, these studies revealed a novel redox-sensitive mitochondrial TXNIP-Trx2-ASK1 signaling cascade by which TXNIP competes with ASK1 for Trx2 binding, thereby releasing ASK1 from its inhibition by Trx2 and resulting in phosphorylation/activation of ASK1, cytochrome c release, and beta cell apoptosis (Fig. 8). Similarly, in endothelial cells, fluid shear stress has been shown to modulate inflammation by affecting a non-mitochondrial TXNIP-Trx1-ASK1 pathway including Trx1 instead of Trx2 (35).
In fact, although the TXNIP-Trx1 interaction has been studied extensively (2–6), TXNIP-Trx2 binding has not been described previously even though Trx1 and Trx2 share the two critical cysteine residues in the active site required for this interaction (10, 24, 25). In contrast, unlike other mammalian thioredoxins, Trx2 does not contain any additional cysteine residues and is therefore believed to be more resistant to oxidative stress (24, 25). Nevertheless, we observed that Trx2 was readily oxidized in response to hydrogen peroxide or to TXNIP overexpression (Fig. 5), and Trx2 has also been shown to undergo oxidation in response to other stimuli such as troglitazone in hepatocytes (36).
Surprisingly, we found that knockdown of Trx2, but not Trx1, resulted in activation of ASK1, cytochrome c release, and activation of caspase-3 (Fig. 4), suggesting that mitochondrial Trx2 is a critical factor for cell survival and underlining the importance of TXNIP-Trx2 signaling and mitochondrial integrity for normal beta cell biology. These conclusions are further supported by work demonstrating that Trx2 is essential for mitochondrion-dependent apoptosis (25); that Trx2 overexpression renders cells more resistant to oxidant-induced apoptosis (37), whereas Trx2 knockdown increases the susceptibility to glucose toxicity (38); and that TXNIP induces beta cell apoptosis through the intrinsic mitochondrial pathway (17). In contrast to Trx1, Trx2 has also been shown to specifically regulate the JNK (c-Jun N-terminal kinase)-independent intrinsic apoptotic pathway in vascular endothelial cells, suggesting that Trx1 and Trx2 control apoptosis by modulating different downstream signaling pathways (26). Our findings therefore do not exclude the possibility that the TXNIP-Trx1 interaction in the cytoplasm also plays an important role in cell biology.
Interestingly, ASK1 was found to be a major binding partner of Trx1 (4, 5, 29). The Trx1-ASK1 interaction is regulated predominantly by TXNIP and specifically by TXNIP binding to the catalytic cysteines of Trx1, which inhibits both Trx1 activity and its ability to bind to ASK1 (3, 39). Indeed, down-regulation of TXNIP by RNA interference increases the association of Trx1 with ASK1 and thereby inhibits the activation of the apoptotic cascade (35). Similarly, when Trx1 is oxidized by reactive oxygen species, the binding of Trx1 and ASK1 is disrupted, and ASK1 is activated to transmit the oxidative stress-induced apoptotic signal (29). Whereas these earlier studies had again focused on Trx1 and cytosolic ASK1, more recent data show that Trx2 also interacts with mitochondrial ASK1, inhibits its activity, and prevents ASK1-induced apoptosis (26, 36). Studies in hepatocytes have suggested that oxidation of Trx2 eliminates these protective effects of Trx2 (36), which is consistent with our findings in beta cells that TXNIP overexpression led to Trx2 oxidation and ASK1 activation (Figs. 5 and 6). The fact that, in contrast, TXNIP knockdown promoted Trx2-ASK binding and prevented ASK1 activation (Fig. 7) further underlines the importance of TXNIP in controlling these critical cellular processes. Pancreatic beta cells are known to be particularly sensitive to oxidative stress due to their low expression level of antioxidant enzymes (40), and recent data provide strong evidence for the critical role of mitochondrial reactive oxygen species production in diabetes (41, 42). In addition, we recently discovered that TXNIP deficiency protects against beta cell apoptosis and prevents type 1 and 2 diabetes, suggesting that inhibition of TXNIP signaling represents a novel approach to diabetes therapy (18). However, the exact signaling pathways had remained largely elusive. Identification of the redox-sensitive mitochondrial TXNIP-Trx2-ASK1 signaling pathway in the present study therefore represents a major breakthrough in this regard. Although the present data indicate that TXNIP promotes ASK1 activity and apoptosis through directly interacting with and inhibiting mitochondrial Trx2, our previous findings also demonstrate that TXNIP inhibits anti-apoptotic Akt (18). Interestingly, Akt has been shown to negatively regulate ASK1 (43), and we therefore cannot exclude the possibility that TXNIP-mediated inhibition of Akt may contribute to the observed activation of ASK1 in response to TXNIP overexpression. Alternatively, it is also conceivable that TXNIP independently modulates Akt and ASK1 signaling and that the two pathways converge at the level of cytochrome c release and mitochondrion-dependent apoptosis. In summary, this study has shed new light on the subcellular localization of TXNIP and its intracellular shuttling in response to oxidative stress and has identified a novel redox-sensitive mitochondrial TXNIP-Trx2-ASK1 signaling pathway.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK-078752 from NIDDK and R21HL-089205 from NHLBI (to A. S.). This work was also supported by American Diabetes Association Grant 7-07-CD-22 and Juvenile Diabetes Research Foundation Grant 1-2007-790) (to A. S.) and is the result of work supported by resources and use of facilities at the William S. Middleton Memorial Veterans Hospital (Madison, WI).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- siRNA
- small interfering RNA.
REFERENCES
- 1.Chen K. S., DeLuca H. F. (1994) Biochim. Biophys. Acta 1219, 26–32 [DOI] [PubMed] [Google Scholar]
- 2.Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H., Takagi Y., Sono H., Gon Y., Yodoi J. (1999) J. Biol. Chem. 274, 21645–21650 [DOI] [PubMed] [Google Scholar]
- 3.Junn E., Han S. H., Im J. Y., Yang Y., Cho E. W., Um H. D., Kim D. K., Lee K. W., Han P. L., Rhee S. G., Choi I. (2000) J. Immunol. 164, 6287–6295 [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka H., Maehira F., Oshiro M., Asato T., Yanagawa Y., Takei H., Nakashima Y. (2000) Biochem. Biophys. Res. Commun. 271, 796–800 [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama A., Masutani H., Nakamura H., Nishinaka Y., Yodoi J. (2001) IUBMB Life 52, 29–33 [DOI] [PubMed] [Google Scholar]
- 6.Patwari P., Higgins L. J., Chutkow W. A., Yoshioka J., Lee R. T. (2006) J. Biol. Chem. 281, 21884–21891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yodoi J., Nakamura H., Masutani H. (2002) Biol. Chem. 383, 585–590 [DOI] [PubMed] [Google Scholar]
- 8.Kim K. Y., Shin S. M., Kim J. K., Paik S. G., Yang Y., Choi I. (2004) Biochem. Biophys. Res. Commun. 315, 369–375 [DOI] [PubMed] [Google Scholar]
- 9.Hirota T., Okano T., Kokame K., Shirotani-Ikejima H., Miyata T., Fukada Y. (2002) J. Biol. Chem. 277, 44244–44251 [DOI] [PubMed] [Google Scholar]
- 10.Schulze P. C., Yoshioka J., Takahashi T., He Z., King G. L., Lee R. T. (2004) J. Biol. Chem. 279, 30369–30374 [DOI] [PubMed] [Google Scholar]
- 11.Minn A. H., Hafele C., Shalev A. (2005) Endocrinology 146, 2397–2405 [DOI] [PubMed] [Google Scholar]
- 12.Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. (2002) Endocrinology 143, 3695–3698 [DOI] [PubMed] [Google Scholar]
- 13.Cheng D. W., Jiang Y., Shalev A., Kowluru R., Crook E. D., Singh L. P. (2006) Arch. Physiol. Biochem. 112, 189–218 [DOI] [PubMed] [Google Scholar]
- 14.Schulze P. C., De Keulenaer G. W., Yoshioka J., Kassik K. A., Lee R. T. (2002) Circ. Res. 91, 689–695 [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka J., Schulze P. C., Cupesi M., Sylvan J. D., MacGillivray C., Gannon J., Huang H., Lee R. T. (2004) Circulation 109, 2581–2586 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., De Keulenaer G. W., Lee R. T. (2002) J. Biol. Chem. 277, 26496–26500 [DOI] [PubMed] [Google Scholar]
- 17.Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. (2008) Diabetes 57, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Hui S. T., Couto F. M., Mungrue I. N., Davis D. B., Attie A. D., Lusis A. J., Davis R. A., Shalev A. (2008) FASEB J. 22, 3581–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halvey P. J., Watson W. H., Hansen J. M., Go Y. M., Samali A., Jones D. P. (2005) Biochem. J. 386, 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damdimopoulos A. E., Miranda-Vizuete A., Pelto-Huikko M., Gustafsson J. A., Spyrou G. (2002) J. Biol. Chem. 277, 33249–33257 [DOI] [PubMed] [Google Scholar]
- 22.Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 23.Nishinaka Y., Masutani H., Oka S., Matsuo Y., Yamaguchi Y., Nishio K., Ishii Y., Yodoi J. (2004) J. Biol. Chem. 279, 37559–37565 [DOI] [PubMed] [Google Scholar]
- 24.Spyrou G., Enmark E., Miranda-Vizuete A., Gustafsson J. (1997) J. Biol. Chem. 272, 2936–2941 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T., Hosoi F., Yamaguchi-Iwai Y., Nakamura H., Masutani H., Ueda S., Nishiyama A., Takeda S., Wada H., Spyrou G., Yodoi J. (2002) EMBO J. 21, 1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R., Al-Lamki R., Bai L., Streb J. W., Miano J. M., Bradley J., Min W. (2004) Circ. Res. 94, 1483–1491 [DOI] [PubMed] [Google Scholar]
- 27.Goldman E. H., Chen L., Fu H. (2004) J. Biol. Chem. 279, 10442–10449 [DOI] [PubMed] [Google Scholar]
- 28.Ichijo H., Nishida E., Irie K., ten Dijke P., Saitoh M., Moriguchi T., Takagi M., Matsumoto K., Miyazono K., Gotoh Y. (1997) Science 275, 90–94 [DOI] [PubMed] [Google Scholar]
- 29.Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S. H., Jeon J. H., Ju H. R., Jung U., Kim K. Y., Yoo H. S., Lee Y. H., Song K. S., Hwang H. M., Na Y. S., Yang Y., Lee K. N., Choi I. (2003) Oncogene 22, 4035–4046 [DOI] [PubMed] [Google Scholar]
- 31.Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., Sadoshima J. (2008) Cell 133, 978–993 [DOI] [PubMed] [Google Scholar]
- 32.Minn A. H., Pise-Masison C. A., Radonovich M., Brady J. N., Wang P., Kendziorski C., Shalev A. (2005) Biochem. Biophys. Res. Commun. 336, 770–778 [DOI] [PubMed] [Google Scholar]
- 33.Dutta K. K., Nishinaka Y., Masutani H., Akatsuka S., Aung T. T., Shirase T., Lee W. H., Yamada Y., Hiai H., Yodoi J., Toyokuni S. (2005) Lab. Invest. 85, 798–807 [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto Y., Saiwaki T., Yamashita J., Yasuda Y., Kotera I., Shibata S., Shigeta M., Hiraoka Y., Haraguchi T., Yoneda Y. (2004) J. Cell Biol. 165, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamawaki H., Pan S., Lee R. T., Berk B. C. (2005) J. Clin. Invest. 115, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim P. L., Liu J., Go M. L., Boelsterli U. A. (2008) Toxicol. Sci. 101, 341–349 [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Cai J., Murphy T. J., Jones D. P. (2002) J. Biol. Chem. 277, 33242–33248 [DOI] [PubMed] [Google Scholar]
- 38.Liang M., Pietrusz J. L. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 77–83 [DOI] [PubMed] [Google Scholar]
- 39.Bodnar J. S., Chatterjee A., Castellani L. W., Ross D. A., Ohmen J., Cavalcoli J., Wu C., Dains K. M., Catanese J., Chu M., Sheth S. S., Charugundla K., Demant P., West D. B., de Jong P., Lusis A. J. (2002) Nat. Genet. 30, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson R. P., Harmon J., Tran P. O., Tanaka Y., Takahashi H. (2003) Diabetes 52, 581–587 [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa T., Araki E. (2007) Antioxid. Redox Signal. 9, 343–353 [DOI] [PubMed] [Google Scholar]
- 42.Anderson E. J., Lustig M. E., Boyle K. E., Woodlief T. L., Kane D. A., Lin C. T., Price J. W., 3rd, Kang L., Rabinovitch P. S., Szeto H. H., Houmard J. A., Cortright R. N., Wasserman D. H., Neufer P. D. (2009) J. Clin. Invest. 119, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim A. H., Khursigara G., Sun X., Franke T. F., Chao M. V. (2001) Mol. Cell. Biol. 21, 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.