Abstract
Ferritins are important iron storage and detoxification proteins that are widely distributed in living kingdoms. Because plant ferritin possesses both a ferroxidase site and a ferrihydrite nucleation site, it is a suitable model for studying the mechanism of iron storage in ferritin. This article presents for the first time the crystal structure of a plant ferritin from soybean at 1.8-Å resolution. The soybean ferritin 4 (SFER4) had a high structural similarity to vertebrate ferritin, except for the N-terminal extension region, the C-terminal short helix E, and the end of the BC-loop. Similar to the crystal structures of other ferritins, metal binding sites were observed in the iron entry channel, ferroxidase center, and nucleation site of SFER4. In addition to these conventional sites, a novel metal binding site was discovered intermediate between the iron entry channel and the ferroxidase site. This site was coordinated by the acidic side chain of Glu173 and carbonyl oxygen of Thr168, which correspond, respectively, to Glu140 and Thr135 of human H chain ferritin according to their sequences. A comparison of the ferroxidase activities of the native and the E173A mutant of SFER4 clearly showed a delay in the iron oxidation rate of the mutant. This indicated that the glutamate residue functions as a transit site of iron from the 3-fold entry channel to the ferroxidase site, which may be universal among ferritins.
Keywords: Metals/Iron, Methods/X-ray Crystallography, Organisms/Plant, Protein/Iron, Protein/Metalloproteins, Protein/Structure
Introduction
Ferritin is a class of iron storage proteins distributed ubiquitously in plants, mammals, and bacteria. This protein class plays an important role in storage and detoxification of excess iron in a living cell. Although the primary sequences of ferritins vary, their three-dimensional structures of 24 structurally equivalent subunits assembled into a cage-like oligomer and related by 4-, 3-, and 2-fold symmetry axes (1–3) are highly conserved. Each subunit has four helices (A-D), each of which is composed of ∼30 amino acid residues, along with a fifth short helix (E) having 10 residues or more, resulting in the formation of a “four-helix bundle.” In general, the outer shell diameters of the spherical oligomeric ferritin composed of 24 subunits are ∼120 Å, whereas the inner cavity diameters are ∼80 Å. Up to 4500 iron atoms are deposited in the inner cavity of ferritin (1–3). To date, three major metal binding sites have been identified in ferritin. These exhibit some common aspects related to the iron storage mechanism: (i) they exist around the 3-fold symmetry channel, (ii) constitute a ferroxidase center in the four-helix bundle, and (iii) constitute a nucleation site facing the inner cavity of ferritin (4–7). It has been suggested that iron atoms pass through the hydrophilic channels that traverse the ferritin shell around the 3-fold symmetry axes of oligomeric ferritin (8–11). This funnel-like channel is surrounded by hydrophilic residues, such as aspartic acid and glutamic acid, which have been demonstrated to serve as ligands for metal ions, such as ferrous, calcium, or zinc ions (4, 5). With respect to the ferroxidase center and nucleation center, two distinct subunits, heavy (H)2 and light (L) chains, are seen in mammalian ferritin (12–14). The H chain possesses the ferroxidase site (15, 16), which is positioned inside the 4-helix bundle of each subunit and is responsible for oxidation of ferrous iron atoms to produce the μ-oxo-bridged Fe(III) species. The ferroxidase site is composed of six residues: Glu27, Tyr34, Glu62, His65, Glu107, and Gln141, which coordinate with metal ions directly or indirectly. Crystallographic analyses of ferritin in vertebrates (4, 5, 17), insects (18), and bacteria (7, 19) revealed an extremely high degree of similarity among the structures of these ferroxidase centers. On the other hand, the L-type ferritin lacks the ferroxidase center, but possesses the nucleation site. The site is composed of three or four glutamate residues facing the inner cavity of the ferritin shell, and facilitates ferrihydrite nucleation (20, 21). In mammals, the H and L chains form a heteropolymer and cooperatively share the role of iron storage in vivo (14).
Recently, multiple copies of the ferritin gene were identified from various plant species (22–30). It has been suggested that their expression was differentially regulated via various environmental stresses or metal overload (25, 31–36), and are sometimes subjected to different types of processing (26, 30). In contrast to the case of the H and L chains of mammalian ferritin, all of the identified plant ferritin subunits have similar primary sequences, especially in their deduced ferroxidase center and nucleation site. Although the primary structures of plant ferritins are basically well conserved with those of mammalian ferritins (Fig. 1), plant ferritins show two further differences: first, plant ferritin possesses the N-terminal extension regions, which can be divided into two parts based on their functions (37). The upstream one is called a “transit peptide” responsible for transportation of the polypeptide chain to plastid, whereas the downstream one is called an “extension peptide” (EP) with an unknown function (38). Several reports have suggested that EP helps to stabilize the oligomeric protein shell (39–41). Second, plant ferritins can incorporate and store iron without the differentiation of subunits. As mentioned above, all plant ferritins possess both a ferroxidase center and a nucleation site. These observations led us to hypothesize that a crystallographic analysis of plant ferritin would offer new insight into the pathway of metal ion transfer from the iron entry channel via the ferroxidase center to the nucleation site and inner cavity of the ferritin shell. With respect to the three-dimensional structures of plant ferritin, Lobréaux et al. (42) presented the deduced structure of ferritin from pea (Pisum sativum) using a computer simulation. They predicted that the main chain structure of plant ferritin can be superimposed onto human H chain ferritin up to 1 Å of the root mean square distance (42). Thus, it can be considered that its overall structure, including structure of the 3-fold symmetry channel, ferroxidase center, and nucleation site, is highly conserved. However, the actual metal binding sites and precise orientation of side chains of metal ligands have remained unclear. On the other hand, we have attempted to crystallize plant ferritin from soybean (SFER1) to obtain the real structure of these active sites; however, the diffraction data from the crystal were not sufficient to provide high resolution structural analysis (43).
FIGURE 1.
Amino acid sequences of the plant ferritin, SFER4, and other ferritin from vertebrate and bacteria. The sequences of SFER4 (GenBank number AB062756), human H (HuHF, M11146), human L (HuLF, M11147), bullfrog H (BfHF, M15655), and two E. coli ferritins (EcBFR (Swiss Prot P0ABD3) and EcFTN (GenBank number X53513)) are shown. Conserved ferroxidase center, putative nucleation center, and transit site discussed in this study are shown boxed. The letters F, N, and T on the boxed sequences indicate the ferroxidase site, nucleation site, and transit site, respectively.
In this study, to address the question of the metal ion pathway in the ferritin oligomer, we have crystallized a recombinant plant ferritin subunit from soybean (SFER4). The high resolution x-ray crystallographic analysis of this soybean ferritin subunit has revealed a novel metal binding site between the 3-fold symmetry channel and ferroxidase center. The deletion of this site resulted in a delay of the iron oxidation/nucleation process, suggesting that this site functions as a transit site for metal translocation in ferritin. Based on these results, coupled with the extremely high conservation of this site, we propose that this site is part of the metal ion pathway widely distributed in ferritin from various species.
EXPERIMENTAL PROCEDURES
cDNA Cloning and Protein Preparation
Cloning of the soybean ferritin SFER4 cDNA (GenBankTM number AB062756) was performed as described previously (44). The SFER4 cDNA fragment containing NcoI and BamHI restriction sites was cloned into pET21d (Novagen, San Diego, CA) through restriction sites NcoI and BamHI using a PCR-based method. The E173A SFER4 variant was obtained by oligonucleotide site-directed mutagenesis using pET_SFER4 as a template. The DNA sequence of the resulting plasmid pET_ SFER4 and pET_SFER4-E173A was verified by automated DNA sequencing (ABI PRISM 3100, Applied Biosystems, Foster City, CA). The constructs were then introduced into Escherichia coli strain BL21(DE3). The positive transformants of each construct were grown at 37 °C on LB medium supplemented with 50 mg/liter of carbenicillin, and protein expression was induced with 100 μm isopropyl β-d-1-thiogalactopyranoside when the cell density reached an A600 of 0.6. Eventually, the cells were harvested by centrifugation after 3 h of induction and resuspended in Buffer A (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.15 m NaCl, 0.1 mm p-amidinophenyl)methanesulfonyl fluoride hydrochloride, 0.2 μm pepstatin, 0.5 g/liter leupeptin) to a concentration of 40 g (fresh weight bacteria)/liter, followed by disruption by sonication. The supernatant of the resulting crude extract was collected by centrifugation and fractionated by 50–60% saturation of ammonium sulfate. The pellet was re-suspended in Buffer B (20 mm Tris-HCl, pH 7.8, 1 mm EDTA) and dialyzed against the same buffer. The protein solution was applied to an ion-exchange column (Q-Sepharose 26/10, GE Healthcare, Piscataway, NJ), followed by elution with 0–0.5 m NaCl gradient. Finally, the protein solution was concentrated and purified on a gel filtration column (Superdex 200pg 16/60, GE Healthcare), equilibrated with Buffer C (10 mm Tris-HCl, 0.15 m NaCl). The resulting SFER4 and SFER4-E173A were electrophoretically pure (data not shown). The protein concentrations were estimated by A280, using molar absorptivity (ϵM) of 17,420 cm−1 m−1, calculated from amino acid composition (45).
Crystallization, Data Collection, and Processing
Initial crystals of native SFER4 were obtained after about 2 weeks (Wizard I; B-Bridge, Mountain View, CA) using the hanging drop vapor diffusion method. The optimized crystallization drops were prepared by mixing equal volumes of mother liquid composed of 15% PEG 1000, 0.2 m calcium acetate, 0.1 m imidazole, pH 7.8, and protein solution containing 10 mg/ml of recombinant SFER4. Orthorhombic crystals of SFER4 (space group P21212; a = 222.610, b = 220.886, c = 122.452 Å; VM = 2.61 Å3/Da for 24 monomers per asymmetric unit) of ∼0.5 × 0.2 × 0.2 mm appeared within 3 weeks at 20 °C. The crystallization condition of the E173A variant was the same as that of the native crystal. Diffraction data of the native and E173A crystals were collected to 1.8- and 1.9-Å resolution at 77 K at the SPring-8 beamline 44XU after flash cooling with 30% of 2-methyl-2,4-pentanediol as a cryoprotectant. Data were processed, merged, and scaled with HKL-2000 (HKL Research, Charlottesville, VA) (46). Data processing statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for the crystals of plant ferritin SFER4 and its variant E173A
| Native SFER4 | SFER4 variant E173A | |
|---|---|---|
| Data collection statistics | ||
| Space group | P21212 | P21212 |
| Lattice parameter (Å) | a = 222.610, b = 220.886, c = 122.452 | a = 223.494, b = 221.798, c = 122.705 |
| Wave length (Å) | 0.90 | 0.90 |
| Resolution (highest shell) (Å) | 50-1.8 (1.83-1.80) | 50-1.9 (1.93-1.90) |
| No. unique reflections | 546,597 | 473,818 |
| Completeness (%) | 98.9 (97.1) | 99.2 (97.0) |
| Data redundancy | 5.8 (4.6) | 6.6 (5.9) |
| Rmerge | 0.058 (0.259) | 0.102 (0.356) |
| 〈I/σ(I)〉 | 18.4 (7.96) | 15.7 (8.34) |
| Refinement statistics | ||
| Resolution (Å) | 50-1.8 | 50-1.9 |
| Number of amino acid residues | 4,642 | 4,629 |
| Number of atoms | ||
| Protein | 39,272 | 39,197 |
| Solvent | 4,570 | 3,429 |
| Ca2+ | 136 | 130 |
| Acetate (No. of molecule) | 72 (18) | 72 (18) |
| R-factor | 0.1423 | 0.1504 |
| Rfree | 0.1727 | 0.1850 |
| Root mean square deviation from ideality | ||
| Bond length (Å) | 0.004 | 0.006 |
| Bond angle (deg.) | 0.771 | 0.870 |
| Average B-factor (Å2) | ||
| Main chain (Å2) | 15.85 | 16.74 |
| Side chain (Å2) | 24.48 | 23.56 |
| Water (Å2) | 26.48 | 23.64 |
| Ca2+ (Å2) | 35.00 | 35.27 |
| Acetate (Å2) | 37.38 | 37.74 |
Structure Determination and Refinement
The structure of native SFER4 was determined by molecular replacement using the Molrep program (47) in CCP4i suit 1.4.4 (48) using the structure of bullfrog M ferritin (PDB code 1MFR) (17) as a search model. All water molecules and calcium atoms were removed from the search model prior to molecular replacement. Refinement was performed with 24-fold non-crystallographic symmetry using the REFMAC5 program (49) in CCP4i suit 1.4.4 (48). The structure was visualized and rebuilt using COOT 0.5.2 (50) and further modified on σ-weighted (2|Fo|−|Fc|) and (|Fo| − |Fc|) electron density maps. Water molecules were added using the COOT autofind function with a lower cutoff of 3 σ in the |Fo| − |Fc| map. Metal ions were positioned into higher (|Fo| − |Fc|) residual densities and according to shorter bond distances with neighboring water molecules or other protein ligands. A Bijvoet difference Fourier map calculated from the final protein model was used to identify the metal binding sites. The final refinement was carried out using PHENIX software (51). The structure of the E173A variant was determined using the final model of the native SFER4, and the refinement strategy was the same as that for the native. Figs. 2–6 and 8 were produced by PyMOL (DeLano Scientific, San Carlos, CA).
FIGURE 2.
Ribbon diagram of the plant ferritin, SFER4. a and b, the whole oligomer viewed down a 3-fold and a 4-fold symmetry axis, respectively. The N-terminal extension peptide forms a loop and a short helix lies on the neighboring 3-fold symmetry mate subunit. c, stereoview of a superimposition of the soybean ferritin SFER4 subunit (blue) on the human ferritin H chain (red, PDB accession code 2FHA).
FIGURE 3.
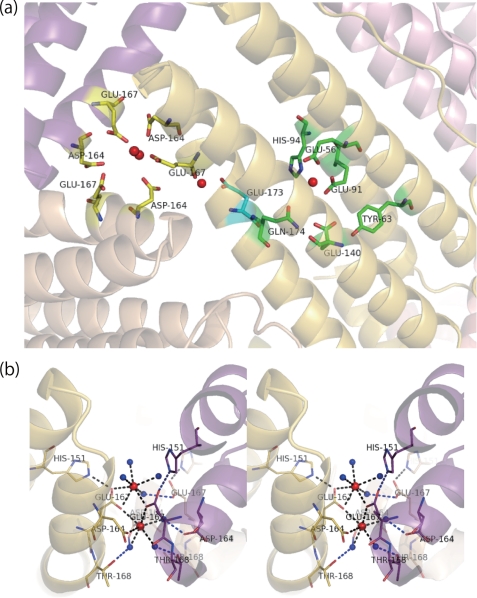
a, positions of the three calcium binding sites, 3-fold symmetry axis, ferroxidase site, and novel intermediate site coordinated by Glu173 in the ferritin shell. The calcium atoms are shown as red balls. Residues forming a metal binding site of the 3-fold axis, a novel “transit site,” and ferroxidase center are shown in yellow, cyan, and green, respectively. b, the structure of the deduced metal ion entry channel penetrating along the 3-fold symmetry axis and metal binding site in the channel. This channel is lined with hydrophilic side chains of Asp164, Glu167, and Thr168, and two calcium ions per channel are seen (in red). Waters ligated to the calcium ions or hydrogen bonded to the side chains are shown in blue. Blue broken lines, hydrogen bonds; black broken lines, metal coordination bonds. The channels formed by chain L (shown in “yellow”), M (purple), and T (beige) are represented from the view perpendicular to the axis (stereo view).
FIGURE 4.
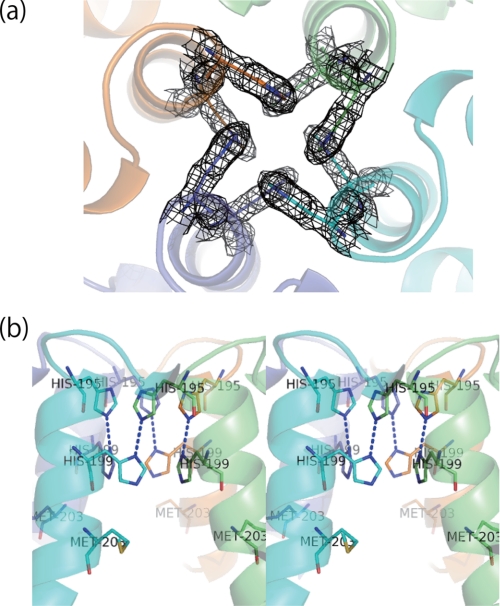
Inter-subunit interactions around the 4-fold symmetry axis of the SFER4. The channels are represented with two different orientations: (a) aligned on the 4-fold axis and (b) perpendicular to the axis (stereo view). The electron densities of the four lined histidines contoured at 1.5 σ are shown as a mesh in a. Blue broken lines, electrostatic interaction of histidine side chains.
FIGURE 5.
Metal binding site in the ferroxidase center of the SFER4 (a) and its variant E173A (b). A metal binding site positioned near the center (described below) is also shown a. The atom color code is black for calcium and gray for oxygen of water. Gray broken lines, hydrogen bonds; black broken lines, metal coordination bonds.
FIGURE 6.

A novel metal binding site coordinated by Glu173. A novel metal binding site is represented by coordination residues, Glu173 and Thr168, with the electron density (2|Fo| − |Fc|) contoured at 1.5σ shown as a mesh. The atom color code is black for calcium and gray for oxygen of water. Black broken lines indicate the metal coordination bonds.
FIGURE 8.
The hydrophilic route from the 3-fold channel to the ferroxidase site of the E173A variant (stereo view). This figure is the view from the inner cavity side. The ferroxidase site (Glu56, Glu91, His94, Glu140, and Gln174) of chain G and the 3-fold channel composed of chains G, F, and O of the E173A variant are illustrated. The 3-fold channel is lined with Asp164 and Glu167 of the three chains (left side). The hydrophilic route emerged because of the substitution (E173A) and the kink of the D-helix in Tyr170. The calcium atoms and water molecules are shown as black and gray balls, respectively.
Iron Uptake
Apoferritins of native SFER4 and the E173A variant were obtained using the methods described by Chasteen and Theil (52) with some modifications. Purified ferritins were dialyzed against 50 mm HEPES-Na buffer, pH 7.0, containing 1% thioglycolic acid, followed by successive changes of HEPES-Na buffer with (0.1%) or without thioglycolic acid. The protein was then dialyzed against Tris-HCl buffer containing 13 g/liter of Chelex 100 (Bio-Rad Laboratories) and 0.15 m NaCl, and finally dialyzed against 10 mm Tris-HCl, pH 7.5, containing 0.15 m NaCl. The concentration of purified proteins was determined using the absorbance at 280 nm.
Reactions examining iron uptake by native and the E173A variant were performed in 10 mm Tris-HCl, pH 7.5, containing 0.15 m NaCl with a Fe2+/ferritin molar ratio of 100:1, 40:1, and 20:1 (100, 40, and 20 μm ferrous ammonium sulfate and 1 μm ferritin) at 25 °C. Iron oxidation/nucleation by each type of ferritin was monitored by measuring the absorbance at 310 nm, the generation of μ-oxo-bridged Fe(III) species (16, 54) using a UV spectrophotometer (UV160, Shimadzu, Kyoto, Japan), and the resulting absorbance every 10 s for kinetic analysis. After each Fe(II) addition, several spectra were taken until the absorbance increased no further. The molar absorptivity of 4570 (cm−1 m−1), which was calculated for μ-oxo bridged Fe(III) in a iron excess condition on horse spleen ferritin by Yang et al. (21), was used to calculate the kinetic parameters. The kinetics data were analyzed with Excel 2007 software (Microsoft, Redmond, WA). The initial rates of iron oxidation were obtained from the linear A1 term of a third-order polynomial fitted to the experimental data (21, 55). The kinetic parameters were calculated by Lineweaver-Burk plots.
Data Deposition
Coordinates of plant ferritin SFER4 and its variant E173A have been deposited at the RCSB Protein Data Bank with accession codes 3A68 and 3A9Q, respectively.
RESULTS
Model Quality and Overall Structure of Plant Ferritin (SFER4) Subunit
The overall coordinate error of the final model, estimated from Luzzati plots, was 0.18 Å. Root mean square deviations of bond lengths and angles from ideality as calculated by the PHENIX program are listed in Table 1. These values are well within accepted limits, indicating that all of the structures have tight stereochemical constraints. A Ramachandran plot of the final structure showed that 98.43 and 1.57% of all residues were in the most favored and allowed regions, respectively. Refinement statistics are shown in Table 1.
The final model contained the whole oligomeric protein shell composed of 24 subunits with 4642 residues, 4570 waters, 136 Ca2+ ions, and 18 acetate ions. Like the structures of ferritins from vertebrates and bacteria, the structure of plant ferritin, SFER4, has a cage-like hollow shell composed of 24 subunits, which are related by 4-, 3-, and 2-fold symmetry (Fig. 2). There are four 3-fold axes and three 4-fold axes traversing the shell of the assembled 24-mer, resulting in eight 3-fold channels and six 4-fold channels. In the present model for SFER4, each subunit excludes the N-terminal 11 to 13 residues and the C-terminal 5 residues, because they have no electron density. Exceptionally, chain E excludes the first 17 residues. The N-terminal EP (from the N terminus to Thr30 in this model) forms a loop and a short helix that lies on the C-helix in a neighboring 3-fold symmetry mate (counterclockwise) (Fig. 2a), although the side chains of amino acids in the region are relatively disordered and have high temperature factors (Fig. 2). The interactions between the EP and the conserved helical region of an adjacent chain were formed mainly by apolar contacts, except for an ion pair formed between Glu20 and Lys146. Table 2 shows an example of side chains and residues related to the interactions between the EP of chain G and the helical region of chain O. The conserved region of each chain is composed of A-helix (from Asp43 to Asp71), B-helix (from Arg78 to Arg105), BC loop (from Gly106 to Gly127), C-helix (from Asp128 to Asn157), D-helix (from Val160 to Val191), and E-helix (from His195 to Glu207). The D-helix contains a typical kink at position Tyr170, which is required to open the 3-fold channel. These backbones are highly conserved among ferritins with known structures. For example, when superimposing a single chain of the SFER4 on a human H chain (PDB code 2FHA) (4), the average root mean square distance between Cα was 0.95 Å. The large deviation, except for the N-terminal EP, was observed at the end of the BC and DE loops due to the insertion and deletion of 4 and 3 residues, respectively (Fig. 2c).
TABLE 2.
Interactions between the extension peptide of the chain G and conserved helical regions of the chain O
The cutoff distances for C-C contacts and a hydrogen bonds are 4.20 and 3.25 Å, respectively.
| Extension peptide | Conserved helical regions | Distances |
|---|---|---|
| Å | ||
| Phe15 CZ | Leu142 (helix C) CD1 | 3.41 |
| Phe15 CE1 | Asn58 (helix A) CB | 3.81 |
| Phe15 CE2 | Lys146 (helix C) CE | 3.83 |
| Pro17 CG | Val62 (helix A) CG3 | 4.13 |
| Pro17 CB | Leu139 (helix C) CD1 | 4.00 |
| Phe18 CE2, CZ | Val62 (helix A) CG2, CG1 | 3.80, 3.59 |
| Phe18 CZ, CD1 | Leu118 (BC loop) CB, CG | 3.71, 3.68 |
| Phe18 CE1 | Leu135 (helix C) CD1 | 3.56 |
| Glu20 CB | Leu142 (helix C) CD2 | 4.19 |
| Val21 CG2 | Leu139 (helix C) CD1 | 3.98 |
| Leu25 CD2 | Leu135 (helix C) CD2 | 3.84 |
| Glu20 OE2 | Lys146 (helix C) NZ | 3.10 (charged hydrogen bond) |
The model showed 136 metal ions. Most had 6–7 coordination bonds with weak significant anomalous signals. Thus, they were interpreted as calcium ions, which were contained in the crystallization solution. These calcium ions can be categorized into 5 groups. First, in the outer surface of the ferritin molecule (22 Ca2+ ions); second, around the 3-fold symmetry channel deduced as an iron entry channel (18 Ca2+ ions); third, in the ferroxidase center located inside the 4-helix bundle (24 Ca2+ ions); fourth, on the inner surface of the ferritin shell containing two binding sites (48 Ca2+ ions); and fifth, in the novel intermediate site coordinated by Glu173 (24 Ca2+ ions). The three calcium ion binding sites, around the 3-fold symmetry axes, ferroxidase sites, and novel intermediate sites coordinated by the Glu173, are shown in Fig. 3a.
Calcium ions on the outer surface of the ferritin shell had seven coordinated waters, two of which were hydrogen bonded to the carboxylate oxygen atoms of Asp43 and Glu46, and one of which was hydrogen bonded to the acetate anion located between the side chains of Arg38 and Lys108. The acetate anions were also usually hydrogen bonded to the main chain nitrogen of Ser35. These outer calcium ions were not related to the inter-molecular contact, in contrast to vertebrate ferritin (4–7). Intermolecular contacts were formed mainly by hydrogen bonds or salt bridges between the main chains or side chains of residues positioned around the end of the C-helix (Ser152, Thr155, Lys156, and Asn157) and the side chains of Lys40, Asp43, Glu44, and Ser47 positioned around the start of the A-helix.
3-Fold Symmetry Axis
The hydrophilic channel penetrating along the 3-fold symmetry axis is considered the iron entry channel to the inner cavity of the ferritin shell (8–11). The channel of soybean ferritin, SFER4, had similar coordination with mammalian ferritins. As for the 3-fold channel of SFER4, the channel is lined with Asp164, Glu167, and Thr168 of three symmetrically related subunits (Fig. 3a, residues shown in yellow), corresponding to Asp131, Glu134, and Thr135 of the human H chain. In the crystal structure of SFER4, two electronic densities of calcium ions are found around the 3-fold symmetry axis in the bottleneck of the funnel-like channel leading to the cavity. One calcium ion positioned deeper inside the 3-fold symmetry channel is coordinated by OEs in the side chains of two symmetrically related Glu167, one OD of Asp164 and three waters (Fig. 3b). On the other hand, the other calcium ion, positioned outside the channel, is coordinated by an OE of Glu167 and three to four waters. The structure around the 3-fold channel composed of chain L, M, and T was unambiguously determined, whereas the outer calcium ions and its ligands in other channels were sometimes disordered. In the channel composed of chains L, M, and T, the inner calcium ion was coordinated by two ODs of Asp164 in chains L and T, two OEs of Glu167 in chains L and M, and three waters. On the other hand, the outer calcium is coordinated by two OEs of Glu167 of chains L and M plus four coordinated waters (Fig. 3b).
4-Fold Symmetry Axis
The E-helices of vertebrate ferritins lie roughly parallel with the 4-fold axis, making a hydrophobic channel traverse the shell. In the homo 24-mer of human H and L chains, this channel is formed by three layers. In the case of the H chain, the inner chain is lined with four histidines, whereas the middle and outer chains are each lined with four leucines. On the other hand, the channel in the human L chain is lined with 12 leucines, making it highly hydrophobic. In contrast, this channel in plant ferritin SFER4 is formed by histidine side chains (Fig. 4a) composed of two layers (Fig. 4b), because the side chain of Met203, which corresponds to the His173 or Leu169 positions in the inner layer of the human H chain or L chain, does not form the inner layer of the 4-fold channel. The two layers of SFER4 are composed of two histidines (His195 and His199) and their 4-fold symmetry related mates (Fig. 4b). Thus, the channel is lined by eight histidines. The NDs in the side chains of outer histidines (His195) are directed to the outer surface of the 4-fold channel, whereas the NEs of His195 electrostatically interact with the NDs of the inner histidine (His199) in a symmetrically related subunit (Fig. 4b). Consequently, the NEs of inner histidines are directed to the inner cavity of the protein shell. The distance between the two nitrogen atoms in histidine side chains is ∼2.9 Å. No electronic peaks of metal ion are seen around the channel, in contrast to other kinds of ferritins (17, 56).
Deduced Ferroxidase Center and Nucleation Center
In general, the ferroxidase center, located inside the four-helix bundle of each subunit, contains two metal binding sites, named A and B. It was suggested that the occupancy of site A is higher than that of site B (4). Concerning the SFER4, the ferroxidase center is composed of Glu56, Tyr63, Glu91, His94, Glu140, and Gln174, corresponding to Glu27, Tyr34, Glu62, His65, Glu107, and Gln141, respectively, of the human H chain ferritin (Fig. 1). In the native crystal structure of SFER4, metal binding was seen only in site A, although all amino acid residues of the ferroxidase center are conserved and the overall structure of this site is nearly identical. The metal binding site A is coordinated by Glu56, Glu91, His94, and three waters (Fig. 5a), showing that the coordination of calcium ion in this site is nearly identical with metal binding site A of the human H chain.
The putative nucleation center of SFER4 is composed of glutamates facing the inner cavity, Glu86, Glu89, Glu90, and Glu93, corresponding to Glu53, Glu56, Glu57, and Glu60 of the human L chain (Fig. 1). In the present model Glu90 has an alternative conformation, and these four residues tend to be disordered, as seen in crystallographic analysis of other L chains. The electron densities with high intensity (8 to 10 σ) are seen in the vicinity of the side chains of Glu93 and Glu96. Some of the peaks exhibited weak anomalous signals; therefore, they were considered calcium ions. In addition to these residues, Asp71, Glu86, Glu89, Glu90, Glu100, Glu169, Glu173, Glu176, and Glu183 are also facing the inner cavity, rendering it acidic. Calcium ions are also seen in the vicinity of Glu183, Asp71, and Lys179, all of which are facing the inner cavity.
Novel Metal Binding Site
A novel metal binding site is seen intermediate between the channel and ferroxidase center binding sites 3-fold. The site, which is positioned at the bottom of the 3-fold symmetry channel and facing the inner cavity of the ferritin shell, is coordinated by the OE of Glu173, carbonyl oxygen of Thr168, and four waters (Fig. 6). The distances from two OEs of Glu173 and the carboxyl oxygen of Thr168 are 2.48, 2.53, and 2.32 Å. This calcium ion binding site is seen clearly in all chains of the SFER4 crystal structure. The distance of this calcium ion from the inner calcium ion in the 3-fold symmetry axis is 9.7 Å, whereas that from the ferroxidase site A is 12.4 Å.
Biochemical Property of the E173A Mutant
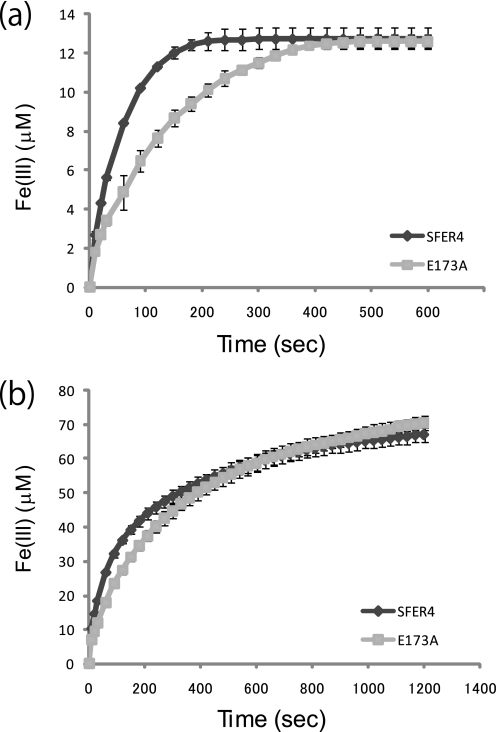
To assess the function of the Glu173 residue of SFER4, the E173A mutant, in which Glu173 was replaced with alanine, was prepared by the E. coli expression system, and ferroxidase/nucleation activity was compared with the native SFER4. The proper oligomeric formation of the E173A mutant was verified by gel filtration (data not shown). UV spectral absorption around 305–330 nm has been traditionally used to monitor the ferroxidase activity of ferritins. When adding ferrous ions to ferritin possessing a ferroxidase center such as the human H chain, the absorbance around 310 nm, which is characteristic of μ-oxo-bridged Fe(III) dimers (21, 57), increases as a result of the Fe(II) oxidation. Fig. 7 shows the time-dependent change in absorbance at 310 nm, when ferrous ions were added to SFER4 and the E173A mutant at molar ratios of 1:20 (20 μm ferrous ions was added to 1 μm of the SFER4 and E173A) (Fig. 7a) and 1:100 (Fig. 7b). The results show delays in the formation of the μ-oxo-bridged Fe(III) species in the E173A mutant compared with SFER4 in all conditions tested here at ferritin/iron ratios of 1:20, 1:40, and 1:100, whereas the final absorbencies were nearly identical (Fig. 7). The kinetic parameters were calculated as follows: Km(SFER4) = 31.9 μm, Km(E173A) = 69.6 μm, Vmax(SFER4) = 0.525 μm/s, Vmax(E173A) = 0.508 μm/s, Kcat(SFER4) = 0.525/s, and Kcat(E173A) = 0.508/s. The E173A mutant exhibited a specificity constant (Kcat/Km) 2.27-fold decrease compared with the value of the SFER4.
FIGURE 7.
Effect of E173A mutation on iron oxidation rates of plant ferritin SFER4. Progress curves of μ-oxo-bridged Fe(III) generation monitored at 310 nm upon the addition of ferrous iron sulfate ammonium to the SFER4 and its E173A variant at a protein concentration of 1 μm and iron concentrations of 20 (a) and 100 (b) μm. The formation of μ-oxo-bridged Fe(III) was calculated from absorbance at 310 nm. Data are given as the mean ± S.D. of at least three individual experiments.
DISCUSSION
Since 1991, when the three-dimensional structure of human H-type ferritin was solved (4), many structures of ferritin from various species have been investigated and iron storage mechanisms have been proposed. In the present study, we reported the three-dimensional structural model and novel metal binding site of plant ferritin from soybean. The results showed that the overall structure of plant ferritin, SFER4, was highly conserved among known structures of ferritins. It was proved that part of the plant-specific EP formed a loop and a short helix, although this region was highly disordered. This observation supports the prediction by Lobréaux et al. (42) that this region of pea ferritin would form a short helix named the P-helix, and it would lie on the outer surface of the assembled ferritin shell. They also predicted that the P-helix was folded back to the surface of the original subunit. However, the present model shows that this N-terminal helix is interacting with a neighboring subunit on the shell surface (see Fig. 2 and Table 2). It has been suggested that the N-terminal EP stabilizes the whole oligomeric conformation of plant ferritin (39–41). The interactions between the EP and the shell surface were formed mainly via apolar contacts. The model demonstrates that seven residues contribute to the interactions, in which four residues (Leu135, Leu139, Leu142, and Lys146) are members of the C-helix (Table 2). Thus, it can be considered that the EP stabilizes the protein shell by maintaining the conformation of the C-helix. It has been suggested that interaction between the conserved Leu110 in the C-helix and Leu134 in the D-helix of the bullfrog H chain, which correspond to Leu147 and Leu171 of SFER4 (Fig. 1), plays a crucial role in stabilization of the 3-fold channel and that the mutation of these residues enhances the iron release from the ferritin mineral core (58–60). The EP may contribute to tight packing of the 3-fold pore conformation by stabilizing the C-helix. This hypothesis is based on the faster rate of iron incorporation of plant ferritin with EP deletion than that of the native protein (40). It is possible that the removal of EP accelerates not only the rate of iron incorporation but also that of iron release from the ferritin shell. Concerning the function of the EP, Li et al. (61) recently suggested that the EP has iron binding capacity and facilitates iron incorporation into the ferritin shell. However, there is no metal binding site in the EP of the present SFER4 model. To elucidate the function of EP in iron binding, structural analysis of a iron derivative of plant ferritin is required. However, we have not yet obtained crystals of the plant ferritins soaked with iron.
Another unique feature of plant ferritin is its 4-fold symmetry channel. The present model of SFER4 indicates that this narrow channel is lined with eight tightly packed histidine side chains, resulting in formation of a hydrogen bond network from inside to outside the ferritin shell (Fig. 4). In contrast, in mammalian ferritins the channel is lined with 12 leucines or 8 leucines plus 4 histidines (4, 5). Takahashi and Kuyucak (62) suggested that the 4-fold channel functions as a proton channel that facilitates hydrogen ion transfer in and out of the human H ferritin shell. As a result of the ferroxidase reaction, the proton concentration in the inner cavity increases, which leads to protonation of NEs of the His199 side chain facing the inner cavity. The proton can be transferred by sequential protonation/deprotonation of His199 and His195 side chains to be exported from the shell. Thus, from a structural point of view, it can be considered that the 4-fold symmetry axis of plant ferritin is more feasible than that of the mammalian ferritin if this channel functions as a pathway for proton transfer. The histidine residues (His195 and His199) lining the channel are highly conserved among plant ferritins, suggesting that the histidine cluster forming the 4-fold channel is a common feature of plant ferritin species.
The structures of the key sites for the iron storage process of ferritin, the 3-fold iron entry channel, the ferroxidase center, and the nucleation center were proven to be highly conserved among the SFER4 and vertebrate ferritins. In the present model of SFER4, metal binding in the ferroxidase site was seen only in site A (see Fig. 5a). In general, the ferroxidase center is composed of two metal binding sites, A and B (4, 5, 63). However, x-ray crystallographic analysis of non-heavy atom derivatives indicated that site B was sometimes vacant and that an alkaline-earth metal such as calcium or magnesium bound only to site A (56, 57). Because many studies concerning ferroxidase activity of plant ferritin have suggested that plant ferritin species have ferroxidase activity comparable with that of mammalian or bacterial ferritin (30, 40, 64), the ferroxidase site of SFER4 can be considered functional.
Recently, the roles of the carboxyl side chains facing the inner cavity of ferritins have been discussed. It was suggested that the acidic side chains play a critical role in Fe(III) nucleation on the cavity surface (14, 65, 66). In contrast, Bou-Abdallah et al. (67) demonstrated that the core formation rate was not affected by deletion of the inner glutamate residues (Glu64 and Glu67), which were considered the nucleation site of human H ferritin. In the present model of SFER4, in addition to the traditional nucleation site Glu93/Glu96, a calcium ion was seen in the vicinity of Asp71 and Glu183 facing the inner cavity. Asp71 and Glu183 of SFER4 correspond to Asp42 and Asp150 of the human H chain (Fig. 1). It is possible that this site functions as the second nucleation site for Fe(III) core formation and that the traditionally identified nucleation site is not the sole nucleation site. In fact, these residues, Asp42 and Asp150, were maintained in the experiments performed by Bou-Abdallah (67). Furthermore, Levi et al. (14) suggested that disruption of Asp42 was required to abolish the iron incorporation activity of human H chain ferritin. These observations also support the hypothesis that the putative nucleation site is not the sole nucleation site. The contribution of each acidic side chain on the cavity surface should be determined.
The pathway from the iron entry channel via the ferroxidase center to the nucleation site has been investigated by x-ray crystallographic analysis and site-directed mutagenesis (4, 7, 56, 68, 69). In the present SFER4 model, two calcium ions were seen around the iron entry channel (Fig. 3b), showing that amino acid residues related to the coordination with calcium ions were Asp164 and Glu167, which correspond to Asp131 and Glu134 of human H chain, and to Asp127 and Glu130 of human L chain, respectively (Fig. 1). Grainer et al. (68) suggested that the alternative conformation of Asp127 and Glu130 in the human L chain were important in assisting the metal movement from outside to inside through the channel. Similarly, Toussaint et al. (56) observed metal binding to Cys130 positioned at the entrance to the 3-fold channel and suggested that the alternative configuration of this residue facilitated metal entry into the channel. As for the movement from the ferroxidase site to the nucleation site, x-ray crystallographic analysis of human H chain has shown that Glu61 had an alternative conformation and served to transport metal ions from the ferroxidase site to the nucleation site (4, 5). Furthermore, Stillman et al. (7) indicated that the alternative conformation of His46 of ferritin from E. coli (EcFTN), which is a highly conserved amino acid residue among prokaryotic ferritin, enabled the metal to move from the ferroxidase center. In the present model of plant ferritin SFER4, a novel metal binding site, Glu173, was discovered between the two well defined metal binding sites: the iron entry channel and the ferroxidase site (Fig. 3a). To date, few studies have described the pathway from the metal binding site in the 3-fold channel to the ferroxidase center. We postulate that this residue, Glu173 of the SFER4, functions as a transit site from the metal entrance to the ferroxidase site. This assumption was supported by the difference in ferroxidase activity of native SFER4 and the E173A mutant, whose ferroxidase site was not disrupted by the mutation (Fig. 5b). The Km value for iron oxidation of the mutant was increased to 2.2-fold compared with that of native SFER4. The delay in iron oxidation in the E173A mutant was more prominent at low iron concentrations (ferritin/iron = 1:20) (Fig. 7a), whereas the difference in activity diminished as the iron concentration increased (ferritin/iron = 1:100) (Fig. 7b). These results, together with the fact that Glu173 is not involved in iron oxidation in the ferroxidase site (Fig. 5a), indicate that the side chain of this residue is required for mechanical transfer of the metal ions from the entry channel to the ferroxidase site, especially under ordinary physiological conditions in vivo. For instance, iron atoms enter from the 3-fold channels and interact with Glu167 and/or Asp164 and are then translocated from Asp164 or Thr168 to Glu173. In the crystal structure of native SFER4, calcium ions are seen coordinated by the side chain of Glu173 and the main chain oxygen of Thr168 (Fig. 6). In another orientation, the side chain of Glu173 can interact with the side chains of Gln174 and His94, which are part of the ferroxidase site of SFER4. On the other hand, in the crystal structure of the E173A variant, no metal ion was seen adjacent to Ala173. But, a hydrophilic route emerged between Thr168 (deep in the entry channel) and the ferroxidase site. This hydrophilic route crossing the D-helix is positioned on the main chain oxygen of Thr168 and Glu169, which do not hydrogen bond with the main chain nitrogen of Gly172 and Ala173 because of the kink in the D-helix (Tyr170) (Fig. 8). This hydrophilic route of the mutant may enable the diffusion of metal ions from the entry channel to the ferroxidase site, especially in the presence of a high concentration of metal ions (Fig. 7b).
The Glu173 of SFER4 is conserved not only among all known members of plant ferritins such as soybean, pea, maize, tobacco, and Arabidopsis (22, 25, 28, 30, 44), but also among ferritins from eukaryotes, except the mouse L chain ferritin (Fig. 1). In the well characterized structure of human H ferritin, conformation around Glu140 (Glu173 of the SFER4) is nearly identical to that of SFER4. Therefore, Glu140 of human H ferritin can also contribute to the mechanical transfer of metal ions. However, further investigation into the function of the glutamate residue using vertebrate ferritins is required. In prokaryotic ferritin, this glutamate residue is also conserved in non-heme binding bacterial ferritin, such as EcFTN (from E. coli) (Fig. 1). In contrast, the corresponding glutamate is substituted with a lysine (Lys140) in the mouse L chain. Santanbrogio et al. (53) suggested that this substitution causes the low iron incorporation activity of the mouse L chain compared with the human L chain, which has a Glu140, corresponding to the Glu173 of SFER4. This observation also supports the significance of the Glu173 side chain in iron traffic in ferritin.
Thus, it is possible that the function of this residue, Glu173 of SFER4, as a transit site from the iron entry channel to the ferroxidase site, is related to a universal mechanism in iron translocation in ferritin. This finding provides evidence that a common pathway does exist from the iron entry channel to the ferroxidase center in ferritin molecules in various living kingdoms.
Acknowledgments
We thank Prof. Evelyn M. Tecson-Mendoza (University of the Philippines, Los Banos) for critical reading of the manuscript. The synchrotron radiation experiments were performed at the BL44XU and BL38B1 in the SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal numbers 2008B6828 and 2009A1249).
This work was supported a grant from the Shorai Foundation for Science and Technology (to T. M.).
The atomic coordinates and structure factors (codes 3A68 and 3A9Q) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- H
- heavy
- L
- light
- EP
- extension peptide.
REFERENCES
- 1.Harrison P. M., Arosio P. (1996) Biochim. Biophys. Acta 1275, 161–203 [DOI] [PubMed] [Google Scholar]
- 2.Theil E. C. (1987) Annu. Rev. Biochem. 56, 289–315 [DOI] [PubMed] [Google Scholar]
- 3.Arosio P., Ingrassia R., Cavadini P. (2009) Biochim. Biophys. Acta 1790, 589–599 [DOI] [PubMed] [Google Scholar]
- 4.Lawson D. M., Artymiuk P. J., Yewdall S. J., Smith J. M., Livingstone J. C., Treffry A., Luzzago A., Levi S., Arosio P., Cesareni G., Thomas C. D., Shaw W. V., Harrison P. M. (1991) Nature 349, 541–544 [DOI] [PubMed] [Google Scholar]
- 5.Hempstead P. D., Yewdall S. J., Fernie A. R., Lawson D. M., Artymiuk P. J., Rice D. W., Ford G. C., Harrison P. M. (1997) J. Mol. Biol. 268, 424–448 [DOI] [PubMed] [Google Scholar]
- 6.Granier T., Comberton G., Gallois B., d'Estaintot B. L., Dautant A., Crichton R. R., Précigoux G. (1998) Proteins 31, 477–485 [PubMed] [Google Scholar]
- 7.Stillman T. J., Hempstead P. D., Artymiuk P. J., Andrews S. C., Hudson A. J., Treffry A., Guest J. R., Harrison P. M. (2001) J. Mol. Biol. 307, 587–603 [DOI] [PubMed] [Google Scholar]
- 8.Stefanini S., Desideri A., Vecchini P., Drakenberg T., Chiancone E. (1989) Biochemistry 28, 378–382 [DOI] [PubMed] [Google Scholar]
- 9.Treffry A., Harrison P. M., Luzzago A., Cesareni G. (1989) FEBS Lett. 247, 268–272 [DOI] [PubMed] [Google Scholar]
- 10.Yablonski M. J., Theil E. C. (1992) Biochemistry 31, 9680–9684 [DOI] [PubMed] [Google Scholar]
- 11.Levi S., Santambrogio P., Corsi B., Cozzi A., Arosio P. (1996) Biochem. J. 317, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arosio P., Adelman T. G., Drysdale J. W. (1978) J. Biol. Chem. 253, 4451–4458 [PubMed] [Google Scholar]
- 13.Leibold E. A., Aziz N., Brown A. J., Munro H. N. (1984) J. Biol. Chem. 259, 4327–4334 [PubMed] [Google Scholar]
- 14.Levi S., Yewdall S. J., Harrison P. M., Santambrogio P., Cozzi A., Rovida E., Albertini A., Arosio P. (1992) Biochem. J. 288, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson D. M., Treffry A., Artymiuk P. J., Harrison P. M., Yewdall S. J., Luzzago A., Cesareni G., Levi S., Arosio P. (1989) FEBS Lett. 254, 207–210 [DOI] [PubMed] [Google Scholar]
- 16.Levi S., Luzzago A., Cesareni G., Cozzi A., Franceschinelli F., Albertini A., Arosio P. (1988) J. Biol. Chem. 263, 18086–18092 [PubMed] [Google Scholar]
- 17.Ha Y., Shi D., Small G. W., Theil E. C., Allewell N. M. (1999) J. Biol. Inorg. Chem. 4, 243–256 [DOI] [PubMed] [Google Scholar]
- 18.Hamburger A. E., West A. P., Jr., Hamburger Z. A., Hamburger P., Bjorkman P. J. (2005) J. Mol. Biol. 349, 558–569 [DOI] [PubMed] [Google Scholar]
- 19.Frolow F., Kalb A. J., Yariv J. (1994) Nat. Struct. Biol. 1, 453–460 [DOI] [PubMed] [Google Scholar]
- 20.Chasteen N. D., Harrison P. M. (1999) J. Struct. Biol. 126, 182–194 [DOI] [PubMed] [Google Scholar]
- 21.Yang X., Chen-Barrett Y., Arosio P., Chasteen N. D. (1998) Biochemistry 37, 9743–9750 [DOI] [PubMed] [Google Scholar]
- 22.Lobreaux S., Massenet O., Briat J. F. (1992) Plant Mol. Biol. 19, 563–575 [DOI] [PubMed] [Google Scholar]
- 23.Wicks R. E., Entsch B. (1993) Biochem. Biophys. Res. Commun. 192, 813–819 [DOI] [PubMed] [Google Scholar]
- 24.Wardrop A. J., Wicks R. E., Entsch B. (1999) Biochem. J. 337, 523–530 [PMC free article] [PubMed] [Google Scholar]
- 25.Petit J. M., Briat J. F., Lobréaux S. (2001) Biochem. J. 359, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda T., Goto F., Yoshihara T. (2001) J. Biol. Chem. 276, 19575–19579 [DOI] [PubMed] [Google Scholar]
- 27.Strozycki P. M., Skapska A., Szcześniak K., Sobiezcruzuk E., Briat J. F. (2003) Physiol. Plant. 118, 380–389 [Google Scholar]
- 28.Jiang T. B. (2005) J. Integr. Plant Biol. 47, 477–486 [Google Scholar]
- 29.Dong X., Sun Q., Wei D., Li J., Li J., Tang B., Jia Q., Hu W., Zhao Y., Hua Z. C. (2007) FEBS Lett. 581, 5796–5802 [DOI] [PubMed] [Google Scholar]
- 30.Li C., Hu X., Zhao G. (2009) Biochimie 91, 230–239 [DOI] [PubMed] [Google Scholar]
- 31.Fobis-Loisy I., Loridon K., Lobréaux S., Lebrun M., Briat J. F. (1995) Eur. J. Biochem. 231, 609–619 [DOI] [PubMed] [Google Scholar]
- 32.Lescure A. M., Proudhon D., Pesey H., Ragland M., Theil E. C., Briat J. F. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8222–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaymard F., Boucherez J., Briat J. F. (1996) Biochem. J. 318, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savino G., Briat J. F., Lobréaux S. (1997) J. Biol. Chem. 272, 33319–33326 [DOI] [PubMed] [Google Scholar]
- 35.Wei J., Theil E. C. (2000) J. Biol. Chem. 275, 17488–17493 [DOI] [PubMed] [Google Scholar]
- 36.Petit J. M., van Wuytswinkel O., Briat J. F., Lobréaux S. (2001) J. Biol. Chem. 276, 5584–5590 [DOI] [PubMed] [Google Scholar]
- 37.Briat J. F., Labouré A. M., Laulhère J. P., Lescure A. M., Lobréaux S., Pesey H., Proudhon D., Wuytswinkel O. (1995) in Iron Nutrition in Soils and Plants (Abadía J., ed) pp. 265–276, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 38.Ragland M., Briat J. F., Gagnon J., Laulhere J. P., Massenet O., Theil E. C. (1990) J. Biol. Chem. 265, 18339–18344 [PubMed] [Google Scholar]
- 39.Laulhere J. P., Laboure A. M., Briat J. F. (1989) J. Biol. Chem. 264, 3629–3635 [PubMed] [Google Scholar]
- 40.van Wuytswinkel O., Savino G., Briat J. F. (1995) Biochem. J. 305, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Wuytswinkel O., Briat J. F. (1995) Biochem. J. 305, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobreaux S., Yewdall S. J., Briat J. F., Harrison P. M. (1992) Biochem. J. 288, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda T., Mikami B., Goto F., Yoshihara T., Utsumi S. (2003) Biochim. Biophys. Acta 1645, 113–115 [DOI] [PubMed] [Google Scholar]
- 44.Masuda T., Goto F., Yoshihara T., Ezure T., Suzuki T., Kobayashi S., Shikata M., Utsumi S. (2007) Protein Expr. Purif. 56, 237–246 [DOI] [PubMed] [Google Scholar]
- 45.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 47.Vagin A., Teplyakov A. (1997) J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 48.Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 49.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 51.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 52.Chasteen N. D., Theil E. C. (1982) J. Biol. Chem. 257, 7672–7677 [PubMed] [Google Scholar]
- 53.Santambrogio P., Cozzi A., Levi S., Rovida E., Magni F., Albertini A., Arosio P. (2000) Protein Expr. Purif. 19, 212–218 [DOI] [PubMed] [Google Scholar]
- 54.Pâgues E., Pâques A., Crichton R. R. (1980) Eur. J. Biochem. 107, 447–453 [PubMed] [Google Scholar]
- 55.Zhao G., Bou-Abdallah F., Arosio P., Levi S., Janus-Chandler C., Chasteen N. D. (2003) Biochemistry 42, 3142–3150 [DOI] [PubMed] [Google Scholar]
- 56.Toussaint L., Bertrand L., Hue L., Crichton R. R., Declercq J. P. (2007) J. Mol. Biol. 365, 440–452 [DOI] [PubMed] [Google Scholar]
- 57.Langlois d'Estaintot B., Santambrogio P., Granier T., Gallois B., Chevalier J. M., Précigoux G., Levi S., Arosio P. (2004) J. Mol. Biol. 340, 277–293 [DOI] [PubMed] [Google Scholar]
- 58.Jin W., Takagi H., Pancorbo B., Theil E. C. (2001) Biochemistry 40, 7525–7532 [DOI] [PubMed] [Google Scholar]
- 59.Liu X., Jin W., Theil E. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3653–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theil E. C., Liu X. S., Tosha T. (2008) Inorg. Chim. Acta 361, 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li C., Fu X., Qi X., Hu X., Chasteen N. D., Zhao G. (2009) J. Biol. Chem. 284, 16743–16751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi T., Kuyucak S. (2003) Biophys. J. 84, 2256–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., Theil E. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8557–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laulhere J. P., Briat J. F. (1993) Biochem. J. 290, 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C. Y., Meagher A., Huynh B. H., Sayers D. E., Theil E. C. (1987) Biochemistry 26, 497–503 [DOI] [PubMed] [Google Scholar]
- 66.Levi S., Santambrogio P., Cozzi A., Rovida E., Corsi B., Tamborini E., Spada S., Albertini A., Arosio P. (1994) J. Mol. Biol. 238, 649–654 [DOI] [PubMed] [Google Scholar]
- 67.Bou-Abdallah F., Biasiotto G., Arosio P., Chasteen N. D. (2004) Biochemistry 43, 4332–4337 [DOI] [PubMed] [Google Scholar]
- 68.Granier T., Langlois d'Estaintot B., Gallois B., Chevalier J. M., Précigoux G., Santambrogio P., Arosio P. (2003) J. Biol. Inorg. Chem. 8, 105–111 [DOI] [PubMed] [Google Scholar]
- 69.Stillman T. J., Connolly P. P., Latimer C. L., Morland A. F., Quail M. A., Andrews S. C., Treffry A., Guest J. R., Artymiuk P. J., Harrison P. M. (2003) J. Biol. Chem. 278, 26275–26286 [DOI] [PubMed] [Google Scholar]