Abstract
Notch is a transmembrane receptor that shares homology with proteins containing epidermal growth factor-like repeats and mediates the cell-cell interactions necessary for many cell fate decisions. In Drosophila, O-fucosyltransferase 1 catalyzes the O-fucosylation of these epidermal growth factor-like repeats. This O-fucose elongates, resulting in an O-linked tetrasaccharide that regulates the signaling activities of Notch. Fucosyltransferases utilize GDP-fucose, which is synthesized in the cytosol, but fucosylation occurs in the lumen of the endoplasmic reticulum (ER) and Golgi. Therefore, GDP-fucose uptake into the ER and Golgi is essential for fucosylation. However, although GDP-fucose biosynthesis is well understood, the mechanisms and intracellular routes of GDP-fucose transportation remain unclear. Our previous study on the Drosophila Golgi GDP-fucose transporter (Gfr), which specifically localizes to the Golgi, suggested that another GDP-fucose transporter(s) exists in Drosophila. Here, we identified Efr (ER GDP-fucose transporter), a GDP-fucose transporter that localizes specifically to the ER. Efr is a multifunctional nucleotide sugar transporter involved in the biosynthesis of heparan sulfate-glycosaminoglycan chains and the O-fucosylation of Notch. Comparison of the fucosylation defects in the N-glycans in Gfr and Efr mutants revealed that Gfr and Efr made distinct contributions to this modification; Gfr but not Efr was crucial for the fucosylation of N-glycans. We also found that Gfr and Efr function redundantly in the O-fucosylation of Notch, although they had different localizations and nucleotide sugar transportation specificities. These results indicate that two pathways for the nucleotide sugar supply, involving two nucleotide sugar transporters with distinct characteristics and distributions, contribute to the O-fucosylation of Notch.
Keywords: Genetics/Drosophila, Signal Transduction, Glycoprotein, Glycosylation, Notch Pathway, Notch Receptor, Subcellular Organelles, Nucleotide Sugar Transporters
Introduction
Notch signaling is an evolutionarily conserved mechanism that regulates a broad spectrum of cell specification events through local cell-cell communication (1, 2). Notch encodes a single-pass transmembrane receptor protein with 36 epidermal growth factor (EGF)3 -like repeats in its extracellular domain. Some of these EGF-like repeats, which contain a consensus sequence, are modified by the O-linked tetrasaccharide Sia-α2,3-Gal-β1,4-GlcNAc-β1,3-Fuc in mammals or the O-linked disaccharide GlcNAc-β1,3-Fuc in Drosophila (3, 4). In Drosophila, O-fucosyltransferase 1 catalyzes this O-linked fucosylation (5). fringe (fng) encodes another glycosyltransferase, β1,3 N-acetylglucosaminyltransferase, which adds GlcNAc specifically to the O-linked fucose of Notch and modulates the binding between Notch and its ligands (3, 6). The modulation of Notch activity through glycosylation by Fng family proteins is largely conserved between Drosophila and vertebrates (7). In contrast, the monosaccharide O-fucose modification of Notch is proposed to have no specific function except to provide a fucose moiety for further modification by Fng in Drosophila (8, 9).
Protein fucosylation requires GDP-fucose as a donor of fucose. Pathways for the synthesis of GDP-fucose are well understood (10, 11). Because fucosyltransferases in the Golgi utilize GDP-fucose, which is synthesized in the cytosol, as a fucose donor, the uptake of GDP-fucose into the Golgi is thought to be a critical step for fucosylation events. The GDP-fucose transporter is a nucleotide sugar transporter, classified as belonging to solute carrier family 35 (SLC35) (12). The transporter is predicted to span the Golgi membrane 10 times and couples the import of GDP-fucose into the Golgi lumen with the export of GMP into the cytoplasm; in the Golgi, GDP-fucose is used by specific fucosyltransferases to add fucose to a variety of glycoproteins and glycolipids. Recently, the gene responsible for congenital disorders of glycosylation (CDG) IIc was cloned by the complementation of cells derived from CDG IIc patients and was found to encode a GDP-fucose transporter, SLC35C1 (13, 14). CDG IIc, also termed leukocyte adhesion deficiency type II (LADII), is a rare recessive syndrome characterized by growth and mental retardation and severe immunodeficiency with marked neutrophilia (15, 16).
Our previous study demonstrated that the O-fucosylation of Notch in Drosophila has a partial requirement for the Drosophila ortholog of SLC35C1, designated as Gfr (Golgi GDP-fucose transporter) (17). Furthermore, a mouse ortholog of SLC35C1 is required for mammalian Notch signaling in vitro (17). Based on these results, it was suggested that a reduction in Notch signaling could contribute to the pathology of CDG IIc/LADII (17). However, mutations of the SLC35C1 genes in Drosophila and mouse yielded much more subtle phenotypes than those expected from the disruption of fng and its homologous gene functions (17, 18). These results suggested the existence of one or more additional GDP-fucose transporter genes in Drosophila and mammals. The identification of a novel GDP-fucose transporter and characterization of its function and intracellular distribution might help to elucidate the pathogenesis of CDG IIc/LADII. Here we describe a novel GDP-fucose transporter in Drosophila that localizes to the endoplasmic reticulum (ER) and participates in the O-fucosylation of Notch.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Genetics
Flies were cultured in a standard medium at 25 °C unless otherwise stated. Canton-S was the wild-type line. patched (ptc)-Gal4 and engrailed (en)-Gal4 were used as Gal4 drivers. Cyan fluorescent protein-endoplasmic reticulum (CFP-ER) (BD Biosciences) and a green fluorescent protein (GFP) variant with an ER retention signal (KDEL) were used as ER markers. UAS-fringe connection (frc)-N-Myc (a gift from S. Goto), UAS-Gfr-C-HA, and UAS-Gfr-C-Myc were the UAS lines used (17). The Gfr1 and GmdH78 mutants were described previously (17, 19).
To generate Efr (ER GDP-fucose transporter) mutants, BG02156 (Bloomington Drosophila Stock Center, Indiana University, Bloomington, IN) was used as a starter P-element line. To excise the P-element, the p[Δ2-3] strain was used as a source of transposase. Mitotic clones were made in Efr1, FRT18A/Ubi-GFP, FRT18A;; MKRS, hs-flp/+ larvae by Flp-mediated mitotic recombination. Recombination was induced in second instar larvae by a 30-min heat shock at 37 °C.
Construction of Plasmids
We amplified the Efr genome fragment from the presumed translation start codon to the stop codon (∼1.0 kb) by genomic PCR and used it as the Efr cDNA, because Efr has no introns in its open reading frame (FBgn0029849). For UAS-Efr-N-HA and UAS-Efr-N-Myc, the hemagglutinin (HA) sequence tag and Myc epitope, respectively, were added to the 5′-end of the Efr cDNA by PCR. For UAS-EfrΔKKVE-N-HA, the HA sequence tag was added to the 5′-end of the Efr cDNA as above, and the 3′-end of the Efr cDNA encoding its dilysine ER retention/retrieval signal (KKVE sequence) was deleted by PCR. pUAS-Efr-N-HA, pUAS-Efr-N-Myc, and pUAS-EfrΔKKVE-N-HA were introduced into the Drosophila genome using a P-element-mediated transformation.
Nucleotide Sugar Transport Assay
Efr cDNA, tagged at the C terminus with HA, was subcloned into the copper-inducible expression vector pYEX-BX and introduced into the Saccharomyces cerevisiae strain YPH500. The in vitro nucleotide sugar transport assay was performed as described previously (17, 20).
Tissue Staining
Immunohistochemistry was performed according to standard protocols, except for the 3G10 antibody staining (see below) (21). The primary antibodies used were mouse anti-Wingless (Wg) (4D4, Developmental Studies Hybridoma Bank; 1:250), mouse anti-120-kDa integral Golgi membrane protein (7H6D7C2, EMD Biosciences; 1:500), mouse anti-Myc (9E10, Developmental Studies Hybridoma Bank; 1:1,000), rat anti-HA (3F10, Roche Applied Science; 1:1,000), rabbit anti- phosphorylated (activated) form of Mother against dpp (p-Mad) (PS1; 1:20,000) (22), and rabbit anti-GFP (MBL, 1:1,000). The secondary antibodies were Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen; 1:500), Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen; 1:500), cyanine 3-conjugated donkey anti-mouse IgG (Rockland; 1:500), cyanine 3-conjugated goat anti-rat IgG (Rockland; 1:500), cyanine 5-conjugated goat anti-rat IgG (Rockland, 1:500), and cyanine 5-conjugated donkey anti-rat IgG (Invitrogen; 1:500).
To detect HS-GAG chains, staining with the 3G10 antibody (Seikagaku, Tokyo, Japan) was performed as described previously (23). Biotin-conjugated ALL (1 μg/ml) (Seikagaku, Tokyo, Japan) and Streptavidin-conjugated Alexa 555 (Invitrogen, 1:500) were used for the Aleuria aurantia lectin (AAL) staining. All images were obtained by confocal microscopy (Pascal, Zeiss).
Western Blotting Analysis
For Western blotting analyses, a rat anti-HA antibody (3F10, Roche Applied Science; 1:500), a rabbit anti-horseradish peroxidase (HRP) antibody (Cappel; 1:12,500), an HRP-conjugated goat anti-rat antibody (KPL Europe; 1:2,000), an HRP-conjugated goat anti-rabbit antibody (1:2,000; Open Biosystems), and enhanced chemiluminescence reagents (GE Healthcare) were used.
RESULTS
Drosophila Efr Encodes Another GDP-fucose Transporter
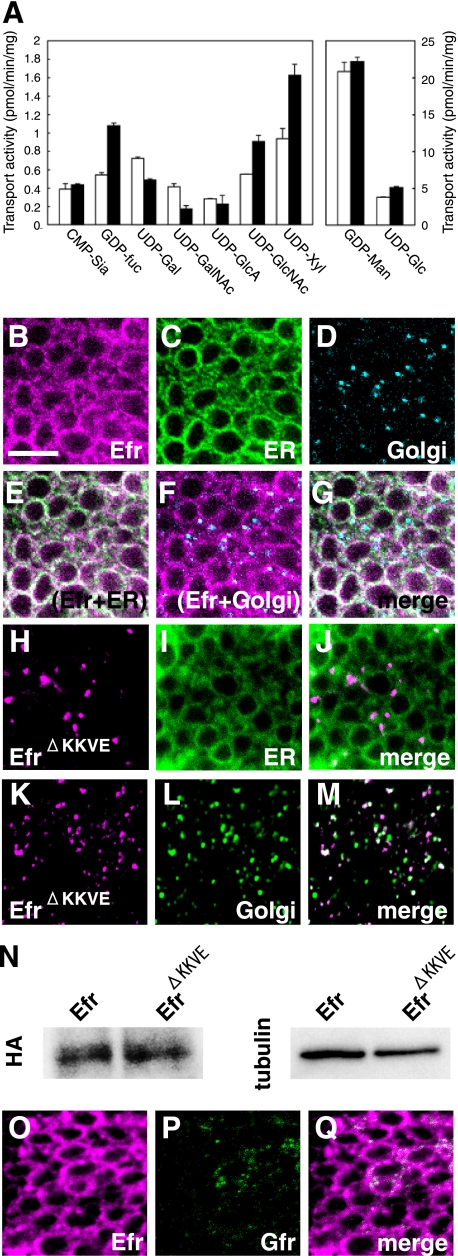
The Drosophila CG3774 gene has been identified as a putative ortholog of human SLC35B4, which encodes a UDP-GlcNAc/UDP-xylose transporter (24). However, a Caenorhabditis elegans SLC35B4 homolog was reported to have some GDP-fucose transport activity (13, 14). Thus, we first tested whether Drosophila CG3774 could transport GDP-fucose. We examined the biochemical activity of CG3774 for transporting various nucleotide sugars, using a yeast in vitro system (20). Microsomes were prepared from transformants carrying vectors with and without a CG3774 cDNA insert, and their ability to transport nucleotide sugars was investigated (Fig. 1A). We found that CG3774 transport of GDP-fucose, UDP-GlcNAc, and UDP-xylose was more than 1.5 times that of control samples, and CG3774 did not transport CMP-sialic acid, UDP-glucuronic acid, GDP-mannose, or UDP-glucose (Fig. 1A). On the other hand, the transport of UDP-galactose and UDP-N-acetylgalactosamine was reduced by the expression of CG3774 (Fig. 1A). This suppression could be explained by possible dimerization between CG3774 and endogenous nucleotide sugar transporters (25, 26), which could lead to inhibition of the activity of the latter, as reported before in similar situations (26, 27). Similarly, it is possible that the increase of GDP-fucose, UDP-GlcNAc, and UDP-xylose transport is due to the activation of endogenous nucleotide sugar transporters via this possible dimerization with CG3774. However, our genetic analyses below supported the idea that CG3774 indeed contributes to the transport of these nucleotide sugars in Drosophila.
FIGURE 1.
The Drosophila homolog of SLC35B4, Efr, transported GDP-fucose, UDP-N-acetylglucosamine, and UDP-xylose and was specifically localized to the ER. A, the transport of CMP-sialic acid (CMP-Sia), GDP-fucose (GDP-Fuc), UDP-Gal, UDP-GalNAc, UDP-GlcA, UDP-GlcNAc, UDP-xylose (UPD-Xyl), GDP-mannose (GDP-Man), and UDP-Glc into vesicles prepared from S. cerevisiae expressing Drosophila SLC35B4 from transfected pYEX-BX-Efr-C-HA (solid bars) or from the mock-transfected equivalent (open bars). Values are the mean ± S.E. from duplicate experiments. B–G, subcellular localization of HA-tagged Drosophila CG3774 (Efr-N-HA) in the epithelium of third instar wing discs. UAS-Efr-N-HA and UAS-CFP-ER were driven by ptc-Gal4. Efr (magenta in B, E, F, and G), CFP-ER (ER) (green in C, E, and G), and Golgi (cyan in D, F, and G) were detected by triple staining with anti-HA, anti-GFP, and anti-Golgi 120-kDa protein antibodies, respectively. E–G are merged images of B and C; B and D; and B, C, and D, respectively. H–M, subcellular localization of HA-tagged Drosophila CG3774 derivative lacking its dilysine ER retention/retrieval signals (EfrΔKKVE-N-HA) in the epithelium of third instar wing discs. UAS-EfrΔKKVE-N-HA and UAS-CFP-ER were driven by ptc-Gal4. EfrΔKKVE (magenta in H, J, K, and M) and CFP-ER (green in I and J) or Golgi (green in L and M) were detected by double staining with anti-HA and anti-GFP or anti-Golgi 120-kDa protein antibodies, respectively. J and M are merged images of H plus I and K plus L, respectively. N, a Western blot analysis of Efr-N-HA and EfrΔKKVE-N-HA proteins. UAS-Efr-N-HA and UAS-EfrΔKKVE-N-HA were driven by da-Gal4, and whole protein extracts were prepared from the embryos. These samples were analyzed by a Western blot using anti-HA antibody (HA). For an internal control, anti-β-tublin antibody (tublin) was used. O–Q, subcellular localization of Myc-tagged Drosophila CG3774 (Efr-N-Myc) and Gfr-C-HA in the epithelium of third instar wing discs. UAS-Efr-N-Myc and UAS-Gfr-C-HA were driven by ptc-Gal4, and Efr (magenta in O and Q) and Gfr (green in P and Q) were detected by double staining with anti-Myc and anti-HA antibodies, respectively. Q is a merged image of O and P. Bar, 5 μm.
The deduced Drosophila SLC35B4 protein is composed of 352 amino acids with 10 putative transmembrane domains and dilysine ER retention/retrieval signals (KKXX or KXKXX). As expected, therefore, we found that CG3774-N-HA co-localized with an ER marker, CFP-ER (Fig. 1, B–D) but not with a generic Golgi marker, the Golgi 120-kDa protein, in wing disc cells (Fig. 1, E–G). CG3774 was predominantly localized to the ER; therefore, we called it Efr (for “ER GDP-fucose transporter”). To exclude the possibility that the localization of Efr to the ER is due to the mislocalization of this protein caused by its overexpression, we also examined the subcellular localization of an Efr derivative (EfrΔKKVE), lacking its putative dilysine ER retention/retrieval signal (KKVE). Under the same conditions, EfrΔKKVE failed to localize to the ER (Fig. 1, H–J) but was detected in the Golgi (Fig. 1, K–M). In addition, a Western blot analysis revealed that almost equal amounts of Efr and EfrΔKKVE proteins were produced from UAS-Efr-N-HA and UAS-EfrΔKKVE-N-HA-driven embryos, suggesting that the distinct subcellular localization of these proteins was not due to the difference in their expression level (Fig. 1N).
These results confirm that Efr is an ER-resident protein, and its dilysine ER retention/retrieval signal localizes Efr to the ER. The localization of Efr did not overlap with that of a previously identified GDP-fucose transporter, Gfr, which specifically localizes to the Golgi (Fig. 1, O–Q).
HS-GAGs Synthesis Partly Requires Efr
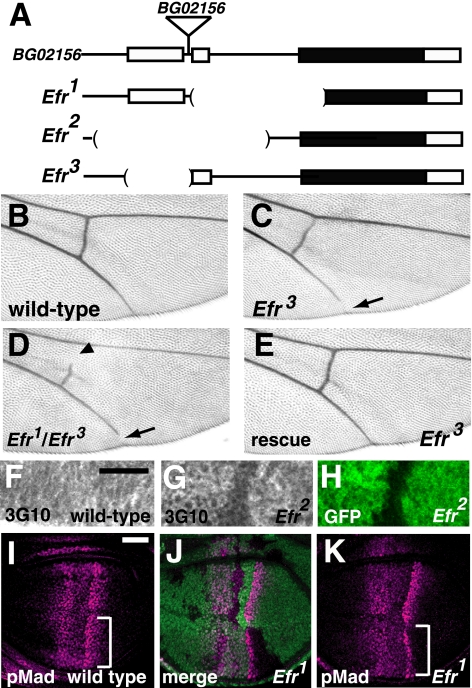
Next, to understand the roles of Efr, we generated mutations of Efr in Drosophila. We used an approach involving imprecise excision of a P-element, a Drosophila transposon. Derivative strains that lost the P-element were established from BG02156, in which the P-element was inserted at the 5′-end of the Efr gene locus (Fig. 2A). The genomic lesions were identified by PCR and genomic sequence analyses, and we obtained three deletion lines, designated Efr1, Efr2, and Efr3 (Fig. 2A). In Efr1, the second exon, second intron, and the 5′ region of the third exon, which includes the predicted initiation codon, were deleted, suggesting that Efr1 was a null allele of Efr (Fig. 2A). In Efr2, the first and second exons and the first and most of the second introns were deleted (Fig. 2A). Homozygotes and hemizygotes of Efr1 or Efr2 were lethal, although some of them survived until the third-instar larval stage (24% of Efr1 and 30% of Efr2 homozygotes). This is possibly due to maternally supplied Efr mRNA, which was detected by in situ hybridization (data not shown). Introduction of a genomic fragment of the Efr locus into the genome rescued this lethality (data not shown). In Efr3, the first exon was deleted (Fig. 2A). In the adult wings of the Efr3 homozygotes and hemizygotes, a gap in the fifth longitudinal vein was observed (Fig. 2C) (data not shown). In Efr3/Efr1, this phenotype became more severe than in the homozygous Efr3, and a gap in the posterior cross-vein was also observed (Fig. 2D). The phenotype of the Efr3 homozygote was completely rescued by introducing a genomic fragment of the Efr locus, indicating that this phenotype was caused by the loss of Efr gene functions (Fig. 2E). This phenotype is reminiscent of that of Dally hypomorphic mutants (28). Dally encodes the core protein of Drosophila glypican, an HS-GAG proteoglycan (28).
FIGURE 2.
Efr was involved in HS-GAG biosynthesis in vivo. A, genomic organization of the Efr locus. The exons of the Efr gene are shown as boxes, and the predicted coding regions are filled in black. The genomic regions deleted in the Efr1, Efr2, and Efr3 mutants are indicated by parentheses. In the Efr1 mutant, a ∼1.2-kb genomic region that includes a predicted initiation codon, was deleted. In the Efr2 mutant, a ∼1.5-kb genomic region that includes the first and second exons was deleted. In the Efr3 mutant, a ∼0.5-kb genomic region that includes the first exon was deleted. BG02156 was the original P-element insertion line, and a triangle indicates the position of the P-element insertion site. B–E, adult wings. B, a wild-type wing. C and D, a wing of Efr3/Efr3 (C) and a wing of Efr3/Efr3 (D), demonstrating the gap in the fifth longitudinal vein (arrow). In D, a gap in the posterior cross-vein was also observed (arrowhead). E, a wing of Efr3/Efr1 carrying a transgene of the Efr genome fragment. The wing vein gap was completely rescued. F–H, anti-heparan sulfate (3G10) staining of the late third instar wing discs. F, a wild-type wing disc. G and H, a chimeric wing disc, including Efr2/Efr2 cells, which are marked by the absence of GFP (H). Note that the intensity of anti-heparan sulfate staining was reduced in the mutant cells (G). I–K, late third instar wing discs stained with an anti-p-Mad antibody (magenta). I, a wild-type wing disc. J and K, a chimeric wing disc, including Efr1/Efr1 cells, which are marked by the absence of GFP (green in J). Anti-p-Mad antibody staining was decreased in Efr1 mutant cells (regions indicated by brackets in I compared with that of K). Bar, 25 μm.
As demonstrated above, Efr transports UDP-GlcNAc and UDP-xylose, which are required for HS-GAG biosynthesis. In addition, the presence of the wing phenotype that is similar to that of Dally could be due to a defect in HS-GAG synthesis. Therefore, we examined the effect of the Efr mutation on the biosynthesis of HS-GAG chains. The wing discs of third instar larvae were stained with the monoclonal antibody 3G10, which recognizes an epitope produced by heparitinase I digestion of HS-GAG chains (29). In the wild-type wing discs, uniform 3G10 antibody staining was detected (Fig. 2F). However, in somatic clones of the Efr2 mutant (marked by the lack of GFP in Fig. 2H) induced by the FLP/FRT system, the 3G10 staining was slightly reduced (Fig. 2, G and H). These results suggest that the biosynthesis of HS-GAGs partly requires an Efr function in Drosophila.
HS-GAGs play important roles in the functions of various morphogens, such as Dpp (Decapentaplegic), Hh (Hedgehog), and Wg (Wingless) (30, 31). Thus, we investigated the functions of Efr in these morphogen-mediated signaling pathways. First, we detected the activation of Dpp signaling by antibody staining for p-Mad, a cytoplasmic signal transducer of Dpp signaling (32). In the wild-type wing disc, the p-Mad level was high in the central region of the wing pouch, because dpp is expressed in a stripe of the anterior compartment along the anterior-posterior compartment boundary (Fig. 2I). However, as shown in Fig. 2, J and K, the p-Mad level was decreased in the Efr mutant cells. This phenotype was similar to that of mutants for genes required for the synthesis of HS-GAGs, which play important roles in the functions of Dpp (23, 33, 34). Therefore, these results suggest that Dpp signaling in the wing disc partly requires Efr function. In contrast, the mutation of Gfr does not affect HS-GAG synthesis or Dpp signaling (17), indicating that Efr and Gfr have distinct activities, at least in part. We also examined the effect of the Efr mutant in Hh and Wg signaling; however, neither was affected in the Efr mutant (data not shown).
In Combination with Gfr, Efr Has an Essential Role in Notch Signaling
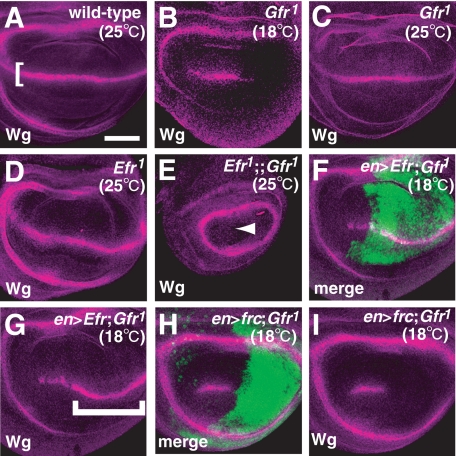
We previously reported that the O-fucosylation of Notch has a partial requirement for Gfr, suggesting that at least one other GDP-fucose transporter is present in Drosophila (17). Given that Efr transported GDP-fucose in vitro, we speculated that Efr function might be involved in the O-fucosylation of Notch. In wild-type wing discs, Notch signaling is activated and induces wg, a target gene of Notch signaling, along the dorsal-ventral compartment boundary in the late third instar (Fig. 3A) (35, 36). wg induction depends on the modulation of Notch ligand binding by Fng, which adds GlcNAc to O-fucose on the EGF-like repeats of Notch (3, 6). Therefore, the O-fucosylation of these EGF-like repeats is also essential for wg induction. As reported previously, in the wing disc of Gfr1 (a null allele) homozygotes, the expression of wg was partially reduced at the non-permissive temperature (18 °C) (Fig. 3B), but this defect was not observed at the permissive temperature (25 °C) (Fig. 3C) (17). However, wg expression was not affected in Efr1 or Efr2 homozygous wing discs at either 18 or 25 °C (Fig. 3D) (data not shown). Therefore, we next tested whether Efr and Gfr function redundantly as GDP-fucose transporters, by examining a double mutant of Efr and Gfr. In the double mutant wing discs of Efr1 and Gfr1 or Efr2 and Gfr1, the Notch signaling-dependent wg expression was completely abolished at the dorsal-ventral boundary at 25 °C (Fig. 3E) (data not shown). This phenotype is similar to that of wing discs homozygous for Gmd (GDP-mannose-4,6-dehydratase), which is essential for GDP-fucose synthesis in Drosophila (8, 19), or homozygous for O-fucosyltransferase 1 (5, 37).
FIGURE 3.
Efr and Gfr functioned redundantly in Notch signaling. A–I, anti-Wg antibody staining (magenta) of late third instar wing discs. A, a wild-type wing disc. The Notch signaling-dependent expression of wg along the dorsal-ventral compartment boundary is indicated by a bracket. B, a wing disc of Gfr1/Gfr1 at 18 °C. C, a wing disc of Gfr1/Gfr1 at 25 °C. D, a wing disc of Efr1/Y at 25 °C. E, a wing disc of Efr1/Y;;Gfr1/Gfr1 at 25 °C. The Notch signaling-dependent expression of wg was completely abolished (arrowhead). F and G, a wing disc of Gfr1/Gfr1-overexpressing Efr under the control of en-Gal4 at 18 °C. F, anti-GFP antibody staining (green) shows the region overexpressing Efr. G, the Notch signaling-dependent expression of wg was restored in the region expressing Efr (bracket). H and I, a wing disc of Gfr1/Gfr1 overexpressing frc under the control of en-Gal4 at 18 °C. H, anti-GFP antibody staining (green) shows the region overexpressing frc. I, the Notch signaling-dependent expression of wg was not restored. Bar, 50 μm.
If Efr acts as a GDP-fucose transporter, the overexpression of Efr should rescue the reduced Notch signaling activity in the wing disc of the Gfr homozygote at the non-permissive temperature (18 °C). In the posterior compartment of wing discs homozygous for Gfr1, Efr was overexpressed by en-Gal4. This restored the expression of wg at 18 °C in all cases examined (n = 15) (Fig. 3, compare F and G with B). On the other hand, the overexpression of frc, which encodes a transporter capable of transporting various UDP-sugars, including UDP-GlcNAc and UDP-xylose, but not GDP-fucose, failed to rescue this wg expression under the same conditions in all cases examined (n = 15) (Fig. 3, H and I). This result suggests that the incorporation of GDP-fucose, but not of UDP-GlcNAc or UDP-xylose, is responsible for the rescue of Notch signaling activity, although Efr also transports these other two nucleotide sugars, as described above.
It was recently found that rumi, which encodes an O-glucose transferase, is essential for Notch signaling in Drosophila (38). Rumi adds O-glucose to a subset of Notch EGF-like repeats, and two xylose moieties are further added to this O-glucose (38). However, Efr and Gfr do not transport UDP-glucose (17). Therefore, it is unlikely that the reduction of Notch signaling in the Gfr mutant is associated with a defect in the O-glucosylation. In addition, the xylose elongation is probably not involved in the rescue of Notch signaling by the overexpression of Efr in Gfr homozygotes either, because Frc, which transports UDP-xylose in vitro, did not rescue the Notch signaling activity in the Gfr homozygote, as mentioned above. Consistent with this idea, in wing discs double homozygous for rumi44, a null mutation of rumi, and either Efr1 or Gfr1, the expression of wg was not reduced further than in the Efr or Gfr homozygotes at 18 °C (data not shown).
Gfr but Not Efr Plays a Major Role in the Fucosylation of N-Glycans
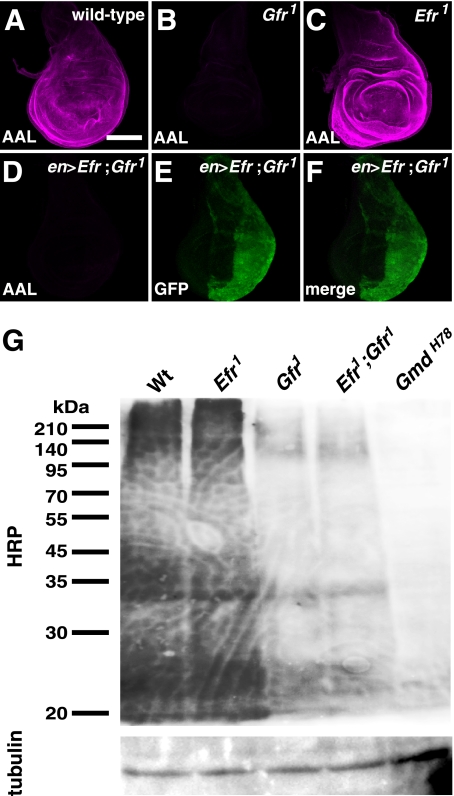
AAL is defined as an l-fucose lectin that recognizes α-1,3- and α-1,6-linked fucose residues (39, 40). In the wild-type wing discs of third instar larvae, AAL staining was ubiquitously detected (Fig. 4A). However, as reported previously, in Gfr1 homozygous wing discs, AAL staining was virtually abolished (Fig. 4B) (17). In contrast, in the wing discs homozygous for Efr1 or Efr2, this staining was not affected under the same conditions (Fig. 4C) (data not shown). Furthermore, the reduced AAL staining in the Gfr1 mutant wing discs was not restored by the overexpression of Efr driven by en-Gal4 (Fig. 4, D–F) (data not shown), although a similar overexpression of Gfr does restore the AAL staining in Gfr1 mutant wing discs (17). Therefore, unlike Gfr, Efr did not play a role in the fucose modifications of N-glycans.
FIGURE 4.
Gfr but not Efr was required for fucose modifications of N-glycans. A–C, AAL staining (magenta) of the late third instar wing discs. A, a wild-type wing disc. B, a wing disc of Gfr1/Gfr1. C, a wing disc of Efr1/Y. All confocal images were obtained under the same conditions. D–F, a wing disc of Gfr1/Gfr1 overexpressing Efr under the control of en-Gal4. D, AAL staining (magenta). E, anti-GFP antibody staining (green) showing the region overexpressing Efr. F, a merged image of D and E. G, Western blot analysis of the total protein extracts prepared from the imaginal discs and central nervous system of wild-type (Wt), Efr1/Y (Efr1), Gfr1/Gfr1 (Gfr1), Efr1/Y;; Gfr1/Gfr1 (Efr1; Gfr1), and GmdH78/GmdH78 (GmdH78) mutant larvae. α-1,3-fucose modifications were detected by anti-HRP antibody staining. Bar, 100 μm.
We next confirmed the influence of the Efr and Gfr mutations on the fucose modifications of bulk proteins by a biochemical approach. The imaginal discs and central nervous system were isolated from wild-type, Efr1/Efr1, Gfr1/Gfr1, the double homozygote of Efr1 and Gfr1, and GmdH78/GmdH78 third instar larvae cultured at 25 °C. The protein extracts of these organs were then analyzed by Western blotting with an anti-HRP antibody, which recognizes the α-1,3-fucose (41). Consistent with the above results obtained by tissue staining with AAL, the Western blot analysis showed that the reactivity of the anti-HRP antibody was not affected in the lysate of the Efr1 mutant (Fig. 4G, Efr1 lane). In contrast, this reactivity was significantly reduced in the lysate of the Gfr1 mutant (Fig. 4G, Gfr1 lane). The reactivity in the lysate of the Efr1 and Gfr1 double homozygote was not reduced further (Fig. 4G, Efr1;Gfr1 lane), although the Notch signaling activity was diminished in this double homozygote compared with the single homozygotes of either of these mutants under the same conditions. Based on these results, we concluded that Efr does not play a crucial role in the fucosylation of N-glycans. These results are consistent with a previous finding that the disruption of N-glycan fucosylations does not disrupt Notch signaling activity, further suggesting that these fucose modifications do not play a crucial role in Notch signaling (17). On the other hand, in the lysate of the GmdH78 mutant, in which GDP-fucose is not synthesized, the reactivity to the anti-HRP antibody was abolished (Fig. 4G, GmdH78 lane). Thus, there is a weak, residual N-glycan fucosylation activity in the double homozygote of Efr1 and Gfr1. This finding suggests the existence of a third GDP-fucose transporter in Drosophila.
Efr and Gfr Have Redundant Roles That Are Essential for the O-Fucosylation of Notch
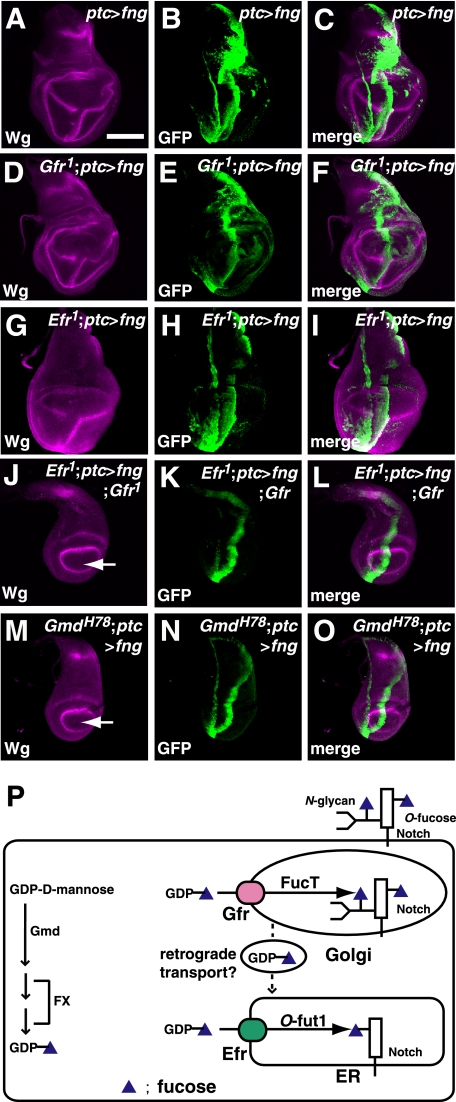
In contrast to the fucosylation of N-glycans, it is difficult to examine the level of protein O-fucosylation directly. Therefore, we used an indirect approach, involving the ectopic expression of fng, to determine whether Efr is required for the O-fucosylation of Notch. The regions expressing GFP indicate where fng is expressed under the control of ptc-Gal4. As reported previously, the ectopic expression of fng driven by ptc-GAL4 resulted in an ectopic induction of wg in a stripe posterior to the region ectopically expressing fng in the ventral compartment of the wild-type wing disc at the third instar (Fig. 5, A–C) (3, 6). The functions of fng depend on the O-fucosylation of Notch; therefore, the absence of Notch O-fucosylation should prevent this ectopic wg induction by fng. We previously demonstrated that the ectopic expression of fng induces a normal level of wg expression in the wing discs of Gfr1 homozygotes at the permissive temperature (25 °C) (Fig. 5, D–F) (17). In the wing discs of Efr homozygotes, the ectopic induction of wg by fng overexpression was also not affected at 25 °C (Fig. 5, G–I). In contrast, in all wing discs isolated from double homozygotes of Efr1 and Gfr1 at the same stage and temperature (n = 17), the ectopic expression of fng did not induce wg. This suggests that Notch O-fucosylation was absent in these wing discs (Fig. 5, J–L). This phenotype was essentially the same as that induced by fng expression in the wing discs of GmdH78 homozygotes under the same conditions (n = 19) (Fig. 5, M–O). Therefore, Efr and Gfr have redundant roles, which are essential for the O-fucosylation of Notch.
FIGURE 5.
Efr and Gfr function redundantly for the O-fucosylation of Notch. A–O, the expression of wg (magenta in A, C, D, F, G, I, J, L, M, and O) in wild-type or various mutant wing discs of late third-instar larvae that ectopically express fng and GFP (green in B, C, E, F, H, I, K, L, N, and O). The expression of fng and GFP was driven by ptc-Gal4. A–C, a wild-type wing disc; D–F, Gfr1/Gfr1; G–I, Efr1/Y; J–L, Efr1/Y;;Gfr1/Gfr1; M–O, GmdH78/GmdH78. GFP shows the regions ectopically expressing fng. J and M show the ectopic expression of fng failed to induce ectopic wg expression (arrows). C, F, I, L, and O are merged images of A and B, D and E, G and H, J and K, and M and N, respectively. P, two pathways for transporting GDP-fucose to the ER, where the O-fucosylation of Notch occurs in Drosophila. GDP-mannose-4,6-dehydratase (Gmd) and GDP-4-keto-6-deoxymannose epimerase/reductase (FX) convert GDP-fucose from GDP-mannose in the cytoplasm (arrows at left). GDP-fucose is then transported into the ER or Golgi by two GDP-fucose transporters, Efr and Gfr, respectively. GDP-fucose is incorporated by Gfr into the Golgi, where it is used for the fucosylation of N-glycans. GDP-fucose that is incorporated into the Golgi by Gfr is also transported to the ER, possibly by retrograde vesicular transport. In the ER, where the O-fucosylation of Notch occurs, GDP-fucose is used as a donor of fucose for this modification of Notch. Bar, 100 μm.
DISCUSSION
Nucleotide sugars are transported into the ER and Golgi, where they become donors of saccharides for the enzymatic reactions of various glycosyltransferases. The properties of several nucleotide sugar transporters, which transport overlapping species of nucleotide sugars, have been studied in detail. Many of these transporters are distributed in the ER and Golgi. The intracellular routes for the nucleotide sugar supplies are not well understood, and they are predicted to be very complex. In this study, we identified two pathways in Drosophila for transporting GDP-fucose into the ER, where the O-fucosylation of Notch occurs.
We identified Efr as a second GDP-fucose transporter in Drosophila and demonstrated that Gfr and Efr have redundant roles for the O-fucosylation of Notch. Despite the similar roles of Gfr and Efr in O-fucosylation, these two nucleotide sugar transporters have distinct biochemical and cell biological characteristics. First, the specificities of the transported nucleotide sugars are quite different between them. Efr transports a broader spectrum of nucleotide sugars than Gfr, including UDP-GlcNAc, UDP-xylose, and GDP-fucose. Thus, Efr simultaneously contributes to HS-GAG synthesis and the O-fucosylation of Notch. In contrast, Gfr transports GDP-fucose almost exclusively (17). Therefore, in the Gfr homozygotes, HS-GAG synthesis was not affected, whereas the fucosylation of bulk protein N-glycans was mostly abolished. Second, the subcellular localizations of Efr and Gfr are different; they are specifically localized to the ER and Golgi, respectively. These results together with the previous finding that O-fucosylation occurs in the ER (42) indicate that GDP-fucose is supplied from two pathways, one in the ER and one in the Golgi, as shown schematically in Fig. 5P. This is the first example of such a dual system for supplying a nucleotide sugar to the ER for a specific glycosylation. In the first pathway, GDP-fucose is directly incorporated into the ER via Efr, although Efr is involved in the incorporation of various nucleotide sugars (Fig. 5P). In the second pathway, GDP-fucose is incorporated into the Golgi via Gfr and transported into the ER, where the O-fucosylation of Notch takes place, possibly via retrograde vesicular trafficking from the Golgi to the ER (Fig. 5P).
Recent studies have demonstrated that the substrate specificities of nucleotide sugar transporters are very difficult to predict based on their primary structure (25). We generated a null mutant of Drosophila CG14971, an ortholog of mouse Slc35c2 whose primary structure is most closely related to Drosophila Gfr. However, our genetic analysis of this mutant did not provide any indication that it plays a role in fucose modifications (data not shown). On the other hand, although the sequence identity of Efr and Gfr is only 10%, both of these nucleotide sugar transporters transport GDP-fucose for the catalytic activity of O-fucosyltransferase 1. Thus, our results provide another example of the difficulty of predicting the substrate specificity of nucleotide sugar transporters. Our results also suggest that collaborative functions of nucleotide sugar transporters that belong to distantly related groups need to be considered to elucidate their redundant roles.
In the mouse, Slc35c1, the ortholog of Gfr, plays a crucial role in fucose modifications in vivo (18). Thus, mice homozygous for a Slc35c1-null mutation show hypofucosylation of glycoproteins, the absence of selectin ligands, growth retardation, and postnatal defects in various organs (18, 43). However, these defects are significantly less severe than those associated with the mutation of FX, which encodes an enzyme that converts GDP-mannose to GDP-fucose, although some of the defects overlap (44). This observation suggests the presence of another GDP-fucose transporter in the mouse. This idea is also supported by the observation that the hypofucosylation in Slc35c1 mutant cells is restored by adding fucose, indicating the presence of a residual activity for GDP-fucose incorporation (18). Furthermore, the oral administration of fucose is an effective therapy for the immunodeficiency of CDG IIc patients, which have mutations in their SLC35C1 locus (45, 46). In cultured fibroblasts from a CDG IIc patient, fucosylation of N-glycans is significantly reduced, whereas O-fucosylation of Notch is not affected (47). This result also suggests the presence of additional GDP-fucose transporters that provide a sufficient level of GDP-fucose for the O-fucosylation of Notch in the absence of SLC35C1 in human. The residual activities of GDP-fucose incorporation in the absence of SLC35C1 or its ortholog (Gfr) could be more predominant in human than Drosophila, because the O-fucosylation of Notch was affected in Drosophila but not in human under this condition. However, this comparison is based on experiments involving very different systems, and there might be other explanations (17). Taken together, these observations suggest that at least one other GDP-fucose transporter is also present in mammals. In this study, we revealed the nature of this redundancy in Drosophila, which could be important for understanding how a low level of GDP-fucose transporting activity is maintained in CDG IIc patients.
Acknowledgments
We thank Y. Takei-Yamaguchi, T. Tabata, M. Kawakita, N. Taniguchi, and Y. Nagai for valuable suggestions. We also thank the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for flies and antibodies.
This work was supported by grants-in-aid from the Japanese Ministry of Education, Culture, Sports and Science (to K. M.) and grants from the Proposal-oriented Research Promotion Program, Japan Science and Technology Agency (to K. M.).
- EGF
- epidermal growth factor
- AAL
- A. aurantia lectin
- CDG
- congenital disorders of glycosylation
- ER
- endoplasmic reticulum
- LADII
- leukocyte adhesion deficiency type II
- CFP
- cyan fluorescent protein
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- HRP
- horseradish peroxidase
- HS-GAG
- heparan sulfate-glycosaminoglycan.
REFERENCES
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Fiúza U. M., Arias A. M. (2007) J. Endocrinol. 194, 459–474 [DOI] [PubMed] [Google Scholar]
- 3.Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., Vogt T. F. (2000) Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 4.Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., Haltiwanger R. S. (2000) J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 5.Okajima T., Irvine K. D. (2002) Cell 111, 893–904 [DOI] [PubMed] [Google Scholar]
- 6.Brückner K., Perez L., Clausen H., Cohen S. (2000) Nature 406, 411–415 [DOI] [PubMed] [Google Scholar]
- 7.Haltiwanger R. S., Stanley P. (2002) Biochim. Biophys. Acta 1573, 328–335 [DOI] [PubMed] [Google Scholar]
- 8.Okajima T., Xu A., Lei L., Irvine K. D. (2005) Science 307, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 9.Okajima T., Reddy B., Matsuda T., Irvine K. D. (2008) BMC Biol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker D. J., Lowe J. B. (2003) Glycobiology 13, 41R–53R [DOI] [PubMed] [Google Scholar]
- 11.Roos C., Kolmer M., Mattila P., Renkonen R. (2002) J. Biol. Chem. 277, 3168–3175 [DOI] [PubMed] [Google Scholar]
- 12.Ishida N., Kawakita M. (2004) Pflugers Arch. 447, 768–775 [DOI] [PubMed] [Google Scholar]
- 13.Lühn K., Wild M. K., Eckhardt M., Gerardy-Schahn R., Vestweber D. (2001) Nat. Genet. 28, 69–72 [DOI] [PubMed] [Google Scholar]
- 14.Lübke T., Marquardt T., Etzioni A., Hartmann E., von Figura K., Körner C. (2001) Nat. Genet. 28, 73–76 [DOI] [PubMed] [Google Scholar]
- 15.Etzioni A., Frydman M., Pollack S., Avidor I., Phillips M. L., Paulson J. C., Gershoni-Baruch R. (1992) N. Engl. J. Med. 327, 1789–1792 [DOI] [PubMed] [Google Scholar]
- 16.Hirschberg C. B. (2001) J. Clin. Invest. 108, 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa H. O., Higashi S., Ayukawa T., Sasamura T., Kitagawa M., Harigaya K., Aoki K., Ishida N., Sanai Y., Matsuno K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18532–18537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellbusch C. C., Sperandio M., Frommhold D., Yakubenia S., Wild M. K., Popovici D., Vestweber D., Gröne H. J., von Figura K., Lübke T., Körner C. (2007) J. Biol. Chem. 282, 10762–10772 [DOI] [PubMed] [Google Scholar]
- 19.Sasamura T., Ishikawa H. O., Sasaki N., Higashi S., Kanai M., Nakao S., Ayukawa T., Aigaki T., Noda K., Miyoshi E., Taniguchi N., Matsuno K. (2007) Development 134, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 20.Aoki K., Ishida N., Kawakita M. (2003) J. Biol. Chem. 278, 22887–22893 [DOI] [PubMed] [Google Scholar]
- 21.Matsuno K., Ito M., Hori K., Miyashita F., Suzuki S., Kishi N., Artavanis-Tsakonas S., Okano H. (2002) Development 129, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 22.Persson U., Izumi H., Souchelnytskyi S., Itoh S., Grimsby S., Engström U., Heldin C. H., Funa K., ten Dijke P. (1998) FEBS Lett. 434, 83–87 [DOI] [PubMed] [Google Scholar]
- 23.Takei Y., Ozawa Y., Sato M., Watanabe A., Tabata T. (2004) Development 131, 73–82 [DOI] [PubMed] [Google Scholar]
- 24.Ashikov A., Routier F., Fuhlrott J., Helmus Y., Wild M., Gerardy-Schahn R., Bakker H. (2005) J. Biol. Chem. 280, 27230–27235 [DOI] [PubMed] [Google Scholar]
- 25.Berninsone P. M., Hirschberg C. B. (2000) Curr. Opin. Struct. Biol. 10, 542–547 [DOI] [PubMed] [Google Scholar]
- 26.Gerardy-Schahn R., Oelmann S., Bakker H. (2001) Biochimie 83, 775–782 [DOI] [PubMed] [Google Scholar]
- 27.Bakker H., Routier F., Oelmann S., Jordi W., Lommen A., Gerardy-Schahn R., Bosch D. (2005) Glycobiology 15, 193–201 [DOI] [PubMed] [Google Scholar]
- 28.Nakato H., Futch T. A., Selleck S. B. (1995) Development 121, 3687–3702 [DOI] [PubMed] [Google Scholar]
- 29.David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin X., Perrimon N. (2000) Matrix Biol. 19, 303–307 [DOI] [PubMed] [Google Scholar]
- 31.Selleck S. B. (2001) Semin. Cell Dev. Biol. 12, 127–134 [DOI] [PubMed] [Google Scholar]
- 32.Tanimoto H., Itoh S., ten Dijke P., Tabata T. (2000) Mol. Cell 5, 59–71 [DOI] [PubMed] [Google Scholar]
- 33.Han C., Belenkaya T. Y., Khodoun M., Tauchi M., Lin X., Lin X. (2004) Development 131, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 34.Bornemann D. J., Duncan J. E., Staatz W., Selleck S., Warrior R. (2004) Development 131, 1927–1938 [DOI] [PubMed] [Google Scholar]
- 35.Williams J. A., Paddock S. W., Carroll S. B. (1993) Development 117, 571–584 [DOI] [PubMed] [Google Scholar]
- 36.de Celis J. F., García-Bellido A. (1994) Mech. Dev. 46, 109–122 [DOI] [PubMed] [Google Scholar]
- 37.Sasamura T., Sasaki N., Miyashita F., Nakao S., Ishikawa H. O., Ito M., Kitagawa M., Harigaya K., Spana E., Bilder D., Perrimon N., Matsuno K. (2003) Development 130, 4785–4795 [DOI] [PubMed] [Google Scholar]
- 38.Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., Bellen H. J. (2008) Cell 132, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lühn K., Laskowska A., Pielage J., Klämbt C., Ipe U., Vestweber D., Wild M. K. (2004) Exp. Cell Res. 301, 242–250 [DOI] [PubMed] [Google Scholar]
- 40.Kochibe N., Furukawa K. (1980) Biochemistry 19, 2841–2846 [DOI] [PubMed] [Google Scholar]
- 41.Kurosaka A., Yano A., Itoh N., Kuroda Y., Nakagawa T., Kawasaki T. (1991) J. Biol. Chem. 266, 4168–4172 [PubMed] [Google Scholar]
- 42.Luo Y., Haltiwanger R. S. (2005) J. Biol. Chem. 280, 11289–11294 [DOI] [PubMed] [Google Scholar]
- 43.Yakubenia S., Frommhold D., Schölch D., Hellbusch C. C., Körner C., Petri B., Jones C., Ipe U., Bixel M. G., Krempien R., Sperandio M., Wild M. K. (2008) Blood 112, 1472–1481 [DOI] [PubMed] [Google Scholar]
- 44.Smith P. L., Myers J. T., Rogers C. E., Zhou L., Petryniak B., Becker D. J., Homeister J. W., Lowe J. B. (2002) J. Cell Biol. 158, 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marquardt T., Lühn K., Srikrishna G., Freeze H. H., Harms E., Vestweber D. (1999) Blood 94, 3976–3985 [PubMed] [Google Scholar]
- 46.Hidalgo A., Ma S., Peired A. J., Weiss L. A., Cunningham-Rundles C., Frenette P. S. (2003) Blood 101, 1705–1712 [DOI] [PubMed] [Google Scholar]
- 47.Sturla L., Rampal R., Haltiwanger R. S., Fruscione F., Etzioni A., Tonetti M. (2003) J. Biol. Chem. 278, 26727–26733 [DOI] [PubMed] [Google Scholar]