Abstract
Pathogenic Yersinia species neutralize innate immune mechanisms by injecting type three secretion effectors into immune cells, altering cell signaling. Our study elucidates how one of these effectors, YopO, blocks phagocytosis. We demonstrate using different phagocytic models that YopO specifically blocks Rac-dependent Fcγ receptor internalization pathway but not complement receptor 3-dependent uptake, which is controlled by Rho activity. We show that YopO prevents Rac activation but does not affect Rac accumulation at the phagocytic cup. In addition, we show that plasma membrane localization and the guanine-nucleotide dissociation inhibitor (GDI)-like domain of YopO cooperate for maximal anti-phagocytosis. Although YopO has the same affinity for Rac1, Rac2, and RhoA in vitro, it selectively interacts with Rac isoforms in cells. This is due to the differential localization of the Rho family G proteins in resting cells; Rac isoforms partially exist as a GDI-free pool at the membrane of resting cells, whereas RhoA is trapped in the cytosol by RhoGDIα. We propose that YopO exploits this basic difference in localization and availability to selectively inhibit Rac-dependent phagocytosis.
Keywords: Cell/Phagocytosis, Cytoskeleton/Actin, G Proteins/Low Molecular Weight, Immunology/Innate Immunity, Methods/Confocal Microscopy, Protein/Protein-Protein Interactions
Introduction
Phagocytosis is a multistep process displayed by a variety of cell types but is particularly efficient in professional phagocytes (e.g. macrophages). It involves recognition, internalization, and degradation of particulate material over 0.5 μm in diameter in a membrane-bound compartment called the phagosome. The first critical step during classical, zipper-like phagocytosis is the ligation of cell surface receptors (1). The best studied receptors are the opsono-receptors, Fcγ receptor (FcγR)2 and complement receptor 3 (CR3), which bind immobilized immunoglobulin and surface-deposited C3bi, respectively. Following ligation, receptors are thought to cluster around the phagocytic target and thereby initiate downstream signaling. This involves recruitment and activation of mediators that ultimately result in actin nucleation and polymerization, through activation of Arp2/3, an event crucial to particle engulfment (2). Receptor-induced actin polymerization drives the wrapping of membrane around the particle in a “zipper-like” manner, generally through the advancement of pseudopodia (3). Membrane delivery from intracellular membrane compartments is also required for completion of internalization (4). Phagocytosis, like most processes reliant upon actin dynamics, is controlled through the action of Rho family small G proteins. Indeed, Rho proteins link receptor ligation and actin polymerization in all phagocytic events studied so far (5). However, different receptors employ distinct Rho proteins; RhoA activity is essential for internalization through CR3, whereas Rac1 and Cdc42 act downstream of FcγR (6).
Rho family members act as molecular switches in eukaryotic cells, cycling between inactive GDP-bound and active GTP-bound states (5). Guanine nucleotide exchange factors facilitate exchange of GDP for GTP, thereby activating small G proteins and allowing their GTP-dependent interaction with downstream effectors. The intrinsically low catalytic ability of Rho GTPases is greatly increased by interaction with GTPase-activating proteins, leading to the recycling of the G protein to its GDP-bound inactive form. A further level of regulation comes from the trafficking of the Rho proteins between different cellular compartments. The current models assign the Rho guanine dissociation inhibitors (RhoGDIs) a key function in keeping the Rho proteins in an inactive cytosolic pool. Upon activation, the G proteins are released from RhoGDI and recruited to membranes through their prenylated C termini (7). All Rho family members are thought to be regulated in a similar manner. Unsurprisingly, many bacterial effectors that manipulate phagocytic signaling do so by deregulating the activity of Rho GTPases (8, 9).
Gram-negative bacteria commonly subvert phagocytic uptake through the action of protein effectors that are directly injected into the host cell via a needle-like multiprotein complex called the type III secretion system (T3SS) (10). Yersinia species (Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica) are important extracellular human pathogens that utilize the T3SS injectosome to translocate a number of effector proteins into phagocytic cells (macrophages, dendritic cells, and neutrophils), which in turn block bacterial uptake (11, 12). The effector YopO, an 82-kDa multidomain protein, was reported to trigger apoptosis, prevent cytokine secretion and nitric oxide production in epithelial cells and yeasts, modulate the actin cytoskeleton (typically with loss of actin stress fibers), and importantly inhibit phagocytosis (13–16). Indeed, unopsonized and IgG-coated yopO Yersinia mutants are internalized at a higher level than wild-type bacteria by mouse macrophages (11, 15).
At a molecular level, YopO can be subdivided into four functional domains. The N-terminal region of YopO functions as the chaperone-binding site prior to translocation through the T3SS and mediates plasma membrane localization after injection into host cells (17, 18). It is flanked by a serine/threonine kinase domain recently shown to phosphorylate the heterotrimeric G protein subunit Gαq (19). The C-terminal region of YopO contains a GDI-like domain able to bind Rac and Rho in vitro, a property supported by the recently solved crystal structure of this domain (20, 21). Finally, a short coronin homology domain is proposed to bind G-actin and promote kinase activity (13, 22).
Despite the extensive analysis of YopO, its mechanism of action and the role of its individual domains remain poorly understood. Herein, we use an in vitro model of phagocytosis to study YopO-mediated anti-phagocytosis and the mechanism thereof. We found that the N-terminal membrane-addressing region in conjunction with the GDI-like domain specifically targets the Rac-mediated phagocytic pathway. Importantly, we found that YopO executes its anti-phagocytic activity by targeting pre-existing GDI-free Rac1 at the plasma membrane.
EXPERIMENTAL PROCEDURES
Expression Vectors
Eukaryotic plasmids encoding wild-type YopO (pEGFP::yopO, encoding YopOwt) and N-terminal truncation mutants (pEGFP::yopOΔ20–77, encoding YopO Δ20–77) were described previously (18). pEGFP::yopO435–729 (encoding YopORBD) was constructed from pEGFP::yopO using Expand Long Template PCR system (Roche Applied Science) and primers 5′-GGGGGGCTCGAGAGGCGGATAACACCCAAG-3′ and 5′-GGGGGGGAATTCTTATCACCATTCCCGCTCCAACCGGTTCAG-3′ and cloned into pEGFP-C3 (Clontech). Similarly, pEGFP::yopOΔ435–720 (encoding YopOΔRBD) and pEGFP::yopOD267AΔ435–720 (encoding YopOKDΔRBD) were constructed from pEGFP::yopO and pEGFP::yopOD267A, respectively, using primers 5′- GGGGGGACGCGTCGACTGAACCGGTTCGAGCGGGAATCG-3′ and 5′-GGGGGGACGCGTCCTTACATCAGTAGATAATGGGCTCATTTC-3′. pGEX::yopO435–729 was constructed using the following primers: forward, 5′-GGGGGGGGATCCAGGCGGATAACACCCAAG-3′, and reverse 5′-GGGGGGGAATTCTTATCACCATTCCCGCTCCAACCGGTTCAG-3′. pEGFP::yopOΔ83–434 (encoding YopO Δkinase) was constructed by inverse PCR from pEGFP::yopOwt using primers 5′-GGGGGGACGCGTAGGCGGATAACACCCAAGAAGCTTCG-3′ and 5′-GGGGGGACGCGTGAGTGTCTCAGTAAGGTTACGAGC-3′ and the Expand Long Template PCR kit according to manufacturer's instructions. All constructs were checked by sequencing (MWG Biotec) and prepared for transfection using the Qiagen Endo-free maxi-prep kit. Eukaryotic expression vectors encoding FcγRIIA and CR3 (αM and β2 integrin chains) have been described previously (6). pGEX plasmids encoding GST-tagged Rac1, Rac2, and RhoA, were the kind gift of K. Rittinger. pGEX plasmids encoding GST-tagged PAK-CRIB and Rhotekin have been previously described (23). Unique domain of Lck-GFP was a kind gift of MJ Bijlmakers (Division of Immunology, Infection, and Inflammatory Disease, King's College London, London, UK) (24).
Cell Culture and Transfection
African green monkey COS-7 fibroblast, J774A.1, and RAW264.7 mouse macrophage cell lines were obtained from the ATCC and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Sigma), penicillin (Sigma, 100 units/ml), and streptomycin (Sigma, 0.1 mg/ml) at 37 °C, 5% CO2. COS-7 cells were transiently transfected using Lipofectamine 2000 (Invitrogen), with a 0.8:2 DNA:Lipofectamine 2000 ratio, according to the manufacturer's instructions. RAW264.7 cells were transiently transfected using a 2:3 DNA:Lipofectamine 2000 (Invitrogen) ratio or by electroporation (AMAXA, Nucleofactor V, program D-32) according to the manufacturer's instructions. For pulldown experiments, 9 μg of DNA was used to transfect 3 × 105 COS-7 cells using the SuperFect (Qiagen) reagent according to manufacturer's instructions.
For small interfering RNA transfection, COS-7 cells were seeded at 5 × 105 cells per 10-cm dish. The following day, cells were transfected with 5 nm luciferase or RhoGDIα duplexes (SMARTpools, Dharmacon) using INTERFERinTM (Polyplus transfection) according to the manufacturer's instructions. Cells were left for 48 h before use.
Phagocytosis Assays
Transfected COS-7 or RAW264.7 cells were first preincubated for at least 1 h in serum-free Dulbecco's modified Eagle's medium with 10 mm Hepes (Sigma). To activate the endogenous CR3 receptors, RAW264.7 cells were pretreated with phorbol 12-myristate 13-acetate (150 ng/ml, Sigma) for 15 min at 37 °C. Cells were challenged with 0.5 μl of opsonized sheep red blood cells (sRBC, TCS Biosciences) per 13-mm coverslips for 30 min, washed with phosphate-buffered saline to remove unbound sRBC, and fixed with 4% paraformaldehyde for 15 min at 4 °C. Opsonization with IgG or C3bi was performed as described previously (6). Coverslips were then washed and fixed as described above.
Immunofluorescence and Scoring
Binding and phagocytosis were distinguished by differential staining of sRBC before or after permeabilization with 0.2% Triton X-100, using combinations of Cy5-, rhodamine-, and 7-amino-4-methylcoumarin-3-acetic acid-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch). Coverslips were inverted onto glass slides on 5 μl of Mowiol 4-88 (Calbiochem) and observed using Olympus BX50 epifluorescence or LSM510 Zeiss Axiovert 100 M confocal microscopes. In total, 100 COS-7 cells (50 RAW264.7 cells) or phagosomes (representing 10–20 cells) were counted for each experiment. Association and phagocytic indices (AI/PI) reflect the total number of sRBC or bacteria associated to or internalized by 100 transfected cells, respectively. % internalization corresponds to (AI/PI) × 100. At least three experiments were performed per condition. Student's t tests for significance were performed as indicated, and p values <0.05 are shown.
Protein Expression and GST Pulldown Assay
Expression of YopO435-729, Rac1, Rac2, and RhoA in fusion with GST was induced in subcultures of Escherichia coli BL21 (DE3) using 0.5 mm isopropyl β-d-thiogalactopyranoside. After 4 h at 30 °C, bacteria were resuspended in 50 mm Tris, pH 8, 40 mm EDTA, 25% sucrose, 100 mm MgCl2, 0.2% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and Complete Protease inhibitor mixture (Roche Applied Science). To purify GST-Rac1, -Rac2, and -RhoA, bacteria were resuspended in lysis buffer (50 mm Tris, pH 7.5, 300 mm NaCl, 4 mm dithiothreitol, and EDTA-free protease inhibitors (Roche Applied Science)) and sonicated. After lysate clearing by ultracentrifugation, fusion proteins were affinity-purified on glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions.
J774A.1 or transfected COS-7 cells were lysed in ice-cold lysis buffer (10% glycerol, 1% Nonidet P-40, 50 mm Tris, pH 7.6, 200 mm NaCl, 10 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, Roche Applied Science, complete tablet). Lysates were incubated with glutathione-Sepharose beads coupled to 25 μg of either GST or GST-RBD for 1 h at 4 °C. Beads were washed three times in cold lysis buffer before analysis by SDS-PAGE and Western blotting. The following primary antibodies were used: anti-Rac (23A8, Upstate), anti-RhoA (119, Santa Cruz Biotechnology), anti-Myc (4A6, Upstate), and β-tubulin (TUB2.1, Sigma). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Amersham Biosciences) were used as appropriate, and bands were detected using the enhanced chemiluminescence kit (ECL, Amersham Biosciences). G protein loading was achieved by addition of GDP or GTPγS (100 μm) and EDTA (5 mm) followed by incubation at 30 °C for 10 min and addition of MgCl2 (50 mm). For mass spectrometry, bead eluates were separated by SDS-PAGE and stained using a compatible silver nitrate protocol as described previously (25).
Cell Fractionation
J774A.1 macrophages were seeded the day before fractionation, washed once with cold phosphate-buffered saline, resuspended in lysis buffer (10 mm Tris, pH 7.5, 0.5 mm dithiothreitol, 5 mm NaCl, Roche Applied Science, complete inhibitor), and sonicated on ice for 30 s. Post-nuclear supernatants were subjected to ultracentrifugation at 100,000 × g for 45 min at 4 °C. The supernatant (cytosol fraction) and the pellet (membrane fraction) were resuspended in equal volumes of lysis buffer supplemented with 1% Triton X-100.
Mass Spectrometry (MS) Analysis
In preparation for mass spectrometry, the desired protein band was excised, lyophilized, and digested with trypsin (EC 3.4.21.4, Promega) overnight. Peptides were extracted from gel pieces, and nano-liquid chromatography was performed on an Ultimate 3000 using a PepMap 100 75 μm × 15-cm fused silica C18 analytical column (LC Packings, Dionex, Sunnyvale, CA), coupled to a Probot for fraction collection and matrix addition with 2,5-dihydrobenzoic acid as the matrix. Matrix-assisted laser desorption/ionization-time of flight (MALDI TOF/TOF) MS was performed using an Applied Biosystems 4800 mass spectrometer (Foster City, CA.) in the positive reflectron mode with delayed extraction. MS precursor acquisition was followed by interpretation and data-dependent MS/MS acquisition with the collision-induced dissociation turned on. Data interpretation was configured to select a maximum of 10 precursor ions per fraction with a minimum signal-to-noise ratio of 50. The data were processed using GPS Explorer (Applied Biosystems, CA) against the Swiss Protein Database. Search parameters were as follows: enzyme = trypsin, fixed modifications = carboxymethyl; variable modifications = oxidation; mass tolerance ± 100 ppm; fragment mass tolerance = 0.3 Da; maximum missed cleavages = 1; mass values = monoisotopic.
Isothermal Titration Calorimetry (ITC) Analysis
Purified recombinant proteins produced in E. coli from pGEX expression vectors were cleaved from GST by incubation with human thrombin (Calbiochem) and then further purified for ITC by gel filtration on an equilibrated (50 mm Tris, pH 7.5, 100 mm NaCl) Superdex S-75 column. Fractions containing pure protein were pooled and concentrated before use. Purified proteins were dialyzed into ITC buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 2 mm MgCl2, 2 mm dithiothreitol) overnight prior to use. Isothermal calorimetric titrations were performed with a MicroCal Omega VP-ITC (MicroCal Inc., Northampton, MA). Typically, 400–700 μm G protein was titrated into 40–70 μm opORBD. Data were fitted by the least squares method using the evaluation software, MicroCal Origin version 5.0. All measurements were repeated at least two times, and the estimated error is the standard deviation between measurements.
RESULTS
YopO Is Present at Phagocytic Cups and Specifically Blocks FcγR Phagocytosis
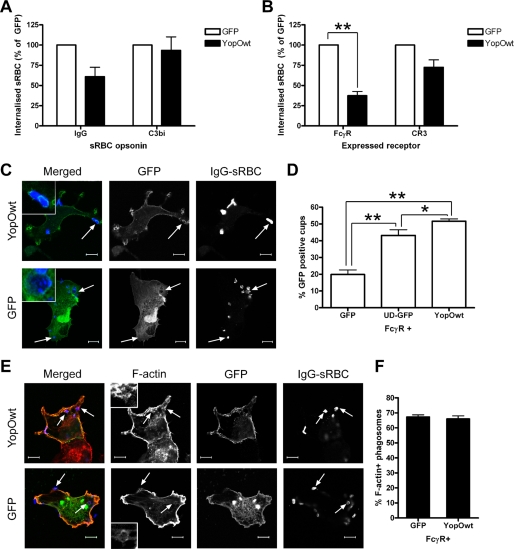
It is established that YopO contributes to anti-phagocytosis mediated by Yersinia type three secretion effectors (11). However, the molecular mechanism involved is unknown. To address this question, we took advantage of the existence of two well defined phagocytic pathways, triggered by ligation of the FcγR and CR3 receptors, which are governed by distinct signaling cascades in macrophages and receptor-transfected fibroblasts (6). We expressed GFP or GFP-tagged YopO in RAW264.7 macrophages (Fig. 1A) and COS-7 fibroblasts co-transfected with constructs encoding either FcγRIIA or CR3 (Fig. 1B). Cells were then challenged with IgG- or C3bi-opsonized RBC to selectively target one phagocytic receptor or the other. In both cell systems, YopO had a greater inhibitory effect on the internalization of IgG- than of C3bi-RBC. In macrophages, FcγR-mediated phagocytosis was reduced by 40%, with no reduction seen for CR3-dependent uptake. In transfected COS-7 cells, FcγR internalization was inhibited by 60%, whereas only a small reduction (∼25%) was observed for CR3 phagocytosis. Importantly, expression of YopO had no effect on the level of RBC attachment (data not shown), suggesting YopO acts on phagocytic signaling rather than on receptor expression or availability. Of note, the selectivity of YopO toward FcγR-dependent uptake parallels the behavior of yopO-deleted Y. enterocolitica, where the presence of YopO was important for phagocytosis of IgG-opsonized bacteria, but loss of YopO had no effect on uptake of complement-opsonized bacteria (11).
FIGURE 1.
Preferential inhibition of FcγR-mediated phagocytosis by YopO. A, RAW264.7 mouse macrophages were transiently transfected with vectors expressing GFP or GFP-tagged YopOwt (YopOwt) and then challenged with IgG- or C3bi-opsonized sRBC for 30 min. B, COS-7 cells were co-transfected with FcγRIIA or CR3 vectors and either GFP or YopOwt and challenged with appropriately opsonized sRBC for 30 min. Phagocytosis assays were performed and scored as described under “Experimental Procedures.” At least 50 GFP-expressing RAW264.7 macrophages or 100 COS-7 cells were scored per experiment. Data represent mean ± S.E. from at least three independent experiments. C, COS-7 cells were transiently transfected with FcγRIIA and either GFP or GFP-YopOwt and then challenged with IgG-opsonized sRBC for 15 min after synchronization at 4 °C. sRBC and F-actin were labeled and observed by confocal microscopy. GFP enrichment at site of sRBC binding was examined. Note the localization of YopO, but not GFP around sRBC (arrows). D, quantification of GFP localization to phagocytic cups in cells co-expressing FcγRIIA and GFP, YopO-GFP, or unique domain of Lck-GFP (membrane-targeted GFP). E, following sRBC challenge, cells expressing GFP or YopOwt were analyzed for F-actin enrichment at sites of sRBC binding as described under “Experimental Procedures.” Representative images are shown, with typical F-actin rings enlarged. F, quantification of actin cup formation. At least 100 sRBC were scored for the presence of F-actin-rich cups per experiment. Data represent mean ± S.E. from at least three independent experiments. Scale bars, 10 μm. * indicates p < 0.05 and ** indicates p < 0.01.
Regardless of the receptor involved, phagocytic uptake is subject to tight spatio-temporal control. Most of its regulators and effectors (e.g. Rho proteins, Arp2/3 complex, and actin filaments) are recruited and are functional at the site of particle binding (9). Because YopO preferentially blocks FcγR-mediated phagocytosis, we tested whether YopO could be found underneath bound IgG-RBC. In COS-7 cells co-expressing FcγRIIA and either GFP or GFP-tagged YopO, then challenged with RBC, YopO was generally membrane-associated and clearly enriched as a ring around 50% of the bound RBC, unlike GFP (Fig. 1, C and D) that showed 20% localization to phagocytic cups. YopO also showed significantly higher enrichment than a membrane-targeted GFP construct, where the unique domain of the Src family kinase Lck mediates association of GFP to the membrane (Fig. 1D, p = 0.0339) implying that YopO enrichment at phagocytic cups is not solely a result of its membrane localization domain. Overall, these data suggest that YopO might interfere with phagocytic pathways downstream of receptor ligation to prevent internalization. As it has been reported that YopO overexpression disrupts the actin cytoskeleton (13, 17), we investigated the possibility that YopO would prevent F-actin polymerization at sites of sRBC binding. YopO-expressing COS-7 cells had no visible defect in actin cup formation (Fig. 1, E and F), indicating that YopO does not drastically affect the de novo actin polymerization initiated during phagocytosis. Interestingly, expression of either dominant negative Rac or Cdc42 has no effect on actin cup formation but reduces phagocytosis; only when both Cdc42 and Rac are simultaneously inactivated is actin cup formation abolished (26). This suggests that Rac and Cdc42 both contribute to actin cup formation at sites of FcγR uptake and that YopO targets one of these Rho proteins, because YopO expression phenocopies inactivation of one of the two Rho proteins in this respect.
YopO Prevents Rac Activation and Not Its Localization to the Phagocytic Cup
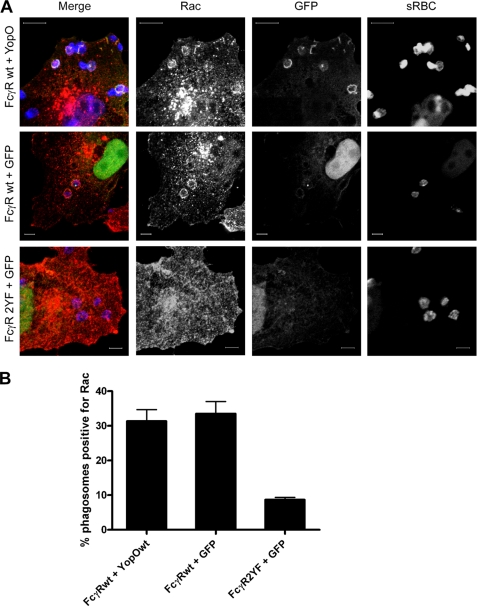
Because YopO does not interact with Cdc42 (21, 27) (supplemental Fig. 2), Rac is the most likely Rho protein target downstream of FcγR signaling. We tested whether YopO blocks FcγR phagocytosis by inhibiting Rac recruitment to the phagocytic cup. COS-7 cells were co-transfected with FcγRIIA and GFP or YopO-GFP and challenged for 10 min after synchronization with IgG-RBC. In parallel, an FcγR mutant (FcγR2YF), which is defective in Rac recruitment (26), was included as a negative control. Endogenous Rac was visualized by immunofluorescence microscopy using a specific antibody, and levels of recruitment to RBC were scored (Fig. 2A). Cells expressing GFP or YopO showed similar levels of endogenous Rac recruitment to cups, and as expected the signaling mutant FcγRY2F showed a much lower level (Fig. 2B). This suggests that the presence of YopO does not exclude Rac from the phagocytic cup.
FIGURE 2.
YopO does not affect Rac recruitment to FcγR-phagocytic cups. A, COS-7 cells were transiently transfected with FcγRIIA wild-type (wt) or mutant FcγRIIA (Y2F) and either GFP or GFP-YopOwt and then challenged with IgG-opsonized sRBC for 10 min after synchronization at 4 °C. sRBC and endogenous Rac were labeled and observed by confocal microscopy. Rac enrichment at site of sRBC binding was examined. Scale bars, 10 μm. B, quantification of Rac localization to phagocytic cups in cells co-expressing FcγRIIA or FcγRIIA2YF and GFP or YopO-GFP. At least 100 sRBC were scored for the presence of Rac at sRBC cups per experiment. Data are mean ± S.E. of three independent experiments.
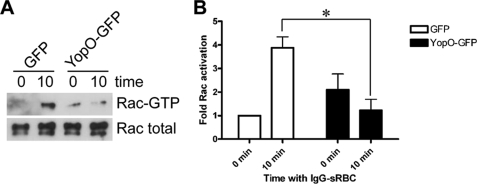
We next investigated if YopO blocks Rac activation. To this end, COS-7 cells co-expressing FcγR and GFP or YopO were subjected to a synchronized IgG-RBC challenge. Cells were lysed at 0 and 10 min after incubation at 37 °C, and cell lysates were incubated with the Rac/Cdc42 binding domain of p21-activated kinase (GST-PAK CRIB) immobilized on beads. Bead eluates and whole cell lysates were analyzed by SDS-PAGE and Western blot, probing for Rac (Fig. 3A), and results were quantified (Fig. 3B). In cells expressing GFP, there was a consistent activation, ∼4-fold, of Rac at 10 min compared with 0 min. Significantly, this activation was not detectable in cells expressing YopO, suggesting that YopO expression blocks activation of Rac that is required for successful phagocytosis downstream of FcγR ligation.
FIGURE 3.
YopO blocks Rac activation during FcγR phagocytosis. A, COS-7 cells were transiently transfected with FcγRIIA and either GFP or GFP-YopOwt and then challenged with IgG-opsonized sRBC for 10 min at 37 °C after synchronization at 4 °C. Cells were lysed at 0 and 10 min and incubated with GST-PAK CRIB immobilized on glutathione-Sepharose beads. Bead eluates and lysates were analyzed by SDS-PAGE and Western blot probing for Rac. A representative experiment is shown. B, fold activation of Rac was quantified and shown relative to activation levels at 0 min (GFP-expressing cells). Data are mean ± S.E. of three independent experiments. * indicates p < 0.05.
GDI-like Region of YopO Is the Preponderant Domain Mediating Anti-phagocytosis
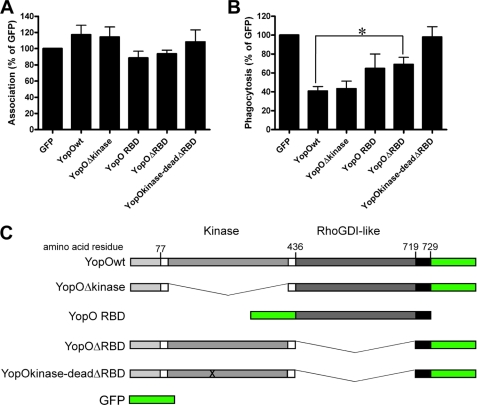
Next, we investigated the role of YopO domains in the inhibition of FcγR-mediated uptake. To this end, several YopO mutants were constructed, co-transfected with a vector encoding FcγRIIA into COS-7 cells, and analyzed for their ability to interfere with phagocytosis of IgG-RBC. A mutant lacking the kinase domain (YopOΔkinase), a deletion mutant lacking the GDI-like region (YopOΔRBD for Rho family G protein binding domain), a kinase-dead and GDI-like region-deleted double mutant (YopO kinase-dead ΔRBD), and the isolated GDI-like domain (YopORBD) were studied in parallel (Fig. 4). No significant difference in protein expression from the different constructs was observed by Western blot or during microscopic analysis (supplemental Fig. 1), and none of the YopO constructs significantly reduced the ability of RBC to bind transfected COS-7 cells (Fig. 4A). The mutant lacking the kinase domain reduced RBC uptake to a similar extent as YopOwt, indicating that this region is not required for activity (Fig. 4B). In contrast, the mutant lacking the GDI-like region showed significantly reduced anti-phagocytic activity compared with YopOwt. The kinase-dead RBD-deleted mutant showed total abrogation of anti-phagocytosis. Consistently, expression of the isolated RBD alone significantly reduced uptake.
FIGURE 4.
Domain analysis of YopO. The GDI-like domain (RBD) is the main contributor to anti-phagocytosis. A, COS-7 cells were co-transfected with FcγRIIA and GFP-tagged constructs as indicated. After 24 h, cells were challenged with IgG-opsonized sRBC for 30 min and processed for immunofluorescence as described under “Experimental Procedures.” Total associated sRBCs for each condition were scored in at least 100 GFP-positive cells per experiment. B, COS-7 cells were transfected as above, and phagocytosed sRBCs were scored for each condition. Data are expressed relative to GFP control, arbitrarily set at 100%. Data represent mean ± S.E. of at least three independent experiments. C, domain organization of the constructs used above. * indicates p < 0.05.
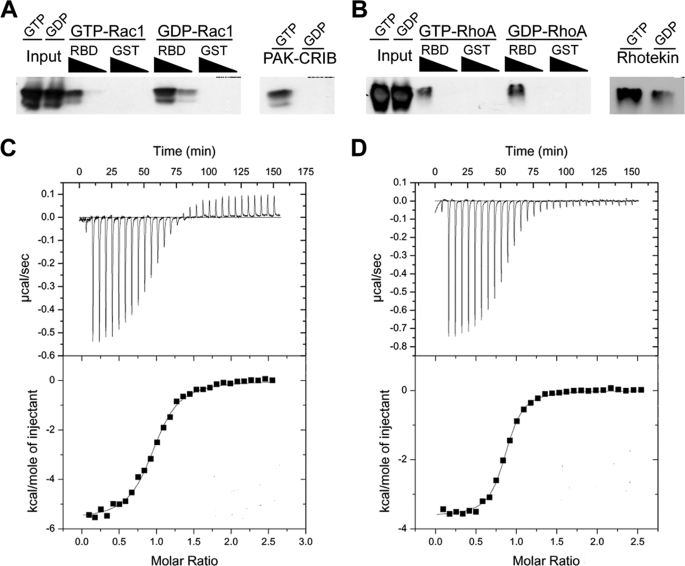
GDI-like Domain of YopO Binds Rac1 and RhoA with Equal Affinity in Vitro
The C-terminal half of YopO was reported to interact with Rac1 and RhoA in vitro (20, 21, 27). Furthermore, recent evidence indicates that YopO GDI-like domain prevents guanine nucleotide exchange on both Rac1 and RhoA in vitro (20). This contrasts with the selectivity (Rac over Rho) we observed in our phagocytosis assays. To further investigate the in vitro interactions of YopO GDI-like region with small G proteins, we performed pulldown assays, incubating purified recombinant Rac1 (Fig. 5A) or RhoA (Fig. 5B), with purified GST-YopO RBD. As the nucleotide dependence of the YopO-small G protein interaction is unclear, we preloaded the purified G proteins with either GDP or GTPγS to mimic the inactive and active state of RhoA and Rac1. Nucleotide loading was confirmed by a parallel pulldown assay using beads coated with GST-PAK-CRIB or GST-Rhotekin, which interact specifically with the active GTP-bound forms of Rac1 and RhoA, respectively (28, 29). Bound proteins were analyzed by SDS-PAGE, followed by immunoblotting. When compared with the input, the levels of bound G proteins were similar, suggesting that YopO RBD binds Rac1 and RhoA equally well in vitro. Furthermore, in both cases, the inactive forms of the G proteins appeared to interact more strongly than the active forms. Additionally, we performed pulldown experiments from lysates of COS-7 cells overexpressing Myc-tagged Cdc42, RhoA, or Rac1 using GST-YopO RBD or GST alone as a control (supplemental Fig. 2). Overexpressed Rac1 and RhoA, but not Cdc42, interacted specifically and equally with the YopO RBD beads. These results suggest that prenylated Rac1 and RhoA behave similarly to the bacterially expressed proteins and that YopO does not bind Cdc42.
FIGURE 5.
YopO RBD shows no preference for Rac1 or RhoA in vitro. Purified recombinant Rac1 (A) and RhoA (B) were loaded with GDP or GTPγS and then incubated at 1 or 0.1 μm with 20 μg of GST-YopO RBD immobilized on glutathione-Sepharose beads. Following 1 h of incubation, eluates were analyzed by SDS-PAGE and Western blotting using antibodies to Rac and Rho. As a nucleotide loading control, GDP or GTPγS-bound Rac1 and RhoA were incubated with the Rac/Cdc42 binding domain of p21-activated kinase (PAK-CRIB) and the Rho binding domain of Rhotekin (Rhotekin) respectively, immobilized on glutathione-Sepharose beads, and analyzed as above. Interactions between purified G proteins and YopO RBD were further analyzed by isothermal titration calorimetry. Representative titrations of purified Rac1 (C) and RhoA (D) are shown. Experiments were repeated on at least two independent occasions with the same results.
We next quantified the YopO RBD-G protein interaction using ITC (Fig. 5, C and D). Increasing concentrations of the small G proteins were titrated into YopO RBD, and the change in heat evoked upon complex formation was measured. Representative titrations of GDP-bound Rac1 and RhoA are shown. Rac1-GDP bound to YopO RBD with an affinity of 1.14 μm and a 1:1 stoichiometry. Similarly, RhoA-GDP formed a 1:1 complex with YopO RBD with an affinity of 0.91 μm. There was no detectable difference between the respective affinities of YopO RBD for Rac1 and RhoA in vitro. ITC experiments using GTP-loaded G proteins were attempted, but there was not sufficient interaction to measure by ITC (data not shown), supporting the idea that YopO RBD has a preference for inactive GDP-bound G proteins.
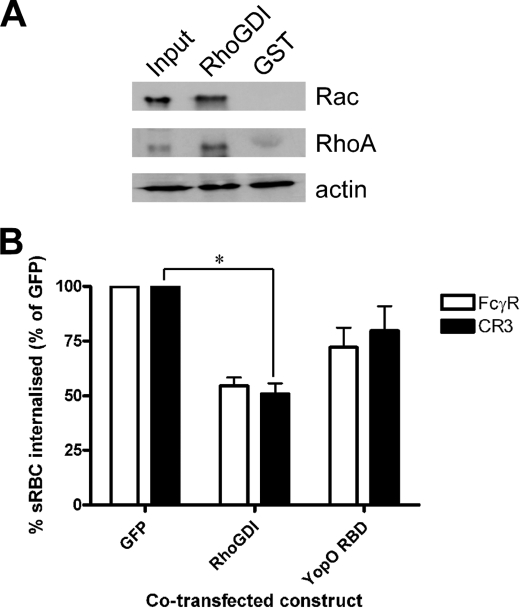
Isolated YopO RBD Behaves Like RhoGDI
The paradoxical difference between the in vitro and in cellulo results, where Rac-dependent FcγR phagocytosis is most sensitive to YopO, prompted us to explore further the biochemical and functional properties of isolated YopO RBD and compare them with those of eukaryotic RhoGDIα. First, the ability of RhoGDIα to interact with Rac1 and RhoA was confirmed (Fig. 6A). Macrophage lysates were incubated with immobilized GST-RhoGDIα, and its interaction with RhoA and Rac1 was tested. As expected (30), RhoGDIα interacted to a similar extent with both G protein isoforms. To directly compare the anti-phagocytic capacities of YopO RBD and RhoGDIα, we co-transfected COS-7 cells with vectors expressing GFP-tagged YopO RBD, RhoGDIα, or GFP alone together with either FcγR or CR3 and measured phagocytosis of RBC (Fig. 6B). Overexpression of RhoGDIα reduced phagocytosis through both receptors by ∼50%. YopO RBD also reduced phagocytosis through both receptors, but to a noticeably lesser extent (by ∼25%) than RhoGDIα. This difference cannot be attributed to different expression levels of YopO RBD and RhoGDIα, as shown by Western blotting (data not shown). Our results suggest a fundamental functional difference between full-length YopO and its isolated RBD domain, because the isolated RBD shows no anti-phagocytic preference for a particular pathway, similar to the effect of overexpressing RhoGDIα.
FIGURE 6.
RhoGDI and YopO RBD block both FcγR- and CR3-mediated phagocytosis. A, J774A.1 lysates were incubated with GST-RhoGDI immobilized on glutathione-Sepharose beads and resolved by SDS-PAGE and Western blot, probing for Rac, RhoA, and actin. A sample of RhoGDI beads and bead eluates was run in parallel. At least 100 GFP-expressing COS-7 cells were scored per experiment. Results shown are representative of three experiments. B, COS-7 cells were co-transfected with FcγRIIA or CR3 and a GFP-tagged construct, either RhoGDIα, YopO RBD, or GFP alone, challenged with appropriately opsonized sRBC for 30 min and scored for sRBC phagocytosis. Data are expressed relative to the GFP control, arbitrarily set at 100%. Data are mean ± S.E. of at least two independent experiments. * indicates p < 0.05.
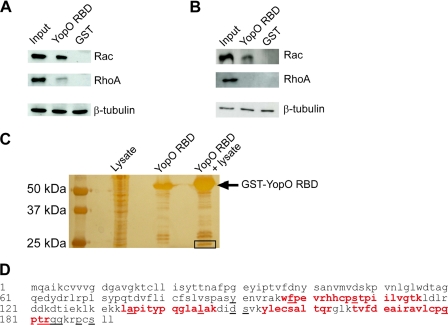
YopO RBD Binds Preferentially to Cellular Rac
Because full-length YopO has a selective anti-phagocytic activity on the FcγR-mediated pathway, we decided to further investigate the interaction of YopO RBD with Rho family proteins in a cellular context. Lysates from J774A.1 macrophages (Fig. 7A) and COS-7 fibroblasts (Fig. 7B) were incubated with GST-YopO RBD; bound proteins were analyzed by SDS-PAGE and immunoblotting to reveal endogenous Rac1 and RhoA. In both cell types, relative to input, YopO RBD repeatedly showed a stronger interaction with Rac1 compared with RhoA. Further evidence that YopO RBD preferentially interacts with Rac in a cellular context comes from mass spectrometry analysis of proteins pulled down with the isolated RBD domain. J774A.1 lysates were incubated with beads coated with GST-YopO RBD, and bead-associated proteins were separated by SDS-PAGE and visualized by silver nitrate staining. A specific YopO-associated band of ∼20–25 kDa (the apparent molecular mass of Rho family proteins) was excised and prepared for nano-liquid chromatography and MS-MS analysis (Fig. 7C). When analyzed using the Swiss Protein Database, the only two significant peptide hits corresponded to mouse Rac2 and Rac1, with scores of 284 and 146, respectively. The sequence coverage of the matched peptides was 31% of the Rac2 protein (Fig. 7D). Importantly, no other Rho family protein, in particular RhoA or Cdc42-related peptides, was identified. Because binding of Rac2 to YopO has never been reported, we quantified their interaction by ITC. The Kd measured was 1 μm, similar to that determined for Rac1 and RhoA (data not shown).
FIGURE 7.
Unlike RhoGDI, YopO RBD preferentially binds Rac from cell lysates. J774A.1 (A) or COS-7 (B) lysates were incubated with GST-YopO RBD immobilized on glutathione-Sepharose beads, and bead eluates were resolved by SDS-PAGE and Western blotting, probing for Rac, RhoA, and β-tubulin. Results shown are representative of three experiments. C, after pulldown and SDS-PAGE, as described for A, bead eluates were stained by an MS-compatible silver nitrate stain. The 25-kDa band (boxed) was analyzed by MALDI-TOF MS. D, mouse Rac2 amino acid sequence, showing the overall sequence coverage of Rac1 and Rac2 peptides identified by MS (bold red). Amino acid differences between Rac1 and Rac2 are underlined.
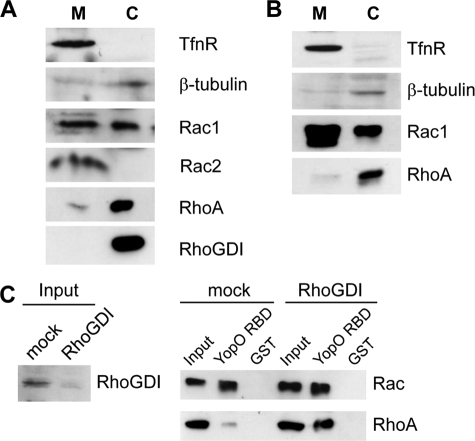
YopO Targets a GDI-free Pool of Rac at the Plasma Membrane
One possible explanation for the observed interaction of YopO for Rac over RhoA is that, within cells, the former is more available to YopO than the latter, which could be linked with the intracellular locale of the proteins involved and/or their interaction with binding partners. It is generally assumed that inactive Rho family proteins are confined to the cytosol and recruited and/or activated locally at membranes, but this has not been systematically analyzed for all Rho proteins. Therefore, we investigated the intracellular localization of RhoA and Rac by separating cells into membrane and cytosolic fractions. Fractions from J774A.1 macrophages (Fig. 8A) and COS-7 fibroblasts (which do not express Rac2 (31), Fig. 8B) were first tested for the presence of transferrin receptor and β-tubulin. As shown in Fig. 8, A and B, there was no significant contamination of these proteins into the cytosolic and membrane fractions, respectively. When these fractions were probed for Rac1, Rac2, and RhoA, the Rac isoforms distributed to the membrane and cytosolic fractions, whereas RhoA was only detected in the cytosolic fraction. In macrophages, Rac2 was found almost exclusively at the membrane, whereas Rac1 was separated into a membrane pool and a larger cytosolic fraction. In COS-7 cells, the majority of Rac1 was found at the membrane, suggesting differences in the localization of Rac1 between different cell types. Interestingly, endogenous RhoGDI was, like Rho, exclusively cytosolic. This suggests that there is a pool of RhoGDI-free Rac at the membrane, and that RhoA is retained in the cytosol, possibly through an interaction with RhoGDI. In such a scenario, YopO RBD would interact better with Rac than RhoA because it cannot efficiently out-compete RhoGDI binding to RhoA. To test this hypothesis, we silenced expression of RhoGDIα in COS-7 cells and compared the ability of YopO RBD to interact with Rac1 and RhoA in pulldown assays (Fig. 8C). Upon loss of RhoGDIα, there was an increased interaction of YopO RBD with both Rac1 and RhoA, compared with control cells (Fig. 8C and Fig. 7A). Strikingly, where previously no interaction of YopO RBD and RhoA could be detected, knockdown of RhoGDIα allowed an interaction suggesting that in control cells RhoA is trapped by RhoGDIα and that loss of this interaction by knockdown allows interaction with YopO RBD.
FIGURE 8.
GDI-free pool of Rac molecules exists at the plasma membrane of resting cells. J774A.1 (A) and COS-7 (B) lysates were separated into membrane (M) and cytosol (C) fractions as described under “Experimental Procedures.” After SDS-PAGE, blots were probed for transferrin receptor (TfnR), β-tubulin, Rac, Rho, and RhoGDI. C, COS-7 cells were treated with luciferase- or RhoGDIα-specific small interfering RNA duplexes, as described under “Experimental Procedures.” After 48 h, cells were lysed and incubated with GST-YopO RBD or GST immobilized on glutathione-Sepharose beads. Bead eluates and samples of lysates were resolved by SDS-PAGE and Western blot, probing for RhoGDI (confirming knockdown), Rac, Rho, and actin (loading control). Data are representative of at least three independent experiments.
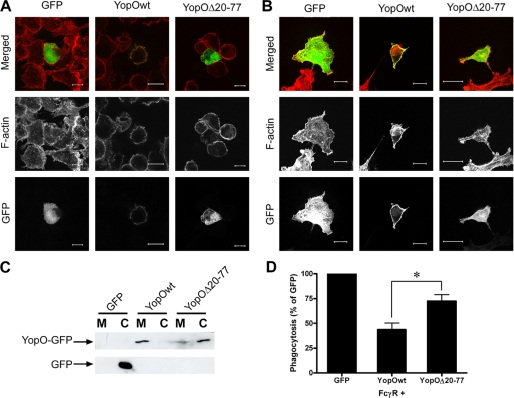
YopO localizes to the plasma membrane of transfected or infected cells through its N-terminal region (17, 18). To test the hypothesis that YopO localization at the plasma membrane is required for anti-phagocytic activity, a GFP-tagged mutant of YopO (YopOΔ20–77) lacking the region required for plasma membrane localization was used. This deletion mutant was almost exclusively cytosolic like GFP but unlike full-length YopO. This was observed both in RAW264.7 macrophages (Fig. 9A) and in COS-7 cells (Fig. 7B) and was confirmed independently by cell fractionation followed by Western blotting. There was no obvious contamination of the fractions, as judged by the separation profile of tubulin (cytosol fraction) and transferrin receptor (membrane fraction) (data not shown). GFP alone was primarily found in the cytosolic fraction, as expected. Conversely, the vast majority of GFP-tagged YopO was in the membrane fraction. Importantly, overexpressed YopOΔ20–77 was found in the cytosol fraction, with only a small proportion still detectable within the membrane fraction. Next, we co-expressed the YopO localization mutant (YopOΔ20–77) with FcγRIIA in COS-7 cells. Upon phagocytic challenge with IgG-sRBC, this mutant (which still harbors an intact RBD) had significantly reduced anti-phagocytic ability (Fig. 9D, p = 0.0489). The remaining anti-phagocytic activity of this mutant is similar to that observed against CR3-mediated phagocytosis (28% for FcγR, 15% for CR3) suggesting it could be a nonspecific effect of overexpression of a GDI-like molecule. Altogether, we conclude that YopO mediates specific anti-phagocytosis toward FcγR, where activity is primarily mediated by its Rho family binding GDI-like domain, through co-compartmentalization with a pool of RhoGDI-free Rac at the plasma membrane of target cells.
FIGURE 9.
Membrane localization of YopO is necessary for anti-phagocytosis. A, RAW264.7 macrophages were transiently transfected with GFP-tagged versions of YopO (wild type or Δ20–77) or GFP alone. After 24 h, cells were stained for F-actin and observed using a Zeiss Axiovert 100 M confocal microscope. Representative examples are shown. Scale bars, 10 μm. B, COS-7 cells were transfected and processed as described in A. Scale bars, 20 μm. C, lysates of COS-7 cells transfected with GFP-tagged constructs as indicated were separated into cytosol (C) and membrane (M) fractions, as described under “Experimental Procedures.” After SDS-PAGE, fractions were probed using anti-GFP antibodies. D, COS-7 cells were co-transfected with FcγRIIA and GFP-tagged YopO (wild type or Δ20–77) and challenged with IgG-sRBC, and at least 100 GFP-expressing COS-7 cells were scored for phagocytosis per experiment. Data are given as mean ± S.E. of at least three independent experiments. * indicates p < 0.05.
DISCUSSION
Herein, we demonstrate that YopO is able to specifically block FcγR-mediated phagocytosis while having no significant effect on CR3-dependent internalization. This is in sharp contrast to the T3SS effector EspJ of enteropathogenic E. coli, which blocks both FcγR-mediated and CR3-dependent phagocytosis, the underlining mechanism of which is not yet known (32). Furthermore, we show that the N-terminal localization domain and the GDI-like Rho family binding domain cooperate in mediating inhibition of Rac-dependent uptake, whereas the kinase domain of YopO is dispensable. Furthermore, this study has revealed differential localization of RhoA, which is only detectable in the cytosol and Rac isoforms that are present, at least in part, as a GDI-free pool at the plasma membrane. We propose that this membrane-associated Rac is the cellular target for YopO anti-phagocytic activity.
Our data establish cooperation between the N-terminal localization domain and the G protein binding regions (RBD) of YopO for anti-phagocytic function. First, we show that YopO deletion mutants lacking only the RBD domain are significantly reduced in their anti-phagocytic potential; this region alone is also sufficient for activity. Second, the N-terminal mutant (Δ20–77) is severely affected in anti-phagocytosis. Third, the selective activity of the RBD toward Rac-dependent phagocytosis is only observed when the N-terminal region, responsible for plasma membrane localization, is present. In contrast, the kinase domain of YopO plays no role in anti-phagocytosis, although our data could suggest a regulatory role for the full kinase domain, because its presence but inactivity seems to totally abrogate YopO activity.
The N-terminal region of YopO has no independent anti-phagocytic activity of its own, because a double mutant of YopO-lacking kinase activity and RBD domain had no effect on FcγR-dependent phagocytosis. The YopO N-terminal region has a dual function as a chaperone-binding site within bacteria and, once translocated, is responsible for plasma membrane localization in host cells (18). As yet, the mechanism by which it mediates this localization is unknown; in particular, it does not harbor any obvious post-translational modification sites, e.g. for prenylation or myristoylation. Interestingly, the N-terminal domain of YopO has a net positive charge, hinting at a possible charge-dependent mechanism by which YopO N terminus may be interacting with the negatively charged inner leaflet of the plasma membrane, as proposed for several Ras family proteins (33).
We show that YopO selectively blocks Rac-dependent uptake pathways. YopO consistently blocked FcγR-mediated uptake both in macrophages and in FcγR-transfected COS-7 cells. We would predict that since Yersinia uptake via Invasin is Rac-dependent, bacterial uptake would also be blocked by YopO. This would confirm that YopO is able to act on phagocytic particles of different sizes, a possibility that we have not addressed in this study. Interestingly, the functional selectivity of YopO for Rac-dependent pathways in cells is not paralleled by an increased affinity of its RBD for Rac over Rho in vitro. As was seen previously, we did not detect any interaction with Cdc42. Importantly, by using pulldown assays followed by mass spectrometry, we have identified Rac1 and Rac2 as the predominant Rho family members able to interact with YopO RBD in macrophages. This suggests that cellular Rho is not available for interaction with YopO RBD in pulldown experiments, most likely because of the tight interaction of RhoA and RhoGDI in the cytosol (7).
A recent report convincingly showed that YopO RBD has a GDI-like structure and activity, although unlike GDI YopO RBD does not interact with the C-terminal prenyl moiety of RhoA and Rac1 (20). In accordance with the idea that YopO RBD is a GDI mimic, overexpression of either RhoGDI or YopO RBD decreased FcγR-mediated phagocytosis. However, our work strongly suggests that the biochemical and structural similarities between YopO RBD and host cell RhoGDIs mask some important functional differences. Indeed, unlike full-length YopO, RhoGDI also blocks CR3-mediated uptake. The main difference between RhoGDI and YopO is their subcellular localization; RhoGDI resides mainly in the cytosol, whereas YopO associates with the plasma membrane when overexpressed (this study and see Ref. 18) or when translocated following infection (17). All known T3SS effectors that impact on the regulation on Rho family G proteins (e.g. SopE from Salmonella typhimurium and ExoS from Pseudomonas aeruginosa) are thought to act purely as mimics (34). Remarkably, our work has revealed that the mechanism of action of YopO depends both on GDI mimicry and a specific subcellular localization.
Our data show that YopO RBD can bind Rac1 and Rac2 in vitro; it targets membrane-associated Rac1 and Rac2 in macrophages but only Rac1 in COS-7 cells. Rac2 expression is restricted to hematopoietic cells, and our results therefore suggest that FcγR phagocytosis is reliant upon Rac1 in COS-7 cells, as suggested before (6). However, our data cannot differentiate, which is the predominant isoform required during FcγR phagocytosis in macrophages. Nevertheless, they provide an explanation for why YopO has an effect on NADPH oxidase activation, a process that is Rac2-dependent (35). Furthermore, our work supports the data that suggest Yersinia target β1-integrin-mediated uptake by phagocytes, a process shown to require Rac and not Rho (36).
Interestingly, our data provide strong experimental evidence for a differential localization of endogenous Rac and RhoA in cells. Overexpressed RhoA was found to localize to the cytosol even when highly overexpressed, whereas overexpressed Rac1 and Rac2 were localized to membrane compartments, in agreement with our results (37, 38). The localization of Rac isoforms to the plasma membrane is thought to be due to the strong polybasic domain of Rac. This charge-dependent membrane association is further utilized by the cell to regulate Rac distribution; when the surface potential is altered, e.g. following activation of phagocytic receptors, Rac is released from the membrane of the forming phagosome (33). By contrast, RhoA has a weak polybasic domain that does not support membrane association. In addition, we show that RhoGDIα helps to retain RhoA in the cytosol, preventing its interaction with binding partners. The functional implications of these fundamental differences are many. The classical GTPase cycle is thought to involve activation and localization to the membrane, followed by inactivation and cytosolic GDI trapping. Our data show that this is not a true representation of the activation cycle of all G proteins. Rac is clearly already present at the membrane in resting cells, and presumably it would be locally activated upon initiation of phagocytosis or other Rac-activating stimuli. Our data show that binding of YopO to membrane Rac interferes with the activation process. By contrast, Rho-dependent phagocytosis and other Rho-dependent processes require trafficking of the RhoGDI-RhoA complex, or RhoA alone, to the membrane. The link between function and subcellular localization of G proteins is only just starting to be understood, and the subtle differences in primary sequence between the different isoforms will have important implications for their regulation and function. The fact that a bacterial virulence factor has evolved to exploit this highlights its importance in cell signaling. Our data also suggest that YopO could be used as a tool to specifically target Rac-dependent pathways.
Acknowledgments
We gratefully acknowledge Professor Gadi Frankel and Drs. J. Mota, M. Bright, and D. Lees for critical reading of this manuscript and Dr. M. J. Bijlmakers and B. Stieglitz for providing reagents.
This work was supported by Medical Research Council studentship (to E. G.) and a core grant (to K. R.). Work in the Caron laboratory was funded by grants from the Medical Research Council and the Biotechnology and Biological Sciences Council.
We dedicate this manuscript to the memory of Emmanuelle Caron, who tragically passed away after suffering from cancer during the preparation of this manuscript. She was an incredibly talented young scientist who brightened the lives of all who knew her and was an inspirational, warm, and caring mentor to her “phago” group, who will always miss her.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- FcγR
- Fcγ receptor
- CR3
- complement receptor 3
- GDI
- guanine-nucleotide dissociation inhibitor
- MS
- mass spectrometry
- RBD
- Rho family binding domain
- T3SS
- type III secretion
- WT
- wild type
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight
- ITC
- isothermal titration calorimetry
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- RBC
- red blood cell
- sRBC
- sheep RBC
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1.Griffin F. M., Jr., Griffin J. A., Leider J. E., Silverstein S. C. (1975) J. Exp. Med. 142, 1263–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May R. C., Caron E., Hall A., Machesky L. M. (2000) Nat. Cell Biol. 2, 246–248 [DOI] [PubMed] [Google Scholar]
- 3.Swanson J. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth J. W., Trimble W. S., Grinstein S. (2001) Semin. Immunol. 13, 357–364 [DOI] [PubMed] [Google Scholar]
- 5.Patel J. C., Hall A., Caron E. (2000) Methods Enzymol. 325, 462–473 [DOI] [PubMed] [Google Scholar]
- 6.Caron E., Hall A. (1998) Science 282, 1717–1721 [DOI] [PubMed] [Google Scholar]
- 7.DerMardirossian C., Bokoch G. M. (2005) Trends Cell Biol. 15, 356–363 [DOI] [PubMed] [Google Scholar]
- 8.Finlay B. B. (2005) Curr. Top. Microbiol. Immunol. 291, 1–10 [DOI] [PubMed] [Google Scholar]
- 9.Groves E., Dart A. E., Covarelli V., Caron E. (2008) Cell. Mol. Life Sci. 65, 1957–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G. R. (2006) Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 11.Grosdent N., Maridonneau-Parini I., Sory M. P., Cornelis G. R. (2002) Infect. Immun. 70, 4165–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marketon M. M., DePaolo R. W., DeBord K. L., Jabri B., Schneewind O. (2005) Science 309, 1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juris S. J., Rudolph A. E., Huddler D., Orth K., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejedlik L., Pierfelice T., Geiser J. R. (2004) Yeast 21, 759–768 [DOI] [PubMed] [Google Scholar]
- 15.Wiley D. J., Nordfeldth R., Rosenzweig J., DaFonseca C. J., Gustin R., Wolf-Watz H., Schesser K. (2006) Microb. Pathog. 40, 234–243 [DOI] [PubMed] [Google Scholar]
- 16.Park H., Teja K., O'Shea J. J., Siegel R. M. (2007) J. Immunol. 178, 6426–6434 [DOI] [PubMed] [Google Scholar]
- 17.Håkansson S., Galyov E. E., Rosqvist R., Wolf-Watz H. (1996) Mol. Microbiol. 20, 593–603 [DOI] [PubMed] [Google Scholar]
- 18.Letzelter M., Sorg I., Mota L. J., Meyer S., Stalder J., Feldman M., Kuhn M., Callebaut I., Cornelis G. R. (2006) EMBO J. 25, 3223–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarro L., Koller A., Nordfelth R., Wolf-Watz H., Taylor S., Dixon J. E. (2007) Mol. Cell 26, 465–477 [DOI] [PubMed] [Google Scholar]
- 20.Prehna G., Ivanov M. I., Bliska J. B., Stebbins C. E. (2006) Cell 126, 869–880 [DOI] [PubMed] [Google Scholar]
- 21.Barz C., Abahji T. N., Trülzsch K., Heesemann J. (2000) FEBS Lett. 482, 139–143 [DOI] [PubMed] [Google Scholar]
- 22.Trasak C., Zenner G., Vogel A., Yüksekdag G., Rost R., Haase I., Fischer M., Israel L., Imhof A., Linder S., Schleicher M., Aepfelbacher M. (2007) J. Biol. Chem. 282, 2268–2277 [DOI] [PubMed] [Google Scholar]
- 23.Wiedemann A., Patel J. C., Lim J., Tsun A., van Kooyk Y., Caron E. (2006) J. Cell Biol. 172, 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bijlmakers M. J., Isobe-Nakamura M., Ruddock L. J., Marsh M. (1997) J. Cell Biol. 137, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 26.Cougoule C., Hoshino S., Dart A., Lim J., Caron E. (2006) J. Biol. Chem. 281, 8756–8764 [DOI] [PubMed] [Google Scholar]
- 27.Dukuzumuremyi J. M., Rosqvist R., Hallberg B., Akerström B., Wolf-Watz H., Schesser K. (2000) J. Biol. Chem. 275, 35281–35290 [DOI] [PubMed] [Google Scholar]
- 28.Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., Narumiya S. (1996) J. Biol. Chem. 271, 13556–13560 [DOI] [PubMed] [Google Scholar]
- 29.Thompson G., Owen D., Chalk P. A., Lowe P. N. (1998) Biochemistry 37, 7885–7891 [DOI] [PubMed] [Google Scholar]
- 30.Gorvel J. P., Chang T. C., Boretto J., Azuma T., Chavrier P. (1998) FEBS Lett. 422, 269–273 [DOI] [PubMed] [Google Scholar]
- 31.Wheeler A. P., Wells C. M., Smith S. D., Vega F. M., Henderson R. B., Tybulewicz V. L., Ridley A. J. (2006) J. Cell Sci. 119, 2749–2757 [DOI] [PubMed] [Google Scholar]
- 32.Marchès O., Covarelli V., Dahan S., Cougoule C., Bhatta P., Frankel G., Caron E. (2008) Cell. Microbiol. 10, 1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung T., Terebiznik M., Yu L., Silvius J., Abidi W. M., Philips M., Levine T., Kapus A., Grinstein S. (2006) Science 313, 347–351 [DOI] [PubMed] [Google Scholar]
- 34.Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., Mattoo S., McMahon S. A., Ghosh P., Hughes T. R., Boone C., Dixon J. E. (2006) Cell 124, 133–145 [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi A., Kim C., Li S., Marchal C. C., Towe J., Atkinson S. J., Dinauer M. C. (2004) J. Immunol. 173, 5971–5979 [DOI] [PubMed] [Google Scholar]
- 36.Alrutz M. A., Srivastava A., Wong K. W., D'Souza-Schorey C., Tang M., Ch'Ng L. E., Snapper S. B., Isberg R. R. (2001) Mol. Microbiol. 42, 689–703 [DOI] [PubMed] [Google Scholar]
- 37.Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. (2001) J. Cell Biol. 152, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamson P., Paterson H. F., Hall A. (1992) J. Cell Biol. 119, 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]