Abstract
Although DNA methylation is critical for proper embryonic and tissue-specific development, how different DNA methyltransferases affect tissue-specific development and their targets remains unknown. We address this issue in zebrafish through antisense-based morpholino knockdown of Dnmt3 and Dnmt1. Our data reveal that Dnmt3 is required for proper neurogenesis, and its absence results in profound defects in brain and retina. Interestingly, other organs such as intestine remain unaffected suggesting tissue-specific requirements of Dnmt3. Further, comparison of Dnmt1 knockdown phenotypes with those of Dnmt3 suggested that these two families have distinct functions. Consistent with this idea, Dnmt1 failed to complement Dnmt3 deficiency, and Dnmt3 failed to complement Dnmt1 deficiency. Downstream of Dnmt3 we identify a neurogenesis regulator, lef1, as a Dnmt3-specific target gene that is demethylated and up-regulated in dnmt3 morphants. Knockdown of lef1 rescued neurogenesis defects resulting from Dnmt3 absence. Mechanistically, we show cooperation between Dnmt3 and an H3K9 methyltransferase G9a in regulating lef1. Further, like Dnmt1-Suv39h1 cooperativity, Dnmt3 and G9a seemed to function together for tissue-specific development. G9a knockdown, but not Suv39h1 loss, phenocopied dnmt3 morphants and G9a overexpression provided a striking rescue of dnmt3 morphant phenotypes, whereas Suv39h1 overexpression failed, supporting the notion of specific DNMT-histone methyltransferase networks. Consistent with this model, H3K9me3 levels on the lef1 promoter were reduced in both dnmt3 and g9a morphants, and its knockdown rescued neurogenesis defects in g9a morphants. We propose a model wherein specific DNMT-histone methyltransferase networks are utilized to silence critical regulators of cell fate in a tissue-specific manner.
Keywords: Brain, Chromatin, Histone Modification, DNA methyltransferase, Neurodevelopment, Zebrafish, DNA Methylation, Dnmt1, Dnmt3, G9a, Histone Methylation
Introduction
In higher eukaryotes, individual tissues exhibit a unique gene expression signature, which contributes toward its identity. These signatures are established during early zygotic development inducing and/or repressing transcriptional programs in a temporal and tissue-specific manner. In some tissues, this regulation is achieved by targeting a master regulator of tissue differentiation to the genes of interest, which then recruits the basal transcriptional regulation machinery. However, transcription factors must contend with the chromatin state of the locus, because different chromatin states can either facilitate transcriptional activity, or instead prevent the binding and activity of transcription factors.
It has been long hypothesized that the process of DNA methylation (a general term for the methylation of cytosine at the 5′ position) is utilized for initiating or maintaining gene silencing during this tissue-specific and temporal transcriptional regulation (1, 2). However, direct evidence for such a role of DNA methylation during development has been sparse. One way to initially test this hypothesis would be to determine if the enzymes that carry out this process function in a tissue-specific manner. Zebrafish provide an attractive model system for tests of this type, because organ development is rapid, easily monitored, and can be manipulated by genetic methodologies. Using zebrafish and antisense morpholino knockdown technology, we have previously shown that Dnmt1, the major maintenance DNA methyltransferase, is required in a tissue-specific manner during zebrafish development (3). Recently, Anderson et al. characterized zebrafish dnmt1 mutants that exhibit defects similar to dnmt1 morphants (4). However, higher eukaryotes harbor three different families of DNA methyltransferases (DNMTs)3: DNMT1/2/3 (5). Whether DNMT2 family enzymes function mainly as DNA or RNA methyltransferases is controversial, although recent data argue in favor of RNA methylation as the primary activity (6, 7). A role for DNMT3 family members as de novo DNA methyltransferases is well established (5, 8), although their tissue-specific roles remain largely unexplored.
The Dnmt3 family consists of three different proteins in mammals: DNMT3A, DNMT3B, and DNMT3L (5). Interestingly, zebrafish harbor six different Dnmt3 orthologs: Dnmt3/4/5/6/7/8 (9). To provide an initial understanding of the scope and function of the Dnmt3 family in zebrafish, our studies here focus on the zebrafish Dnmt3 protein, which is orthologous to DNMT3B in mammals.
Cytosine methylation can repress a gene in two ways; either by blocking binding of a transcription factor, if the methylated cytosine lies in a transcription factor binding site, or by creating a repressive environment through inducing other repressive chromatin modifications (10). For example, DNA methyltransferases themselves or specialized proteins that bind to methylated cytosines (e.g. MBDs) could physically interact with either histone deacetylases or repressive histone methyltransferases (HMTs) and recruit them to the promoter (10). In support of a DNMT-HMT interaction in zebrafish, we have shown that Dnmt1 depended on Suv39h1, a repressive histone H3K9 methyltransferase, for its function in terminal differentiation during zebrafish development (3). This is consistent with previous results showing an interaction between human DNMT1 and SUV39H1 (11). However, recent evidence suggests that DNMT1 can interact with another H3K9 methyltransferase, G9A, raising the question whether all DNMTs generally interact with all HMT enzymes, or whether specific DNMT-HMT relationships exist (12).
A number of questions remain regarding the role of DNMT3 in development and its relationship to DNMT1. For example, does Dnmt3 function in a tissue-specific manner during zebrafish development, and if so, are these functions distinct from Dnmt1? Additionally, does Dnmt3 depend on a repressive HMT for its function and if so, which HMT? Are there regulatory genes that are silenced by DNA methylation and depend on specific DNA methyltransferase? Herein, we used an antisense morpholino (henceforth referred to as “morpholino”) knockdown approach to show that Dnmt3 is required for proper development of specific organs, such as brain and retina. These organs were unaffected by knock down of Dnmt1 suggesting different functions for the two enzymes. Interestingly, in tissues where Dnmt1 and Dnmt3 were both required, Dnmt3 was required at an early stage of tissue differentiation, whereas Dnmt1 was required for terminal differentiation. Moreover, Dnmt1 overexpression in dnmt3 morphants didn't rescue any phenotypes. Similarly, Dnmt3 overexpression in dnmt1 morphants could not reverse terminal differentiation defects. We further show that Dnmt3 function depends on G9a, but not Suv39h1, and that Dnmt1 depends both on G9a and Suv39h1. Finally, we provide evidence that Dnmt3 and G9a cooperatively suppress lef1 to ensure proper neurogenesis.
EXPERIMENTAL PROCEDURES
Zebrafish Morpholino, mRNA, and Plasmid Injections
Zebrafish stocks and embryo culture were performed as described previously (13). Morpholinos, mRNA, and plasmids were injected at the one-cell stage. Morpholino oligonucleotides were ordered from Gene Tools LLC. The morpholino sequences were as follows: dnmt3 Mo1: 5′-TTGTATTTTTACCGGATATGCTGCT-3′; dnmt3 Mo2: 5′-CTCCGATCTTTACATCTGCCACCAT-3′; g9a Mo1: 5′-GACACACACTGACCTGCAGATG ATC-3′; g9a Mo2: 5′-TGTGTAAGTTTGACCTGTACGAGCA-3′; lef1 Mo2: 5′-TTTTTAAGATACGAACCCTCCGGCC-3′; Control Mo: 5′-CCTCTTACCTCAGTTACAATTTATA-3′; and p53 Mo: 5′-CCCTTGCGAACTTACATCAAATTCT-3′. Dnmt1 morpholino has been described earlier (3). Lef1 Mo1 was also described earlier (14). Morpholinos were dissolved in 1× Danieau buffer (58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca(NO3)2, 5.0 mm HEPES, pH7.6). Zebrafish dnmt3 (GenBankTM accession number: NM_131386) and zebrafish g9a (GenBankTM accession number: EU070918) were cloned in pcDNA4/HisMax and pDEST53 (N-terminal GFP gateway destination) vectors, respectively (Invitrogen), for injections of plasmids. For mRNA injections, zebrafish dnmt3, zebrafish g9a, and human DNMT1 and DNMT3B were cloned into pCRII-TOPO vector (Invitrogen). pcDNA3.1-HA-G9A was obtained from Dr. Kenneth L. Wright. Messenger RNA was transcribed using mMessage Machine kit (Ambion). The amounts of zebrafish Dnmt3 mRNA and human DNMT3B injected to rescue dnmt3 morphants were 10 and 25 pg, respectively. 25 pg of zebrafish g9a mRNA and 5pg of human G9A plasmid rescued g9a morphants, whereas 50 pg of zebrafish g9a mRNA was required to rescue dnmt3 morphants and dnmt1 morphants.
Whole Mount in Situ Hybridizations and Histological Analyses
Whole mount in situ hybridizations were carried out as described previously (13) using digoxigenin-labeled Riboprobes for dnmt3, ascl1a, ascl1b, ngn1, dhand, foxa3, cmlc2, insulin, trypsin, pcna, pax6.2, pax2.1, dhand, crx, neurod, irbp, gata6, fabp10, and fabp2. For histological analyses, embryos were fixed in 10% neutral buffered formalin, rinsed in phosphate-buffered saline, and embedded in glycol methacrylate (Polysciences). Five-micron sections were cut using a Leica microtome and stained in toluidine blue. Sections were analyzed using a Zeiss Axiovert 100 microscope, and pictures were taken using an Olympus Magnafire color camera.
5′ and 3′ RACE
For 5′ and 3′ RACE experiments, mRNA made from zebrafish embryos at different stages (2–96 hpf) was pooled together, and RACE ready cDNA was generated using a SMART RACE cDNA amplification kit (Clontech), and PCR was performed using Advantage PCR kit (Clontech).
Methylated DNA Immunoprecipitation
This procedure was previously described (15). Briefly, Dynabeads (conjugated with sheep anti-mouse secondary antibody, Invitrogen) were incubated with 10 μg of MeC antibody (Eurogentec) for 2 h, washed, and then incubated with 4 μg of sonicated DNA (300- to 1000-bp fragments, denatured after sonication) overnight in methylated DNA immunoprecipitation (MeDIP) buffer (20 mm Tris (pH 7.5), 140 mm NaCl, 0.05% Triton X-100). On the next day, immunocomplexes were washed three times in MeDIP buffer and then eluted by Proteinase K digestion at 55 °C.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described earlier (16). Embryos were injected with dnmt3 Mo2 (8 ng) or g9a Mo1 (6 ng) at one cell stage, and extracts were made at 24 hpf. Antibodies used were H3K9me3 (Active Motif, 39161), H3 (Abcam, 1791), and rabbit IgG (Sigma).
RT-PCR
RNA from embryos or human adenoma tissues was isolated using TRIzol (Invitrogen). A cDNA library was prepared using Superscript III kit (Invitrogen). RT-PCR was performed on a Roche Applied Science light cycler. Primer information is available upon request.
Alcian Blue Staining
Embryos were fixed at 96 hpf in 4% paraformaldehyde, washed, and then bleached in 30% hydrogen peroxide for 2 h. After rinsing in PBST (1× phosphate-buffered saline and 0.1% Tween 20) they were stained in filtered Alcian Blue solution (1% concentrated HCl, 70% EtOH, and 0.012% Alcian Blue) overnight. Once stained they were cleared in acidic ethanol (5% concentrated HCl, 70% ethanol) for 4 h and then dehydrated in increasing amounts of EtOH. Finally, embryos were stored in glycerol and pictures were taken.
RESULTS
Zebrafish Dnmt3 Is Required for Neurogenesis in the Brain
Zebrafish Dnmt3 is an ortholog of mammalian DNMT3B and contains a well conserved catalytic motif within its C terminus (9). In addition, Dnmt3 bears an unusually long N terminus, which harbors three domains: a CXXC-like motif (a redox-sensitive metal binding motif), a PWWP domain (for nonspecific DNA interaction), and a calponin homology domain (9). The CXXC-like motif and the PWWP domain are shared with other mammalian Dnmt3 proteins. However, the presence of the calponin homology domain in the zebrafish Dnmts is unusual in that it is not observed in any other DNA methyltransferase-related protein in other organisms. The calponin homology domain is usually found in proteins that have cytoskeletal functions (17), raising the possibility that this domain provides an additional function to Dnmt3.
We began our studies by characterizing the expression pattern of dnmt3 by whole mount in situ analysis. Dnmt3 is expressed in eyes, brain, somites (at 24, 48, and 72 hpf), and the gut (at 48 and 72 hpf; supplemental Fig. S1). To delineate the role of Dnmt3 during development, we knocked down Dnmt3 levels using an antisense morpholino that blocks the splicing of the exon6-intron6 junction (Fig. 1A). We refer to animals injected with this morpholino as dnmt3 morphants, and animals injected with a control morpholino as control morphants. Dnmt3 morphants displayed 71% (at 8 pg) decrease in the levels of correctly spliced dnmt3 transcript at 80 hpf. Similar knockdown was seen at earlier developmental stages as well (data not shown). Consistent with the study of DNMT3B knock-out in HCT116 colon carcinoma cells (18), we did not observe any difference in global 5-methylcytosine levels in dnmt3 morphants (at 71% reduction of dnmt3 transcripts) compared with wild-type zebrafish embryos at either 24 or 72 hpf (supplemental Fig. S2A). To examine this further, we also determined the methylation status of the no tail gene, which is known to undergo developmental methylation (9). Bisulfite sequence analysis of a no tail CpG island spanning the transcription start site, showed a cumulative 43% methylation in wild-type embryos, whereas dnmt3 morphants showed reduced cumulative methylation (18%) (supplemental Fig. S2B). In agreement with loss of methylation, expression of no tail was up-regulated specifically in the notochord in the tail of dnmt3 morphants at 24 hpf (supplemental Fig. S2C). These data indicate that Dnmt3 functions as a DNA methyltransferase in vivo and may specifically target genes during zebrafish development.
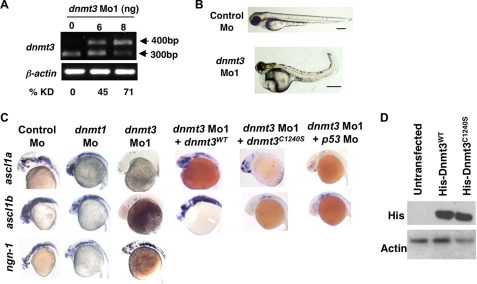
FIGURE 1.
Dnmt3 knockdown in zebrafish embryos results in neurogenesis defects. A, splice blocking by dnmt3 splice-blocking morpholino (dnmt3 Mo1) was monitored by RT-PCR in mRNAs from control and dnmt3 morphants at 80 hpf. PCR was performed using a forward primer in exon 6 and a reverse primer in exon 7. Note that the dnmt3 Mo1 stabilizes the unspliced transcript containing introns 6 and 7 (400 bp), whereas the spliced version (300 bp) is detected in control injected embryos. Percent knockdown (% KD) shown is the ratio of intensity of unspliced product to combined intensity of unspliced and spliced products normalized over β-actin intensity. B, morphology defects in dnmt3 morphants at 80 hpf. Note the smaller head, a pericardial edema, and a curled tail in dnmt3 morphants compared with control morphants. Bar equals 0.5 mm. C, whole mount in situ analysis of ngn-1, ascl1a, and ascl1b expression in dnmt3 morpholino alone or with p53 morpholino-, dnmt1 morpholino-, or control morpholino-injected embryos at 30 hpf. Whereas ngn-1 expression was normal in dnmt3 and dnmt1 morphants, ascl1a and ascl1b were selectively is absent. This defect can be complemented by co-injection of the wild-type (Dnmt3WT) but not by a catalytically inactive derivative (Dnmt3C1240S). D, protein expression levels of Dnmt3 wild-type (Dnmt3WT) and catalytically inactive (Dnmt3C1240S) derivatives. HEK293 cells were transfected with Dnmt3WT and Dnmt3C1240S, and Western blots were performed using antibodies against His tag. Both of these show equal expression of these derivatives. β-Actin was used as a loading control.
The phenotype associated with the dnmt3 morphants was highly penetrant. They displayed multiple abnormalities throughout early embryonic development and died at 96 hpf (Fig. 1B). The most obvious defects occurred in the brain, which was much smaller when compared with brains of age-matched control morphants (Fig. 1B).
Patterning of the brain appeared normal in dnmt3 morphants as evidenced by the normal expression of the following markers: krox20 for rhombomere 3 and 5 (19), dlx2 for diencephalon and telencephalon (20, 21), pax6.2 for dorsal diencephalons (22), and pax2.1 for hindbrain neurons and midbrain-hindbrain boundary (23) (supplemental Fig. S3 and supplemental Table SII). Therefore we examined brain neurogenesis. Interestingly, expression of later pro-neural markers, ascl1a and ascl1b (orthologous to mouse Mash1) (24), was completely absent (or drastically reduced) in dnmt3 morphants from the 19-somite stage to 48 hpf (Fig. 1C, supplemental Table SI, and data not shown). Some examples of our assessment of positive and negative staining are shown in supplemental Fig. S4. In contrast, an earlier marker of brain neurons, neurogenin-1 (25–27), was expressed normally (Fig. 1C) suggesting that Dnmt3 was essential for proper development of brain neurons. Importantly, these defects could be complemented by co-injection of the wild-type zebrafish Dnmt3 (Dnmt3WT) but not by a derivative which has the conserved catalytic cysteine mutated to serine (Dnmt3C1240S) despite their equal expression (Fig. 1, C and D). Also, the defects were rescued by co-injection of wild-type human DNMT3B, but not by a catalytically inactive human DNMT3B derivative (DNMT3BC651S) (supplemental Table SI). Finally, embryos injected with another morpholino (dnmt3 Mo2, an ATG blocker) against Dnmt3 exhibited similar defects in brain neurogenesis (supplemental Fig. S5). Taken together, these experiments establish the specificity of the phenotype, its dependence on catalytic activity, and conservation of function between zebrafish and humans.
As dnmt3 morphants displayed severe cell death in the brain, we tested whether the loss of expression of ascl1a and ascl1b (and hence absence of neurogenic differentiation) was due to selective apoptosis of cells expressing these genes. We sought to determine whether reduction of apoptosis by knocking down expression of p53 (a critical mediator of apoptosis) would result in re-expression of these markers in dnmt3 morphants. To this end, we co-injected p53 splice blocking morpholino along with dnmt3 morpholino and assessed expression of ascl1a and ascl1b at 30 hpf. Although knockdown of p53 resulted in less cell death in dnmt3 morphants (even though knockdown was partial), it did not compensate for the loss of ascl1a or ascl1b expression suggesting p53-independent block in neurogenesis in dnmt3 morphants (Fig. 1C, supplemental Table SI, and supplemental Fig. S6). These data establish p53-independent neural differentiation defects upon loss of Dnmt3.
dnmt1 and dnmt3 Morphants Display Different Tissue-specific Defects
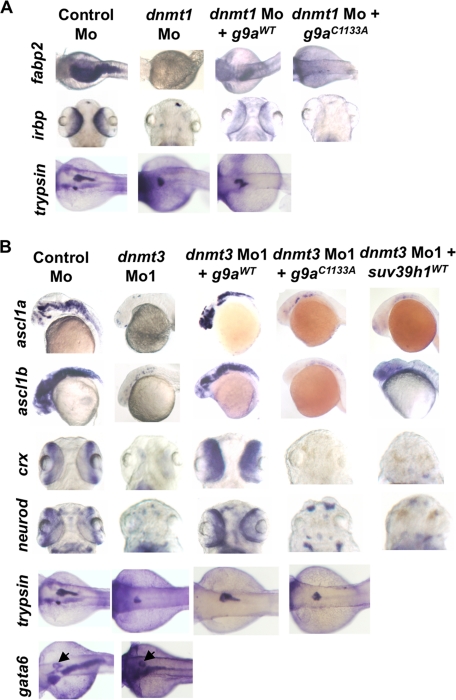
Interestingly, these neurogenic differentiation defects seen in dnmt3 morphants were not present in dnmt1 morphants in that dnmt1 morphants expressed ngn-1, ascl1a, and ascl1b relatively normally (Fig. 1C). This suggested to us that these two DNA methyltransferases do not have redundant functions at least during neurogenesis in the brain and might have different tissue-specific roles during zebrafish development. To test this hypothesis we analyzed other developing tissues. We first looked at the formation of pharyngeal pouches by analyzing expression of dhand at 48 hpf. Dnmt3 morphants were defective in pharyngeal arch formation, as marked by the loss of expression of dhand in the two posterior arches (Fig. 2A and supplemental Table SII) (28). However, dnmt1 morphants formed these pharyngeal arches correctly (Fig. 2A) supporting their different roles in zebrafish development. Next, we looked at the organs that show developmental defects in dnmt1 morphants. We first analyzed jaw cartilage formation in dnmt3 morphants by Alcian Blue staining at 96 hpf. Unlike dnmt1 morphants, jaws formed correctly in dnmt3 morphants (Fig. 2B), again suggesting their differential requirement in jaw development. Next, we analyzed development of the organs in gastrointestinal tract. We have earlier shown that dnmt1 morphants were defective in terminal differentiation of intestine. The specificity seen in jaw and pharyngeal pouch defects in these morphants extended to the intestine in that unlike dnmt1 morphants, terminal differentiation of intestine took place relatively normally in dnmt3 morphants as judged by the expression of intestinal fatty acid binding protein (fabp2) (29) (Fig. 2C). These data suggest that Dnmt3, but not Dnmt1, is dispensable for terminal differentiation of intestine. Next, we tested the expression of markers of terminal differentiation of liver and endocrine pancreas in dnmt3 morphants. Both dnmt1 and dnmt3 morphants express fabp10 (liver) (30) and insulin (endocrine pancreas) (31) normally, suggesting that that the liver and endocrine pancreas do not require these proteins for their terminal differentiation, by the criteria tested (Fig. 2C and supplemental Table SI). Finally, we analyzed expression of terminal differentiation marker of exocrine pancreas (trypsin) (31), whose expression was drastically reduced in dnmt1 morphants (Fig. 2C). Here, similar to dnmt1 morphants, dnmt3 morphants showed reduction in levels of trypsin. However, an earlier marker of pancreatic differentiation, gata6, was intact in dnmt3 morphants (Fig. 2C). Taken together, all these data suggest that Dnmt1 and Dnmt3 have largely different (and some overlapping) tissue-specific functions.
FIGURE 2.
Distinct tissue-specific developmental defects in dnmt1 and dnmt3 morphants. A, C, and E, whole mount in situ analysis of dhand (at 48 hpf) (A), fabp2, trypsin, fabp10, insulin, and trypsin (C), and irbp (E) (all at 96 hpf) in dnmt1, dnmt3, and control morphants. In panel A, arrows show loss of last two pharyngeal pouches in dnmt3 morphants. In panel C, arrows point to expression of gata6 in the pancreas. B, Alcian Blue staining of Control, dnmt1, or dnmt3 morphants at 96 hpf shows loss of jaw structure in dnmt1 morphants but not in dnmt3 morphants. D, toluidine blue staining of histological cross-sections within the dnmt3 morphant retinas at 80 hpf. Note the loss of organization of different retinal layers in dnmt3 morphants. F, quantitative RT-PCR for Opsin Shortwave 1 and Opsin Shortwave 2 in control, dnmt1, or dnmt3 morpholino-injected embryos at 96 hpf.
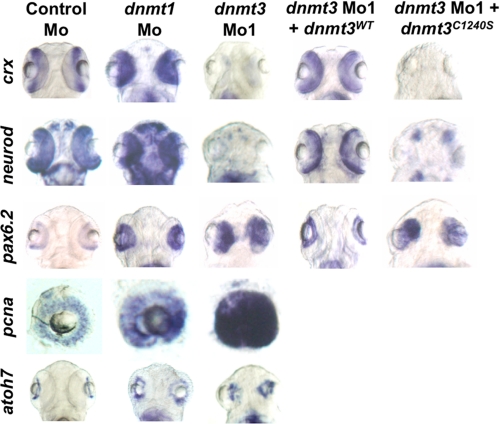
Next, we tested whether Dnmt3 was required for proper development of the eyes, because dnmt3 is expressed in the eyes at various embryonic stages (supplemental Fig. S1 and data not shown), and dnmt3 morphants had significantly smaller eyes than control morphants (Fig. 1B). Cross-sections of eyes at 80 hpf revealed that dnmt3 morphants lacked the proper retinal lamination in comparison to the control morphants (Fig. 2D). When the cross-sections of dnmt3 morphant eyes were compared with those of dnmt1 morphants, several differences were observed. First, loss of retinal pigmented epithelium (RPE) was observed in both morphants but in different anatomical positions. Dnmt1 morphants lack RPE from the dorsal side, whereas dnmt3 morphants are devoid of RPE from the ventral side. Both of these morphants lacked irbp expression, a marker of terminally differentiated RPE (Fig. 2E) (32). Second, the lens morphology was more differentiated in dnmt1 morphants compared with dnmt3 morphants. Finally, on the whole, eyes of dnmt1 morphants looked more differentiated compared with dnmt3 morphants in that dnmt1 morphant neuroepithelial retina consisted of multiple types of cells, whereas in dnmt3 morphants all cells seemed to be of one origin. Both of these morphants lacked terminal markers of photoreceptor cells (Fig. 2F) (opsins are markers of cones (33)). Although terminally differentiated cells were lost in both of these morphants, based on the presence of different types of cells we hypothesized that they were stalled at different stages of eye development. dnmt3 morphant eyes consist of more progenitor-like cells, whereas dnmt1 morphants seemed to be defective at a later stage of differentiation. This hypothesis was supported by the fact that all the cells in the eyes of dnmt3 morphants expressed pax6.2 and pcna, two genes expressed in the earliest retinal progenitors (34, 35) (Fig. 3). This suggests that retinal cells are specified in dnmt3 morphants but retain their progenitor identity and fail to differentiate. However, when compared with dnmt3 morphants, pcna and pax6.2 were less robustly expressed in the eyes of dnmt1 morphants (although their expression was more than that in control morpholino-injected embryos). To more precisely characterize this defect in these morphants, we analyzed the expression of various differentiation markers of neuroepithelial cell types. As mentioned previously, terminally differentiated retinal neuroepithelial cell types were absent in both dnmt1 and dnmt3 morphants. Therefore, we assessed the expression of markers of precursors of these specific cell types. Interestingly, dnmt3 morphants lacked the expression of crx, neurod, and ascl1a (36, 37) (markers of photoreceptors and amacrine cells) but retained expression of atoh7, a marker of ganglion precursors (Fig. 3). These defects were linked to the loss of Dnmt3 activity, because these defects could be complemented by wild-type zebrafish Dnmt3 or human Dnmt3B but not by catalytically inactive derivatives (Fig. 3 and supplemental Table SI). These data suggest that Dnmt3 is required for differentiation of retinal progenitors into specific precursors of horizontal, bipolar, and photoreceptor cells, but not of ganglion cells. In contrast, dnmt1 morphants harbor defects in terminal differentiation of these cell types and showed normal expression of crx, neurod, and atoh7 (Fig. 3). These data suggest that Dnmt3 is required for early steps of retinal neuroepithelial differentiation, whereas Dnmt1 is required during later stages.
FIGURE 3.
Dnmt3 morphants are defective in retinal differentiation. Whole mount in situ for pax6.2, crx, and neurod in dnmt3 and control morphants at 80 hpf. Whereas all the retinal cells expressed pax6.2, a marker of retinal progenitors, markers of specific progenitors (crx and neurod) were absent in dnmt3 morphants. These defects can be complemented by co-injection of the wild-type Dnmt3 (Dnmt3WT) but not with the catalytically inactive Dnmt3 derivative (Dnmt3C1240S).
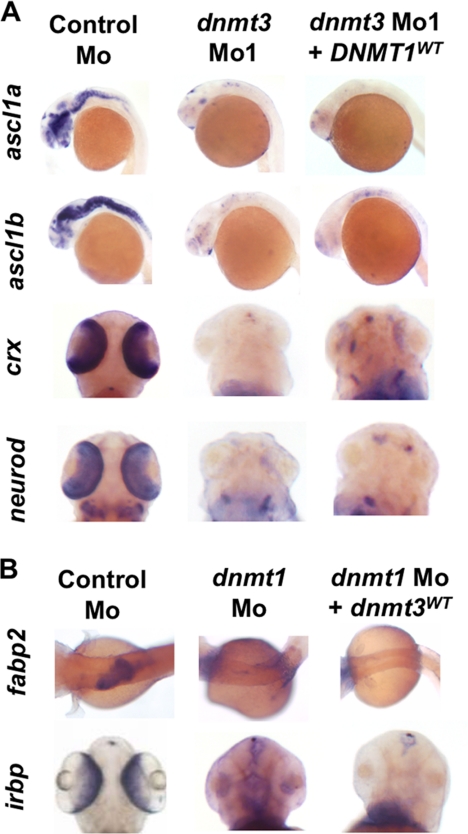
Our extensive analysis of expression of developmental markers of different tissues and different differentiation stages revealed that Dnmt1 and Dnmt3 have largely non-overlapping and tissue-specific functions. Another test of this hypothesis would be to examine if one protein can substitute for the loss of the other. To this end, we tested whether overexpression of Dnmt1 in dnmt3 morphants could rescue its developmental defects and vice versa. Notably, we saw that overproduction of Dnmt1 was unable to make up for loss of Dnmt3 during neurogenic (assessed by ascl1a and ascl1b expression) and retinal differentiation (assessed by crx and neurod expression) (Fig. 4A). Similarly, Dnmt3 was insufficient to rescue intestinal (assessed by fabp2 expression) and retinal defects (assessed by irbp expression) in dnmt1 morphants (Fig. 4B).
FIGURE 4.
Dnmt3 overexpression in dnmt1 morphants and Dnmt1 overexpression in dnmt3 morphants does not rescue developmental defects. A and B, whole mount in situ expression for ascl1a, ascl1b, crx, and neurod (A) and fabp2 and irbp (B) in embryos injected with control morpholino, dnmt3 morpholino alone, or with DNMT1 mRNA (A) and dnmt1 morpholino alone or with dnmt3 mRNA (B).
Aberrantly Hypomethylated and Expressed lef1 Confers Neurogenesis Defects in dnmt3 Morphants
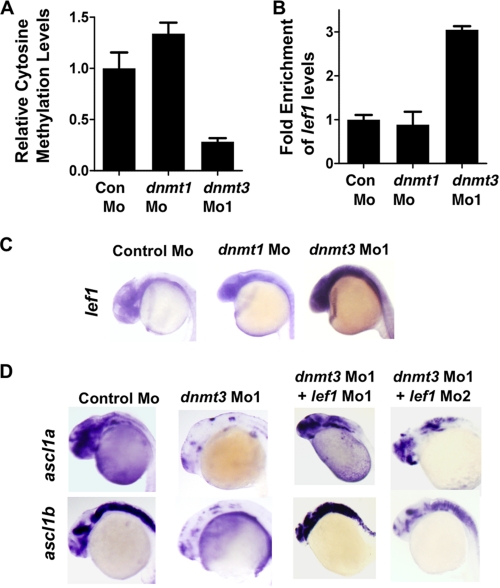
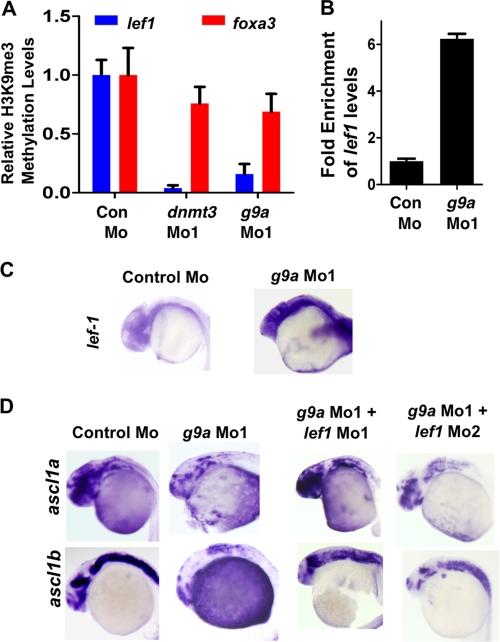
Because we observed a neurogenesis defect in dnmt3 morphants but not in dnmt1 morphants, we decided to investigate the methylation status of lef1 promoter in dnmt3 morphants. Lef1 is a transcription factor that has been shown to be critical for neurogenesis in hypothalamus (14). It was shown that knockdown of Lef1 levels results in loss of expression of ascl1a and ascl1b, but not ngn-1, specifically from hypothalamus, suggesting its requirement at a particular stage in a specific region in the brain (14). As noted above, we see defects in exactly the same stage in dnmt3 morphants in the entire brain (Fig. 1D), not only in hypothalamus. We considered a possibility that Lef1 is normally kept low in other parts of brain through promoter DNA methylation by Dnmt3. In dnmt3 morphants, lef1 promoter may become demethylated and aberrantly expressed in the entire brain, and this misexpression could lead to blockage of neural differentiation. To test this hypothesis, we first examined the methylation status of lef1 promoter by methylated DNA immunoprecipitation (using an antibody specific to 5-methylcytosine) coupled to quantitative PCR in WT, dnmt3 Mo, and dnmt1 Mo embryos to find a demethylated region. We found that a region ∼2300 bp upstream of the transcription start site showed dramatic demethylation in dnmt3 morphants but not in dnmt1 morphants at 24 hpf (Fig. 5A). Consistent with that, we found up-regulation of lef1 transcripts in the brains of dnmt1 morphants but not dnmt1 morphants at 24 hpf (Fig. 5, B and C). Then we asked if this up-regulation of lef1 is the cause of neurogenesis defects in dnmt3 morphants. To this end, we injected two different lef1 morpholinos (lef1 Mo1, 0.8 ng, and lef1 Mo2, 2 ng) along with dnmt3 morpholino, and we observed a robust rescue of ascl1a and ascl1b expression clearly suggesting that demethylation and up-regulation of lef1 in dnmt3 morphants results in neurogenesis defects (Fig. 5D and supplemental Table SI).
FIGURE 5.
Lef1 repression by Dnmt3 is critical for proper neurogenesis. A, MeDIP-PCR based quantification of methylation status of lef1 promoter in control, dnmt1, and dnmt3 morphant embryos at 24 hpf. The y-axis represents values at lef1 promoter first normalized to a negative region (with an insignificant number of CpGs in 1000 bp vicinity) and then normalized to control Mo values as 1. B, graph showing quantitative RT-PCR results for zebrafish lef1 normalized to 28 S values in control morpholino-, dnmt1 morpholino-, or dnmt3 morpholino-injected embryos at 24 hpf. Results are represented in a -fold change format where the lef1/28S ratio from control morphants was normalized to 1. C, expression of lef1 in control, dnmt1, and dnmt3 morphant embryos at 24 hpf as detected by whole mount in situ hybridization. D, whole mount in situ analysis of ascl1a and ascl1b expression in embryos (30 hpf) injected with control morpholino and dnmt3 morpholino co-injected with either lef1 Mo1 or lef1 Mo2.
G9a Functions Cooperatively with Dnmt3
Previously, we showed that suv39h1 morphants largely phenocopy dnmt1 morphants and that overexpression of suv39h1 rescues dnmt1 morphant phenotypes (3) suggesting that Dnmt1 depends on Suv39h1 for its function during zebrafish development. Therefore, we wanted to test whether Dnmt3 also depends on a histone H3K9 methyltransferase for its function during zebrafish development. First, we tested if Dnmt3 functionally interacts with Suv39h1. To test this possibility, we examined if suv39h1 morphants phenocopy dnmt3 morphants, and whether suv39h1 overexpression would rescue dnmt3 morphant defects. We found that suv39h1 morphants did not display defects in neurogenesis in the brain (as measured by ascl1a and ascl1b expression) or eyes (as measured by expression of pcna, pax6.2, crx, and neurod) (Fig. 6C and supplemental Table SI). In agreement with these data, overexpression of suv39h1 in dnmt3 morphants did not rescue the neurogenesis defects in brain or eyes as measured by expression of ascl1a, ascl1b, crx, and neurod (Fig. 7B and supplemental Table SI). These two pieces of data suggest that Dnmt3 does not depend on Suv39h1 for its function during neuronal differentiation.
FIGURE 6.
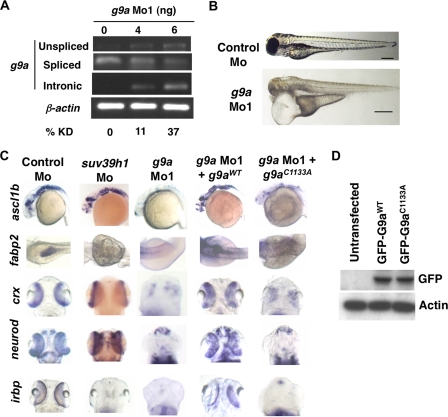
G9a morphants largely phenocopy dnmt3 morphants. A, splice blocking by g9a splice-blocking morpholino (g9a Mo1) was monitored by RT-PCR in mRNAs made from control and g9a morphants at 80 hpf using a forward primer in the exon and reverse primer in the next exon. Note that the g9a morpholino stabilizes the unspliced transcript containing the intermediate intron, whereas the spliced version is detected in control-injected embryos. The percent knockdown shown is the ratio of intensity of unspliced product to combined intensity of unspliced and spliced products normalized to intensity of the β-actin band. Note that a product can be amplified with an intronic primer in the g9a morphants; however, this was not taken into account while calculating percent knockdown. B, morphology defects in g9a morphants at 80 hpf. Note the smaller head in g9a morphants, as with dnmt3 morphants. Bar equals 0.5 mm. C, whole mount in situ analysis of ascl1b, crx, neurod, fabp2, and irbp expression in g9a, suv39h1, and control morphants. ascl1b was analyzed at 30 hpf, whereas all others were analyzed at 80 hpf. The defects present in g9a morphants were complemented by co-injection of the wild-type G9a (G9aWT), but not by the catalytically inactive derivative G9aC1133A. D, expression of exogenous G9a constructs. HEK293 cells were transfected with the wild-type or catalytically dead derivatives of GFP-tagged zebrafish G9a (G9aWT and G9aC1133A). Westerns using antibodies against the GFP tag show equal expression of these derivatives. β-Actin was used a loading control.
FIGURE 7.
G9a overexpression rescues both dnmt3 morphants and dnmt1 morphants. Whole mount in situ analysis of ascl1a, ascl1b, crx, and neurod (A) and fabp2, irbp, and trypsin (B) expression in embryos injected with control morpholino or dnmt3 or dnmt1 morpholino co-injected with wild-type or catalytically null G9a or Suv39h1. ascl1a and ascl1b were analyzed at 30 hpf, whereas all others were analyzed at 80 hpf. Note that co-injection of wild-type G9a (G9aWT) but not of either catalytically inactive G9a (G9aC1133S) or wild-type Suv39h1 rescues the ascl1a and ascl1b expression in dnmt3 morphants. Overexpression of wild-type G9a, but not catalytically inactive G9a, can also rescue fabp2 and irbp expression in dnmt1 morphants. Of note, wild-type G9a could not rescue trypsin expression in dnmt1 morphants. The panel showing Suv39h1 rescue of dnmt1 morphants has been reported previously (7) and is shown for comparative purposes.
To further investigate the dependence of Dnmt3 on a H3K9 methyltransferase we next focused on another major histone H3K9 methyltransferase, G9a. Because zebrafish G9a had not been characterized, we first needed to identify and characterize zebrafish G9a. TBLASTN searches of the zebrafish genome using human and mouse G9a protein sequences revealed a contig containing a partial match to the catalytic domain of human/mouse G9A. We then obtained the complete G9a sequence by 5′- and 3′-RACE (NCBI accession no. EU070918). The predicted protein encoded by this gene was 51% similar to human G9A. We found that g9a is maternally supplied and shows tissue-specific expression during later stages, with prominent expression in the eye, brain, anterior somites, and intestine (supplemental Fig. S7). Notably, these are the tissues where dnmt1 and dnmt3 are also expressed (supplemental Fig. S1).
To test whether G9a cooperates with DNA methyltransferases in promoting gene silencing during development we determined the extent to which g9a morphants phenocopy dnmt1 or dnmt3 morphants, and whether G9a overexpression will rescue the defects present in dnmt1 or dnmt3 morphants. We first knocked down g9a levels by a splice blocking morpholino. An examination of organ development required the titration of this morpholino to reduce G9a levels by 37% at 6 pg (Fig. 6A). Any further knockdown of G9a resulted in lethality within the first 24 h. At 37% knockdown, we did not see any obvious changes in the global H3K9m2 or H3K9me3 levels in the g9a morphants at 24 hpf (supplemental Fig. S2F). However, these morphants were severely compromised throughout early embryonic development and died around 80 hpf. Strikingly, like dnmt3 morphants, g9a morphants exhibited a drastic reduction in brain size suggesting that G9a and Dnmt3 may cooperate to promote brain development (Fig. 6B).
To further test this idea, we examined the status of molecular markers in g9a morphants that were absent in dnmt3 morphants. As observed in the case of dnmt3 morphants, expression of the neuronal markers ascl1a and ascl1b was severely reduced in g9a morphants (Fig. 6C and supplemental Table SI). Similarly, expression of retinal neuroepithelial markers neurod and crx was absent in g9a morphants. The expression of these genes was rescued by co-injection of wild-type zebrafish g9a RNA (G9aWT) with g9a morpholino but not by co-injection of an RNA encoding a catalytically inactivated G9a derivative (G9aC1133A), despite their equal expression (Fig. 6, C and D and supplemental Table SI). Finally, embryos injected with a second morpholino against G9a exhibited similar neurogenesis defects in the brain (supplemental Fig. S8). These data suggest that the neuronal and retinal differentiation defects present in g9a morphants were specific to this gene and dependent on its catalytic activity. Furthermore, wild-type human G9A, but not catalytically inactive human G9A mRNA (G9AC1114A), rescued these defects suggesting conservation of function during evolution (supplemental Table SI).
Next we tested the extent to which g9a morphants phenocopy dnmt1 morphants. Interestingly, g9a morphants partially phenocopied dnmt1 morphants, as they lacked expression of fabp2, irbp, and trypsin; genes whose expression is lost in dnmt1 morphants (Fig. 6C and data not shown). Again these markers were complemented by co-injection of wild-type but not catalytically inactive zebrafish or human G9A (Fig. 6C and supplemental Table SI). As with dnmt1 morphants, g9a morphants harbored normal expression of gata6, a marker of early endoderm (38), suggesting that the intestinal and pancreatic differentiation defects were later defects (data not shown and supplemental Table SI). These data suggest that G9a has diverse roles, whereas Suv39h1 is more specific; g9a morphants have broad defects and phenocopy both dnmt1 and dnmt3 morphants, whereas suv39h1 morphants only phenocopy dnmt1 morphants.
Next we tested whether G9a functions downstream of Dnmt3 or Dnmt1. Interestingly, overexpression of wild-type zebrafish or human G9a, but not catalytically inactive derivatives, was highly effective in rescuing certain dnmt1 and dnmt3 morphant defects; expression of ascl1a, ascl1b, crx, and neurod in dnmt3 morphants and fabp2 in dnmt1 morphants (Fig. 7 and supplemental Table SI). Interestingly, we noticed that overexpression of G9a rescued trypsin expression in dnmt3 morphants but not in dnmt1 morphants suggesting Dnmt3-G9a specificity during pancreatic differentiation. Finally, overexpression of Dnmt3 in g9a morphants did not rescue the neural differentiation defects (data not shown). Taken together, these data strongly suggest specific epistatic relationship and cooperativity between HMTs and DNMTs; G9a functions primarily downstream of Dnmt3 and to some extent with Dnmt1, whereas Suv39h1 acts downstream of Dnmt1 only.
G9a and Dnmt3 Control lef1 Expression by H3K9me3 Methylation of Its Promoter
These data suggested that Dnmt3 and G9a may cooperate to regulate the expression of genes critical for neural development. DNA methylation of Lef1 promoter by Dnmt3 and subsequent repression was found to be critical for ensuring proper neurogenesis (Fig. 5). We hypothesized that DNA methylation of Lef1 by Dnmt3 results in H3K9me3 methylation by G9a (which is recruited either directly or indirectly by Dnmt3), and that sets up repressive environment in the promoter and subsequent silencing of the gene. To test this idea, we checked the status of H3K9me3 in the promoter regions of Lef1 (at the regions that are demethylated) in dnmt3 morphants and g9a morphants. Chromatin immunoprecipitation experiments revealed that H3K9me3 levels were drastically reduced specifically in lef1 promoter but not at foxa3 promoter (a gene whose expression remain unchanged; data not shown) in both dnmt3 and g9a morphants at 24 hpf (Fig. 8A). Consistent with the hypothesis, lef1 expression was increased in g9a morphants, and its knockdown rescued ascl1a and ascl1b expression in g9a morphants (Fig. 8, B–D).
FIGURE 8.
Lef1 repression by G9a is critical for proper neurogenesis. A, quantitative PCR for chromatin immunoprecipitation for H3K9me3 marks on lef1 promoter (Blue) or on foxa3 promoter (Red) in control morphant, dnmt3 morphant, and g9a morphant embryos. Values shown represent enrichment on the experimental region normalized to values on control region. B, graph showing quantitative RT-PCR results for zebrafish lef1 normalized to 28S values in control morpholino or g9a morpholino-injected embryos at 24 hpf. Results are represented in a -fold change format where the lef1/28S ratio from control morphants was normalized to 1. C, expression of lef1 in control and g9a morphant embryos at 24 hpf as detected by whole mount in situ hybridization. D, whole mount in situ analysis of ascl1a and ascl1b expression in embryos (30 hpf) injected with control morpholino and g9a morpholino co-injected with either lef1 Mo1 or lef1 Mo2.
DISCUSSION
Our work provides new information regarding DNMT-HMT cooperativity during zebrafish development. First, we establish that Dnmt3 is required in a tissue-specific manner. Second, we show that Dnmt1 and Dnmt3 have largely distinct tissue-specific and temporal functions during zebrafish development. Third, we identify a critical fate regulator, Lef1, that harbors differential methylation in dnmt3 but not dnmt1 morphants and is responsible for neurogenesis defects seen in dnmt3 and g9a morphants. Fourth, histone H3K9 methyltransferase G9a is also required for tissue-specific development in the early embryo and operates in a manner distinct from Suv39h1. Fifth, we provide genetic evidence that Dnmt3 interacts with G9a to promote zebrafish development. Importantly, Dnmt3 function depends selectively on G9a, but not Suv39h1. Finally, dnmt1 morphants can be partially rescued by G9a overexpression. Taken together, our work suggests that Dnmt1 and Dnmt3 have distinct tissue-specific targets and thus are required in tissue-specific manner during embryonic development. Moreover, there exists specific DNMT-HMT cooperativity to repress critical regulators of development, like Lef1.
Dnmt3 has tissue-specific functions because it is required for proper differentiation of neurons in the brain, exocrine pancreas, pharyngeal arches, and certain retinal tissues. However, Dnmt3 is dispensable for the development of the jaw, intestine, endocrine pancreas, or liver (by the methods tested). The data presented here, along with our previous data that Dnmt1 is also required for the development of particular tissues, strongly argue that DNA methylation has critical roles in establishing tissue-specific gene expression patterns. Also, some of the defects seen in dnmt1 morphants were present in a recently characterized Dnmt1 mutant providing further genetic basis to the idea of tissue specificity of DNMTs (4). Although it is possible that tissue-specific defects arise in a particular dnmt morphant because of selective depletion of the maternal mRNA and/or protein in highly proliferating tissues, several pieces of data suggest that this is unlikely. First, dnmt morphants display defects in both highly proliferative tissues like eyes and exocrine pancreas, as well as other less proliferative tissues like jaw and fins. In addition, highly proliferating tissues like liver are unaffected in both dnmt1 and dnmt3 morphants. It has also been suggested that selective apoptosis in some organs might underlie developmental defects resulting from DNMT loss. For example, Dnmt1 loss in fibroblasts led to p53-dependent death and specific gene expression changes (39). Anderson et al. recently showed that some developmental defects (e.g. in pancreas), but not all, depend on p53-mediated death in Dnmt1 mutant zebrafish (4). However, in dnmt3 morphants, p53 knockdown did not rescue neurogenesis defects despite rescuing some cell death suggesting that p53-mediated cell death might not explain all the defects seen upon DNMT loss. Consistent with this notion of tissue-specific roles of DNA methylation, several recent studies have shown the existence of tissue-specific cytosine methylation in the promoters of regulatory genes in the non-expressing tissues. Some of these genes include testis-specific MAGE genes (40), erythrocyte specific β-globin genes (41), hippocampal neuron-specific Shank3 gene (42), and liver-specific Abcc6 gene (43). Differential existence of DNA methylation in the promoters of genes has also been validated by recent genome-wide DNA methylation analyses. For example, differential patterns of DNA methylation exist between mouse stem cells, progenitor cells, and differentiated cells. Also, germ cell-specific genes were shown to be heavily methylated in somatic tissues (15, 44). Our studies extend these observations and show that the differential methylation of regulatory genes might be critical in governing normal tissue-specific and temporal development.
Of particular interest was our observation that the tissues affected in dnmt3 morphants were different than those affected in dnmt1 morphants suggesting that certain tissues are more reliant on one of these enzymes versus another. For example, dnmt3 morphants display brain neurogenesis defects, which are not observed in dnmt1 morphants. On the other hand, dnmt1 morphants lack terminal intestinal differentiation, whereas normal differentiation is observed in dnmt3 morphants. However, Dnmt3 is indeed expressed in intestine starting at 48 hpf. This suggests that Dnmt3 is not required for morphogenesis or differentiation of this organ, although redundancy between members of the DNMT3 family of enzymes for function in the intestine cannot be ruled out. Other examples of non-overlapping functions of Dnmt1 and Dnmt3 were seen in pharyngeal arch formation and jaw development. Dnmt1 morphants specifically lacked jaw development, whereas dnmt3 morphants harbored abnormal pharyngeal arches. Finally, in retina these enzymes are required for formation of different cell types at different developmental stages (discussed below). Consistent with these data, we note that, although dnmt3 and dnmt1 are co-expressed in some tissues (supplemental Fig. S1) (3), spatial and temporal Dnmt expression coincides well with the defects seen in morphant tissues. Confirming tissue-specific roles of these enzymes Dnmt1 overexpression failed to rescue defects present in dnmt3 morphants. Similarly Dnmt3 overproduction failed to compensate for Dnmt1 loss. We conclude that Dnmt1 and Dnmt3 have partially non-overlapping and tissue-specific functions during zebrafish development.
It is also interesting to note that most of the defects observed in dnmt3 morphants occurred during early steps of organ differentiation (for example in the early neuronal differentiation in brain and early specification of amacrine cells in the eyes) (Figs. 1 and 3). This is in contrast to dnmt1 morphants, which largely harbor terminal differentiation defects (for example in terminal intestinal and retinal differentiation) (3). These data suggest that different DNA methyltransferases regulate different transcriptional programs, which define the differentiation stage of the organ. Our data are consistent with the notion of Dnmt3b acting as a de novo methyltransferase and Dnmt1 functioning primarily as a maintenance methyltransferase. It is plausible that de novo activity could be dominant early during development to establish specific methylation patterns, whereas maintenance activity would function later. The tissue-specific responses following loss of Dnmt1 and Dnmt3 suggest the potential for specific activities of members of the Dnmt3 family present in zebrafish. Of note, zebrafish harbor five additional DNA methyltransferases related to Dnmt3, each of which show distinct spatial and temporal regulation during development. It remains possible (though untested) that these Dnmts likewise have organ-specific roles.
Our previous studies showed that dnmt1 morphants harbor reduced global levels of both DNA methylation and H3K9 trimethylation (3). In counter distinction, dnmt3 morphants retained global levels of methylation. However, no tail (ntl), the only gene that has been documented to be differentially methylated during zebrafish development (9), was seen to be hypomethylated in dnmt3 morphants. Of note, we found that the lack of ntl promoter methylation resulted in up-regulation of its expression. These data differ from Shimoda et al. in that dnmt7 morphants, which also showed loss of ntl methylation in the same promoter region as dnmt3 morphants, did not show differential ntl expression. However, this difference cannot be reconciled at present due to the unavailability of bisulfite sequencing data of ntl promoter in dnmt7 morphants (9). Apart from no change in global cytosine methylation levels, dnmt3 morphants also harbored wild-type levels of global H3K9me3 and H3K9me2 marks unlike dnmt1 morphants (supplemental Fig. S2, A and D). Because overexpression of wild-type, but not catalytically inactive Dnmt3 or G9a, rescued dnmt3 morphants, it is possible that the phenotypic defects observed in dnmt3 morphants result from reduced DNA methylation and H3K9me2/HeK9me3 levels at specific loci rather than at repetitive elements. Strong support for this comes from specific cytosine demethylation of Lef1 promoter in dnmt3 morphants (but not in dnmt1 morphants). Similarly, the same region of the Lef1 promoter showed reduction in H3K9me3 levels in both dnmt3 and g9a morphants. In agreement with these changes in the repressive mark on chromatin, Lef1 was up-regulated in both dnmt3 and g9a morphants, and its knockdown led to rescue of neurogenesis defects in both morphants. This clearly demonstrates that Dnmt3 and G9a cooperate to repress specific developmental regulators through DNA methylation followed by H3K9me3.
In our studies, we note that g9a morphants did not display alterations in global DNA methylation, H3K9me2, or H3K9me3 levels (when monitored at a 37% reduction in G9a) (supplemental Fig. S2, E and F). This could be due to redundancy with other G9a-like genes present in zebrafish, or simply due to the importance of G9a at specific genes, because significant defects in tissue development are observed with only modest reductions in G9a. Indeed, mice and humans harbor another G9a-like transcript, GLP. Also, zebrafish might harbor additional G9a-like genes as many genes are duplicated in this organism. These data suggest that the developmental defects occurring in dnmt3 and g9a morphants are likely due to a lack of repressive methylation occurring at specific genes/loci rather than the bulk genome.
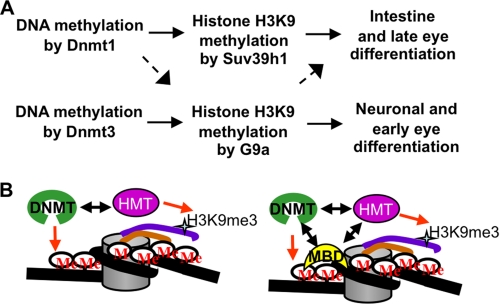
A key result in our work is the demonstration that G9a, but not Suv39h1, cooperates functionally with Dnmt3 in vivo during zebrafish development. Furthermore, this work along with our previous work that Dnmt1 cooperates with suv39h1 morphants points out specific DNMT-HMT networks that might be important in gene regulation during organ development (Fig. 9A). One could argue that the rescue of dnmt morphants by overexpression of an HMT could be an artifact of gene overexpression and need not necessarily reveal the cellular mechanism. We emphasize that our argument, that Dnmt3-G9a and Dnmt1-Suv39h1 cooperativity exists in vivo, does not rely only on the rescue of dnmt morphants by HMT overexpression. These data along with the observation that g9a morphants strikingly phenocopy dnmt3 morphants, whereas suv39h1 morphants strikingly phenocopy dnmt1 morphants, support the notion of selective DNMT-HMT cooperativity.
FIGURE 9.
Models for DNMT-HMT suppression relationships and cooperativity. A, DNMTs methylate the cytosines in the CpG island present in the promoter of the critical regulator of differentiation (for example, Lef1), which is required to be silenced to promote differentiation. DNA methylation marks lead to recruitment of histone H3K9 methyltransferases (HMTs), which then mark the histone tails to help establish a repressed state. Different DNMTs recruit different HMTs; Dnmt3 depends specifically on G9a, whereas Dnmt1 can recruit both Suv39h1 and G9a (but relies more on Suv39h1). This specific DNMT-HMT network is responsible for promoting specific differentiation stages in different organs. B, two physical models can be proposed for the dependence of a DNMT on an HMT. First (left panel), a DNA methyltransferase could directly recruit specific HMT on the promoter of a given gene, and this specific interaction between DNMT and HMT is sufficient to generate specificity. Second (right panel), there exists a ternary complex composed of a DNMT, a methyl binding domain-containing protein (MBD), and an HMT. Specific interactions between these three proteins govern specificity toward their gene targets. Initial recruitment may rely on gene-specific DNA-binding proteins (not shown).
One clear question is how can overexpression of a histone H3K9 methyltransferase rescue the differentiation defects conferred by a DNA methyltransferase hypomorph? Here, we emphasize that morpholinos knock down, but do not eliminate, DNMT protein, providing a dosage hypomorph rather than a genetic null mutation. One possibility is that DNMTs recruit the HMT directly through a physical interaction, with the suppression observed following HMT overexpression resulting from the complete “loading” of all remaining DNMT protein with the HMT, and increased histone methylation of DNMT targets despite their lower DNA methylation levels (Fig. 9B). Notably, studies in mammals have shown direct physical interactions between Dnmt1 and Suv39h1 or G9a (11, 12). Our data suggest that both zebrafish Dnmt3 and human DNMT3B can interact with both of these HMTs when overexpressed in HEK293 cells (data not shown). However, the genetic suppression data clearly show that G9a, but not Suv39h1, can rescue the defects present in dnmt3 morphants. Therefore, a simple model of selective physical association between these two proteins cannot explain the functional data.
An alternative model for suppression can account of the observations without invoking the requirement for a direct DNMT-HMT interaction (Fig. 9B). Here, the DNMT may interact with a specific MBD protein, which then provides a bias for the MBD that occupies a particular methylated site. It is the MBD that then interacts with a particular HMT to provide the suppression selectivity. MBD proteins have been shown to interact with both DNMTs (45, 46) and HMTs (47, 48). Here, selectivity might arise from the use of particular ternary complexes consisting of a DNMT, an MBD, and an HMT; the initial recruitment of these factors to the promoter involves a tissue-specific DNA-binding transcription factor that interacts directly with either the HMT or the DNMT (Fig. 9B). Indeed, the Dnmt1 and Dnmt3 family of proteins contains very different N-terminal motifs, which could be responsible for such selectivity. Further experiments will be needed to test these ideas.
In summary, we provide evidence for specialization of DNMT and HMT function during zebrafish development and selective cooperativity between Dnmt3 and G9a. Because zebrafish include a large family of Dnmt3-related enzymes, it will be of interest to examine whether this notion of selectivity applies to other members of this family and whether the smaller DNMT3 family of enzymes in mammals displays similar DNMT-HMT relationships.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth L. Wright for the kind gift of human G9a plasmid and Drs. Richard Dorsky, Tatjana Piotrowsky, H. Joseph Yost, Brent Bisgrove, and Mariko Sato for helpful suggestions. We thank the DNA peptide resource, DNA sequencing resource, and the centralized zebrafish animal resource at the University of Utah.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HD058506 (to D. A. J. and B. R. C.) and by NCI Grants R01 CA116468 and R01 CA11674 (to D. A. J. and A. R. K.). This work was also supported by the Howard Hughes Medical Institute (to B. R. C.), the Huntsman Cancer Foundation (to D. A. J. and B. R. C.), and Cancer Center Support Grant CA042014.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Tables SI and SII.
- DNMT
- DNA methyltransferase
- HMT
- histone methyltransferase
- MBD
- methyl-binding domain
- MeDIP
- methylated DNA immunoprecipitation
- RACE
- rapid amplification of cDNA ends
- RT
- reverse transcription
- RPE
- retinal pigmented epithelium
- hpf
- hours post- fertilization.
REFERENCES
- 1.Razin A., Cedar H. (1991) Microbiol. Rev. 55, 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A. P. (1986) Nature 321, 209–213 [DOI] [PubMed] [Google Scholar]
- 3.Rai K., Nadauld L. D., Chidester S., Manos E. J., James S. R., Karpf A. R., Cairns B. R., Jones D. A. (2006) Mol. Cell. Biol. 26, 7077–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson R. M., Bosch J. A., Goll M. G., Hesselson D., Dong P. D., Shin D., Chi N. C., Shin C. H., Schlegel A., Halpern M., Stainier D. Y. (2009) Dev. Biol. 334, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 6.Goll M. G., Kirpekar F., Maggert K. A., Yoder J. A., Hsieh C. L., Zhang X., Golic K. G., Jacobsen S. E., Bestor T. H. (2006) Science 311, 395–398 [DOI] [PubMed] [Google Scholar]
- 7.Rai K., Chidester S., Zavala C. V., Manos E. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2007) Genes Dev. 21, 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 9.Shimoda N., Yamakoshi K., Miyake A., Takeda H. (2005) Dev. Dyn. 233, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 10.Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 11.Fuks F., Hurd P. J., Deplus R., Kouzarides T. (2003) Nucleic Acids Res. 31, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estève P. O., Chin H. G., Smallwood A., Feehery G. R., Gangisetty O., Karpf A. R., Carey M. F., Pradhan S. (2006) Genes Dev. 20, 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadauld L. D., Sandoval I. T., Chidester S., Yost H. J., Jones D. A. (2004) J. Biol. Chem. 279, 51581–51589 [DOI] [PubMed] [Google Scholar]
- 14.Lee J. E., Wu S. F., Goering L. M., Dorsky R. I. (2006) Development 133, 4451–4461 [DOI] [PubMed] [Google Scholar]
- 15.Weber M., Hellmann I., Stadler M. B., Ramos L., Pääbo S., Rebhan M., Schübeler D. (2007) Nat. Genet. 39, 457–466 [DOI] [PubMed] [Google Scholar]
- 16.Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimona M., Djinovic-Carugo K., Kranewitter W. J., Winder S. J. (2002) FEBS Lett. 513, 98–106 [DOI] [PubMed] [Google Scholar]
- 18.Rhee I., Bachman K. E., Park B. H., Jair K. W., Yen R. W., Schuebel K. E., Cui H., Feinberg A. P., Lengauer C., Kinzler K. W., Baylin S. B., Vogelstein B. (2002) Nature 416, 552–556 [DOI] [PubMed] [Google Scholar]
- 19.Schneider-Maunoury S., Topilko P., Seitandou T., Levi G., Cohen-Tannoudji M., Pournin S., Babinet C., Charnay P. (1993) Cell 75, 1199–1214 [DOI] [PubMed] [Google Scholar]
- 20.Ellies D. L., Stock D. W., Hatch G., Giroux G., Weiss K. M., Ekker M. (1997) Genomics 45, 580–590 [DOI] [PubMed] [Google Scholar]
- 21.Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. (1994) J. Neurosci. 14, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krauss S., Johansen T., Korzh V., Fjose A. (1991) Nature 353, 267–270 [DOI] [PubMed] [Google Scholar]
- 23.Püschel A. W., Westerfield M., Dressler G. R. (1992) Mech. Dev. 38, 197–208 [DOI] [PubMed] [Google Scholar]
- 24.Allende M. L., Weinberg E. S. (1994) Dev. Biol. 166, 509–530 [DOI] [PubMed] [Google Scholar]
- 25.Korzh V., Sleptsova I., Liao J., He J., Gong Z. (1998) Dev. Dyn. 213, 92–104 [DOI] [PubMed] [Google Scholar]
- 26.Kim C. H., Bae Y. K., Yamanaka Y., Yamashita S., Shimizu T., Fujii R., Park H. C., Yeo S. Y., Huh T. L., Hibi M., Hirano T. (1997) Neurosci. Lett. 239, 113–116 [DOI] [PubMed] [Google Scholar]
- 27.Blader P., Fischer N., Gradwohl G., Guillemot F., Strähle U. (1997) Development 124, 4557–4569 [DOI] [PubMed] [Google Scholar]
- 28.Thomas T., Kurihara H., Yamagishi H., Kurihara Y., Yazaki Y., Olson E. N., Srivastava D. (1998) Development 125, 3005–3014 [DOI] [PubMed] [Google Scholar]
- 29.Mudumana S. P., Wan H., Singh M., Korzh V., Gong Z. (2004) Dev. Dyn. 230, 165–173 [DOI] [PubMed] [Google Scholar]
- 30.Denovan-Wright E. M., Pierce M., Sharma M. K., Wright J. M. (2000) Biochim. Biophys. Acta 1492, 227–232 [DOI] [PubMed] [Google Scholar]
- 31.Biemar F., Argenton F., Schmidtke R., Epperlein S., Peers B., Driever W. (2001) Dev. Biol. 230, 189–203 [DOI] [PubMed] [Google Scholar]
- 32.Stenkamp D. L., Cunningham L. L., Raymond P. A., Gonzalez-Fernandez F. (1998) Mol. Vis. 4, 26. [PubMed] [Google Scholar]
- 33.Forsell J., Ekström P., Flamarique I. N., Holmqvist B. (2001) J. Exp. Biol. 204, 2517–2525 [DOI] [PubMed] [Google Scholar]
- 34.Li H. S., Yang J. M., Jacobson R. D., Pasko D., Sundin O. (1994) Dev. Biol. 162, 181–194 [DOI] [PubMed] [Google Scholar]
- 35.Nornes S., Clarkson M., Mikkola I., Pedersen M., Bardsley A., Martinez J. P., Krauss S., Johansen T. (1998) Mech. Dev. 77, 185–196 [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama J., Kageyama R. (2004) Semin. Cell Dev. Biol. 15, 83–89 [DOI] [PubMed] [Google Scholar]
- 37.Hatakeyama J., Tomita K., Inoue T., Kageyama R. (2001) Development 128, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 38.Field H. A., Ober E. A., Roeser T., Stainier D. Y. (2003) Dev. Biol. 253, 279–290 [DOI] [PubMed] [Google Scholar]
- 39.Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., Jaenisch R. (2001) Nat. Genet. 27, 31–39 [DOI] [PubMed] [Google Scholar]
- 40.De Smet C., Lurquin C., Lethé B., Martelange V., Boon T. (1999) Mol. Cell. Biol. 19, 7327–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu M., Mabaera R., Lowrey C. H., Martin D. I., Fiering S. (2007) Mol. Cell. Biol. 27, 5047–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beri S., Tonna N., Menozzi G., Bonaglia M. C., Sala C., Giorda R. (2007) J. Neurochem. 101, 1380–1391 [DOI] [PubMed] [Google Scholar]
- 43.Douet V., Heller M. B., Le Saux O. (2007) Biochem. Biophys. Res. Commun. 354, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schilling E., Rehli M. (2007) Genomics 90, 314–323 [DOI] [PubMed] [Google Scholar]
- 45.Kimura H., Shiota K. (2003) J. Biol. Chem. 278, 4806–4812 [DOI] [PubMed] [Google Scholar]
- 46.Tatematsu K. I., Yamazaki T., Ishikawa F. (2000) Genes Cells 5, 677–688 [DOI] [PubMed] [Google Scholar]
- 47.Sarraf S. A., Stancheva I. (2004) Mol. Cell 15, 595–605 [DOI] [PubMed] [Google Scholar]
- 48.Fujita N., Watanabe S., Ichimura T., Tsuruzoe S., Shinkai Y., Tachibana M., Chiba T., Nakao M. (2003) J. Biol. Chem. 278, 24132–24138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.