Abstract
The solution structure of the IIA-IIB complex of the N,N′-diacetylchitobiose (Chb) transporter of the Escherichia coli phosphotransferase system has been solved by NMR. The active site His-89 of IIAChb was mutated to Glu to mimic the phosphorylated state and the active site Cys-10 of IIBChb was substituted by serine to prevent intermolecular disulfide bond formation. Binding is weak with a KD of ∼1.3 mm. The two complementary interaction surfaces are largely hydrophobic, with the protruding active site loop (residues 9–16) of IIBChb buried deep within the active site cleft formed at the interface of two adjacent subunits of the IIAChb trimer. The central hydrophobic portion of the interface is surrounded by a ring of polar and charged residues that provide a relatively small number of electrostatic intermolecular interactions that serve to correctly align the two proteins. The conformation of the active site loop in unphosphorylated IIBChb is inconsistent with the formation of a phosphoryl transition state intermediate because of steric hindrance, especially from the methyl group of Ala-12 of IIBChb. Phosphorylation of IIBChb is accompanied by a conformational change within the active site loop such that its path from residues 11–13 follows a mirror-like image relative to that in the unphosphorylated state. This involves a transition of the φ/ψ angles of Gly-13 from the right to left α-helical region, as well as smaller changes in the backbone torsion angles of Ala-12 and Met-14. The resulting active site conformation is fully compatible with the formation of the His-89-P-Cys-10 phosphoryl transition state without necessitating any change in relative translation or orientation of the two proteins within the complex.
Keywords: Biophysics, Methods/NMR, Phosphorylation, Protein/Protein-Protein Interactions, Protein/Structure, Phosphoryl Transfer
Introduction
The bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS)3 couples a phosphorylation cascade involving a sequential series of bimolecular protein-protein complexes to active sugar translocation across the membrane and to regulation of an array of cellular processes, including carbon catabolite repression (1–6). The first two steps of the PTS, involving autophosphorylation of enzyme I (EI) by phosphoenolpyruvate and subsequent phosphoryl transfer to the histidine phosphocarrier protein (HPr), are common to all branches of the pathway. The downstream components of the PTS comprise four major classes of sugar-specific enzymes II corresponding to the glucose (Glc), mannitol (Mtl), mannose (Man), and lactose/chitobiose (Chb) branches of the PTS. The enzymes II are generally organized into two cytoplasmic domains (IIA and IIB), and one transmembrane domain (IIC), which may or may not be covalently linked to one another. The phosphoryl group is transferred from HPr to IIA, from IIA to IIB and finally from IIB onto the incoming sugar bound to IIC. Despite their similar organization, the IIA and IIB domains of the different sugar-specific branches of the PTS bear no sequence similarity to one another and, with the exception of IIBMtl (7, 8) and IIBChb (9–11), no structural similarity either. Whereas structures of many of the individual cytoplasmic components of the PTS have been solved either by crystallography (9, 12–25) or NMR (7, 8, 10, 11, 26–32), the complexes of the PTS have proved refractory to crystallization, presumably because of their weak and transient nature. Weak binding, however, is not an impediment to NMR spectroscopy, and over the last 10 years we have solved the solution structures of the N-terminal domain of enzyme I (EIN) complexed to HPr (33), and the IIA-HPr and IIA-IIB complexes of the glucose, mannitol, and mannose branches of the PTS (30, 31, 34–37). These complexes provide a paradigm for understanding the structural basis of protein-protein interactions and how individual proteins can recognize multiple, structurally dissimilar, partners.
In the present report, we present the solution structure of the IIA-IIB complex of the Escherichia coli N,N′-diacetylchitobiose-specific enzyme II (IIChb), a representative of the lactose/chitobiose branch of the PTS (38–41). The A, B, and C domains of IIChb are encoded by a single operon and expressed as three individual proteins. The solution NMR structure of E. coli IIAChb (26) and the crystal structure of the related family member IIALac from Lactobacillus lactis (21) are symmetric trimers of ∼35 kDa with three equivalent IIB binding sites. The active site residue, His-89, is located deep within a crevice formed by the interface of two helices from adjacent subunits. IIBChb is a small 11-kDa protein that has been studied by both x-ray crystallography (9) and NMR (10, 11) and is structurally similar to IIBMtl (7, 8), despite the absence of any significant sequence similarity (<10%). The active site residue, Cys-10, of IIBChb is located within an 8-residue protruding loop (residues 9–16) whose conformation is very similar to that of the low molecular weight protein-tyrosine phosphatases (42), including hydrogen-bonding interactions in the phosphorylated state between the phosphoryl group and backbone amide protons (11). (Note, throughout the text residues of IIBChb are shown in italics.)
Wild-type IIAChb is highly prone to nonspecific aggregation promoted by a disordered 13-residue N-terminal tail and by metal ions that coordinate three buried aspartic acid residues (Asp-92), one from each subunit, at the center of the trimer interface (26). It has previously been shown that aggregation can be completely eliminated by removing the N-terminal tail and mutating Asp-92 to Leu to generate a mutant, which we refer to hereafter as IIAChb* (26). In the present work, we made use of an active site H89E mutation introduced into IIAChb* to mimic the phosphorylated state. For IIBChb we introduced a C10S mutation to prevent intermolecular disulfide bridge-mediated dimer formation (9, 10). The IIAChb*-IIBChb complex is transient and weak with an equilibrium dissociation constant (KD) in the millimolar range. The affinity of IIAChb*(H89E) for IIBChb(C10S) is a factor of ∼1.5 higher than that of IIAChb* (KD ∼1.3 versus ∼2 mm) making the phosphomimetic mutant more suitable for NMR structural studies by increasing the population of the complex at the concentrations used in the NMR experiments. The structure of the IIAChb*(H89E)-IIBChb(C10S) complex reveals the structural basis of specific recognition and the interactions involved in phosphoryl transfer.
EXPERIMENTAL PROCEDURES
Protein Expression and Mutagenesis
Genes encoding IIAChb* (corresponding to a NΔ13/D92L mutant of wild-type IIAChb) and IIBChb (kindly provided by Dr. Saul Roseman, Johns Hopkins University, Baltimore) were cloned into the pET-11 vector. Additional H89E and C10S mutations of the active site residues of IIAChb* and IIBChb, respectively were introduced using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The H89E mutation in IIAChb* was designed to mimic the charge effects of phosphorylation of His-89, and the C10S mutation of IIBChb was introduced to prevent any potential complications arising from possible intermolecular disulfide bridge formation.
The IIAChb*, IIAChb*(H89E), IIBChb, and IIBChb(C10S) plasmids were introduced into E. coli BL21(DE3) (Novagen) cells for protein expression and induced at an A600 ∼0.8 with 1 mm isopropyl-β-d-thiogalactopyranoside at 37 °C. Cells were grown in either Luria-Bertani medium or minimal medium (in either H2O or D2O) with 15NH4Cl or 14NH4Cl as the main nitrogen source, and U-[13C/1H], U-[12C/1H], U-[13C/2H]- or U-[12C/2H]glucose as the main carbon source. Because Leu, Val, Ile, Met, Gly, Tyr, and Ser residues are involved in the IIAChb*/IIBChb interface, selective labeling was also employed in the preparation of NMR samples. For 2H/13C/15N-(Ile/Leu/Val)-methyl-protonated (but otherwise fully deuterated) protein samples, 100 mg of α-[13C5,3-2H1]ketoisovalerate and 50 mg of α-[13C4,3,3–2H2]ketobutyrate (Cambridge Isotopes) were added to 1 liter of D2O medium 1 h prior to induction (43). 2H/12C/14N-(Ile/Met/Thr-protonated)-IIAChb*(H89E) and 2H/12C/14N-(Gly/Met/Pro/Tyr-protonated)-IIBChb(C10S) samples were prepared by supplementing 1 liter of D2O medium with 300 mg of Gly/Ile/Met/Pro/Thr/Tyr (Sigma- Aldrich) at natural isotopic abundance 1 h prior to induction. After induction (4 and 7 h for growths in H2O and D2O, respectively), cells expressing IIAChb*(H89E) or IIBChb(C10S) were harvested by centrifugation at 15,900 × g for 25 min.
IIAChb* and IIAChb*(H89E) Purification
The cell pellet was resuspended in 50 ml of buffer A (20 mm Tris, pH 8.0, 1 mm EDTA, 0.2 mm sodium azide) with 1 mm phenylmethylsulfonyl fluoride. The cell suspension was lysed by two passages through a microfluidizer at 15,000–23,000 psi and centrifuged at 75,600 × g for 30 min. The supernatant was loaded onto a 5-ml HiTrap QFF column (Amersham Biosciences), and IIAChb* was eluted with a gradient of buffer B (20 mm Tris, pH 8.0, 1 mm EDTA, 0.2 mm sodium azide, 1 m NaCl). The eluted protein was subsequently denatured with 4 m guanidine-HCl for 15 min. The protein solution was then dialyzed against 2 liters of buffer A overnight. After centrifugation of the dialyzed solution to remove precipitated proteins, the supernatant was purified by size exclusion chromatography on a Superdex-75 column (Amersham Biosciences) in buffer C (20 mm Tris, pH 8.0, 1 mm EDTA, 0.2 mm sodium azide, 0.5 m NaCl). The fractions containing IIAChb* were exchanged into buffer A using an Amicon Ultra-15 (Millipore) filter, loaded onto a mono Q 10/100GL column (Amersham Biosciences), and eluted with a gradient of buffer B.
IIBChb and IIBChb(C10S) Purification
The cell pellet was resuspended in 50 ml of buffer D (20 mm sodium phosphate, pH 4.5, 0.2 mm sodium azide) with 1 mm phenylmethylsulfonyl fluoride. The cell suspension was lysed by two passages through a microfluidizer at 15,000–23,000 psi and centrifuged at 75,600 × g for 30 min. The supernatant was loaded onto a 5-ml HiTrap SP FF column (Amersham Biosciences), and IIBChb(C10S) was eluted with a gradient of buffer E (20 mm sodium phosphate, pH 4.5, 0.2 mm sodium, 1 m NaCl). The eluted protein was further purified by size exclusion chromatography in buffer F (20 mm sodium phosphate, pH 6.5, 0.2 mm sodium azide) on a Superdex-75 column (Amersham Biosciences).
Phosphorylation of IIBChb
15N-labeled IIBChb was phosphorylated by the addition of 5 μm enzyme I, 5 μm HPr, 5 μm IIAChb*, 5 mm MgCl2, and 20 mm phosphoenolpyruvate in 20 mm sodium phosphate, pH 6.5, 100 mm NaCl, 0.2 mm sodium azide, and 90% H2O/10% D2O. Phosphorylation was confirmed by two-dimensional 1H-15N correlation spectroscopy, which showed large chemical shift perturbations of residues 10–16 comprising the active site loop (11), relative to unphosphorylated IIBChb. Full-length Enzyme I and HPr were expressed and purified as described previously (44).
NMR Data Collection and Analysis
All NMR samples were prepared in a buffer of 20 mm sodium phosphate, pH 6.5, 100 mm NaCl, 0.2 mm sodium azide, and either 90% H2O/10% D2O or 99.99% D2O. IIAChb* is a symmetric trimer with three equivalent binding sites for IIBChb (26). To achieve optimal linewidths for NMR spectroscopy, a 1:1 mixture of IIAChb*(H76E) trimer to IIBChb(C10S) monomer was employed. NMR spectra were recorded at 20 and 35 °C on Bruker DMX500, DMX600, DRX600, DRX800, and DRX900 spectrometers equipped with either x-, y-, and z-shielded gradient triple resonance probes or z-shielded gradient triple resonance cryoprobes. Spectra were processed with the NMRPipe package (45) and analyzed using the program PIPP (46).
Sequential and side chain assignments of IIAChb*(H89E) and IIBChb(C10S) were derived from the following three-dimensional double and triple resonance through-bond correlation experiments (47–49): HNCA, HN(CO)CA, HNCACB, CBCA(CO)HN, HAHN, HNCA-TROSY, HN(CO)CA-TROSY, HNCB-TROSY, HN(CO)CB-TROSY, C(CCO)NH, H(CCO)NH, and HCCH-TOCSY. Three-dimensional 15N-separated, 13C-separated, and 13C/13C-separated nuclear Overhauser enhancement (NOE) experiments were used to facilitate side chain assignments (47, 48).
Backbone 1DNH residual dipolar couplings (RDC) were obtained from the difference in 1JHN scalar couplings measured in dilute liquid crystalline medium (phage pf1 (50, 51)) and isotropic (water) medium, measured using two-dimensional in-phase/anti-phase (IPAP) 1H-15N heteronuclear single quantum coherence (HSQC) spectra (52).
Intermolecular NOEs were observed on the IIAChb*(H89E)-IIBChb(C10S) complex in D2O buffer using three-dimensional 12C-filtered(F1)/13C-separated(F2) or 13C-separated(F2)/12C-filtered(F3) NOE experiments, and in H2O buffer using two-dimensional 15N-separated/13C-edited and 13C-separated/15N-edited NOE experiments (53, 54). Nine different combinations of isotope-labeled complexes were used for analysis of intermolecular NOEs (Table 1).
TABLE 1.
Labeling schemes for samples used for intermolecular NOE measurements on the IIAChb*(H89E)-IIBChb(C10S) complex
| Sample | Isotope labeling |
|
|---|---|---|
| IIAChb*(H89E) | IIBChb(C10S) | |
| 1 | [13CH3-ILV]/[2H/13C/15N] | [1H-Gly,Ser]/[2H/12C/14N] |
| 2 | [13CH3-ILV]/[2H/13C/15N] | [1H-Pro,Tyr]/[2H/12C/14N] |
| 3 | [13CH3-ILV]/[2H/13C/15N] | [1H-Met]/[2H/12C/14N] |
| 4 | [1H-Thr]/[2H/12C/14N] | U-[1H/13C/15N] |
| 5 | [1H-Met]/[2H/12C/14N] | U-[1H/13C/15N] |
| 6 | [1H-Met]/[2H/12C/14N] | [13CH3-ILV]/[2H/13C/15N] |
| 7 | [1H-Ile]/[2H/12C/14N] | [13CH3-ILV]/[2H/13C/15N] |
| 8 | U-[1H/13C/15N] | [13CH3-ILV]/ [2H/12C/14N] |
| 9 | U-[1H/13C/15N] | [1H-Met,Tyr]/[2H/12C/14N] |
Structure Calculations
NOE-derived interproton distance restraints were classified into loose approximate distance ranges of 1.8–2.7, 1.8–3.5, 1.8–5.0, and 1.8–6.0 Å corresponding to strong, medium, weak, and very weak NOE cross-peak intensities, respectively (55); an empirical correction of 0.5 Å was added to the upper distance bounds of distance restraints involving methyl groups to account for the higher apparent intensity of methyl resonances (56). NOEs involving non-stereospecifically assigned methyl, methylene, and aromatic protons were represented by a (Σr−6)−1/6 sum (57). Backbone torsion angle restraints for the active site loop of IIBChb(C10S) and phospho-IIBChb, and for the mobile loop (residues 75–84) of IIAChb*(H89E) were derived from chemical shifts using the program TALOS+ (58).
Structures were calculated using conjoined rigid body/torsion angle-simulated annealing (59) with the program Xplor-NIH (60, 61). The minimized target function comprises NOE-derived interproton distance restraints, torsion angle restraints, RDC restraints (62), 13Cα/13Cβ chemical shift restraints (63), a quartic van der Waals repulsion term for the non-bonded contacts (64), a multidimensional torsion angle data base potential of mean force (65), and a gyration volume potential to ensure optimal packing (66). Structure figures were generated using the programs VMD-XPLOR (67) and GRASP (68). Reweighted atomic probability density maps were calculated as described previously (69).
The atomic coordinates and NMR experimental restraints (accession codes 2WWV for the unphosphorylated complex and 2WY2 for the phosphoryl transition state complex) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ.
RESULTS AND DISCUSSION
Equilibrium Binding of IIAChb*(H89E) and IIBChb(C10S)
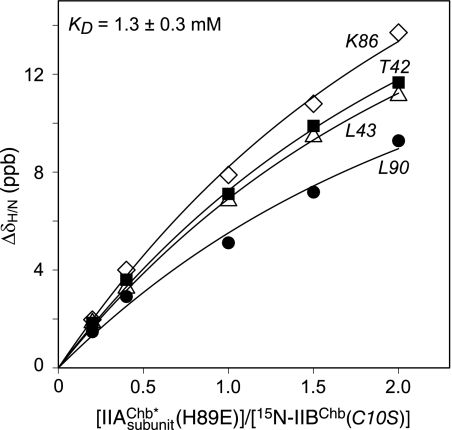
The binding of IIAChb* and the active site IIAChb*(H89E) phosphomimetic mutant (at natural isotopic abundance) to U-15N-labeled IIBChb(C10S) was monitored by 1H-15N correlation spectroscopy. Exchange between the complex and free proteins is fast on the chemical shift time scale. The pattern of 1HN/15N chemical shift perturbations observed for the binding of IIAChb* and IIAChb*(H89E) to IIBChb(C10S) is very similar but the magnitude of the perturbations is smaller for IIAChb* than IIAChb*(H89E). This is caused by the fact that the binding of IIAChb* to IIBChb(C10S) is weaker (KD ∼2.1 ± 0.5 mm) than that of IIAChb*(H89E) (KD ∼1.3 ± 0.3 mm; see Fig. 1), and hence the fraction of complex formed under the same experimental conditions (protein concentrations ≤ 1 mm) is considerably reduced. For this reason, all structural studies were conducted with the IIAChb*(H89E) phosphomimetic mutant.
FIGURE 1.
Binding of IIAChb*(H89E) and IIBChb(C10S). Backbone amide chemical shift perturbations upon titrating unlabeled IIAChb*(H89E) into a solution of 15N-labeled IIBChb(C10S) at 20 °C. The chemical shifts were monitored using 1H-15N HSQC spectroscopy at a spectrometer 1H frequency of 600 MHz. ΔδH/N = [(Δδ15N)2/25 + (Δδ1H)2)/2]1/2 (78). The IIAChb*(H89E):IIBChb(C10S) molar ratios, expressed in terms of subunit concentration of IIAChb*(H89E), are 0, 0.2, 0.4, 1.0, 1.5, and 2.0, with corresponding subunit concentrations of IIAChb*(H89E) of 0, 0.10, 0.20, 0.47, 0.68, and 0.88 mm, respectively, and concentrations of IIBChb(C10S) of 0.5, 0.49, 0.49, 0.47, 0.45, 0.44 mm, respectively. The solid lines represent the results of a global non-linear least squares best-fit to all the titration data simultaneously, using a simple equilibrium binding model. The optimized KD value is 1.3 ± 0.3 mm.
Structural studies were carried out on samples comprising 1 mm IIAChb*(H89E) trimer and 1 mm IIBChb(C10S). Under these conditions, 51.6% of IIAChb*(H89E) and 64.4% of IIBChb(C10S) are in the bound state. Because IIAChb* is a symmetric trimer with three equivalent binding sites for IIBChb, one can calculate that the percentage of IIAChb*(H89E) with one, two, and three IIBChb(C10S) molecules bound is 39.7, 10.9, and 1.0%, respectively; the corresponding percentages, expressed in terms of IIBChb(C10S) are 39.7, 21.7, and 3.0%, respectively. Given molecular masses of 33.6 kDa and 11.4 kDa for free IIAChb*(H89E) and IIBChb(C10S), respectively, and the fact that all species are in fast exchange with one another, the linewidths of IIAChb*(H89E) and IIBChb(C10S) in the NMR sample are determined by population weighted-average molecular masses of 41.0 and 36.2 kDa, respectively.
Structure Determination
Because the chemical shift perturbations observed upon complex formation are small, one can conclude that there are no significant backbone structural changes (within the limits of the NMR method) induced within either IIAChb*(H89E) or IIBChb(C10S). We, therefore, proceeded to solve the structure of the complex using conjoined rigid body/torsion angle dynamics simulated annealing (59, 70), largely on the basis of intermolecular NOE data. In this approach, the backbone (except for certain selected regions, see below) and non-interfacial side chain coordinates are treated as rigid bodies with rotational and translational degrees of freedom, whereas the interfacial sidechains are given full torsional degrees of freedom.
The chemical shift differences between IIAChb* and IIAChb*(H89E) are limited to the immediate vicinity of the mutation, and 1DNH RDC measurements on free IIAChb*(H89E) indicate excellent agreement between observed RDCs for helical residues and those calculated from the NMR structure (restrained regularized mean coordinates) of IIAChb* (26). The RDC R-factor is 8.7% (defined as {<(Dobs − Dcalc)2>/2<Dobs2>)}1/2 (71), where Dobs are the observed RDCs, and Dcalc are the calculated RDCs obtained by singular value decomposition against the coordinates of the protein (72)). The alignment tensor of IIAChb*(H89E) is axially symmetric, as expected for a symmetric trimer, with a magnitude of −11.3 Hz for the axial component (DaNH). We, therefore, used the NMR structure of IIAChb* in the conjoined rigid body/torsion angle dynamics simulated annealing calculations. However, because the loop connecting helices 2 and 3 is partially disordered (i.e. highly mobile) in solution and contributes to the interface with IIBChb, the backbone of residues 75–84 was also give torsional degrees of freedom.
IIBChb(C10S) strongly interacts with the alignment medium (phage pf1) in the absence of salt; RDC measurements were therefore carried out in 100 and 400 mm NaCl using 10 and 17 mg/ml phage pf1, yielding values of 25.5 and −8.5 Hz for DaNH, respectively, and a rhombicity η of ∼0.4. The RDC R-factors are summarized in Table 2 and allow one to conclude the following: (a) both the 1.8-Å resolution x-ray structure of IIBChb(C10S) (9) and the NMR structure of phospho-IIBChb (11) provide a much better representation of the actual solution structure of IIBChb(C10S) than does the NMR structure of IIBChb(C10S) (10), reflecting the lower coordinate accuracy of the latter; (b) there are very large discrepancies between observed and calculated RDCs within the active site loop (residues 9–16) for both the x-ray structure of IIBChb(C10S) and the NMR structure of phospho-IIBChb, reflected in very high RDC R-factors for this region; (c) removing the RDCs for the active site residues results in excellent agreement between observed and calculated RDCs with lower RDC R-factors for the x-ray structure of IIBChb(C10S). Therefore, the coordinates of the x-ray structure of IIBChb(C10S) (9) were used in the calculations, with the backbone of the active site loop (residues 9–16), given torsion degrees of freedom. The observed RDC R-factors of 8–9% observed for the x-ray structure of IIBChb(C10S), excluding the active site loop, are as expected for a crystal structure solved at 1.5 to 2 Å resolution (73, 74). It is worth noting that phosphorylation of IIBChb is accompanied by a large conformational change within the active site loop as manifested both by RDCs (Table 2) and by significant differences in backbone (N, Cα, Hα, and Cβ) chemical shifts (11). Excluding the active site loop, the crystal structure of IIBChb(C10S) still displays lower RDC R-factors than the NMR structure of phospho-IIBChb (Table 2). However, the RDCs within the active site loop are now in excellent agreement with the NMR structure of phospho-IIBChb but exhibit very large discrepancies with respect to the x-ray structure of IIBChb(C10S) (Table 2).
TABLE 2.
RDC R-factors for IIBChb
| RDC R-factor (%)a |
|||
|---|---|---|---|
| X-ray |
NMR |
NMR |
|
| IIBChb(C10S)b | IIBChb(C10S)b | phospho-IIBChbb | |
| 1DNH RDCs measured on free IIBChb(C10S)c | |||
| 100 mm NaCl and 10 mg/ml pf1 | |||
| All data (73)/excluding residues 9–16 (67) | 12.1/8.0 | 44.7/43.8 | 18.8/14.4 |
| Active site loop (residues 9–16) (6)d | 70.1 | 79.5 | 88.2 |
| 400 mm NaCl and 17 mg/ml pf1 | |||
| All data (80)/excluding residues 9–16 (74) | 14.7/8.4 | 40.2/34.8 | 16.2/11.5 |
| Active site loop (residues 9–16) (6)d | 50.9 | 88.4 | 50.2 |
| 1DNH RDCs measured on free phospho-IIBChbc | |||
| 100 mm NaCl and 10 mg/ml pf1 | |||
| All data (66)/excluding residues 9–16 (59) | 38.8/16.8 | 55.4/49.3 | 19.9/20.8 |
| Active site loop (residues 9–16) (7)e | 73.2 | 70.0 | 15.5 |
| 200 mm NaCl and 15 mg/ml pf1 | |||
| All data (76)/excluding residues 9–16 (69) | 34.1/16.0 | 52.2/46.6 | 16.6/16.8 |
| Active site loop (residues 9–16) (7)e | 70.6 | 70.9 | 15.3 |
a The RDC R-factor is defined as 100×[<(Dobs − Dcalc)2>/2<Dobs2>)]1/2 (71), where Dobs are the observed RDCs, and Dcalc are the calculated RDCs obtained by singular value decomposition against the coordinates of the indicated protein (72). The values for the magnitude of the principal component of the alignment tensor (DaNH) and the rhombicity (η) are as follows. For the RDCs measured on free IIBChb(C10S), the values are 25.6 Hz and 0.4, respectively, in 100 mm NaCl and 10 mg/ml pf1, and −8.9 Hz and 0.3, respectively, in 400 mm NaCl and 17 mg/ml phage pf1; the normalized scalar product between the alignment tensors (80) at the two salt concentrations is −0.1, indicating that the two alignment tensors are effectively independent of one another. For the RDCs measured on free phospho-IIBChb, the values are 21.2 Hz and 0.53, respectively in 100 mm NaCl and 10 mg/ml phage pf1, and −9.6 Hz and 0.64, respectively in 200 mm NaCl and 15 mg/ml phage pf1, with a normalized scalar product of 0.96 between the two alignment tensors.
b The PDB accession codes are 1IIB and 1E2B for the x-ray (9) and NMR (10) structures of IIBChb(C10S), respectively; and 1H9C for the NMR structure of phospho-IIBChb (11). The active site residue is located at position 10, and the active site loop comprises residues 9–16.
c The number of experimentally measured 1DNH RDCs is shown in parentheses.
d RDCs were measured for residues 9, 10, 11, 12, 13, and 16. The cross-peaks for the backbone amide groups of Met-14 and Ser-15 were too broad to permit measurement of the 1JNH splitting in the alignment media.
e RDCs were measured for residues 9, 10, 11, 12, 13, 14, and 16. The cross-peak for the backbone amide group of Ser-15 was too broad to permit measurement of the 1JNH splitting in the alignment medium.
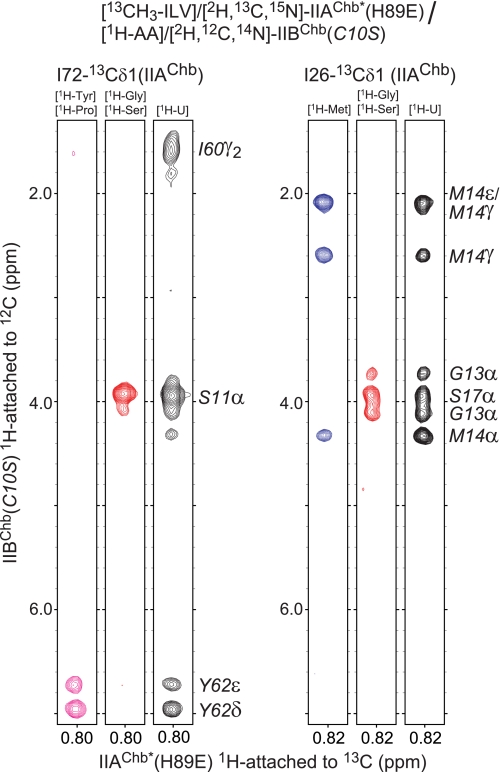
The intermolecular NOE data were derived from a large series of isotope-filtered/isotope-separated intermolecular NOE experiments (53). Because of the relatively large size of the complex and extensive chemical shift overlap, nine different labeling combinations (Table 1), including amino acid specific labeling, were used to eliminate any ambiguities in intermolecular NOE assignments. Examples of the quality of the intermolecular NOE data are shown in Fig. 2.
FIGURE 2.
Intermolecular NOEs in the IIAChb*(H89E)-IIBChb(C10S) complex. NOEs in a three-dimensional 12C-filtered/13C-separated NOE experiment recorded in D2O are specifically observed from protons attached to 12C (in the F1 dimension) to protons attached to 13C (in the F3 dimension). Strips are shown for NOEs involving the 13Cδ methyl groups of Ile-72 and Ile-26 of IIAChb*(H89E). The amino acid specific labeling scheme used for [1H-AA]/[2H, 12C, 14N]-IIBChb(C10S) is shown above each strip.
In protein-protein complexes of the PTS that we have solved previously (30, 31, 33–37), it is usually possible to derive sidechain torsion angle restraints for interfacial side chains based on heteronuclear 3J scalar couplings and short mixing time NOE data (53). In this instance, the complex is not fully saturated because of weak binding (KD ∼ 1.3 mm), and there is a significant proportion of each component in the free state. We therefore employed a heuristic approach in which the interfacial side chains were given torsional degrees of freedom but restrained within the χ1 and, where appropriate, χ2 rotamers occupied in the free structures, unless these were inconsistent with the intermolecular NOE data.
Unfortunately we were not able to use RDC data to provide information on the relative orientation of the two components within the complex. In a simple case of a weak binding binary complex in fast exchange on the chemical shift scale, the observed RDCs are weighted averages of the RDCs in the free and bound state, so that it is possible in principle to back-calculate the RDCs for the pure complex providing one knows exactly the fraction of the bound species (35, 75). However, for the IIAChb*(H89E)-IIBChb(C10S) complex, the bound species comprises a mixture of three states with one, two and three IIBChb(C10S) molecules bound to the IIAChb*(H89E) trimer, each with its own alignment tensor. Deconvolution of the alignment tensors for the individual bound states is not feasible, thereby precluding the use of RDCs in this system. Moreover, the alignment media that we explored (pf1, strained gels and PEG/hexanol (72)) all displayed differential interaction with one of the partners, making any extrapolation of average RDCs for the bound states unreliable.
A summary of the structural statistics is given in Table 3, and a best-fit superposition of the final 90 simulated annealing structures is shown in Fig. 3A. The NOE-derived interproton distance restraints comprised 40 intermolecular NOEs (per bound IIBChb molecule), as well as intramolecular NOEs related to those portions of the IIBChb(C10S) backbone that were given torsional degrees of freedom. The agreement of the RDCs within the active site loop of IIBChb(C10S) is comparable to that of the rest of the protein (Table 4). The relative orientation of IIBChb(C10S) to IIAChb*(H89E) is well defined with a precision of 0.3 ± 0.1 Å for the backbone of the complete complex (IIAChb* and IIBChb best-fitted overall), 0.9 ± 0.3 Å for the backbone of IIBChb with best-fitting to IIAChb*, and 1.2 ± 0.4 Å for IIAChb* with best-fitting to IIBChb. The coordinate precision for the interfacial side chains is 0.9 ± 0.1 Å.
TABLE 3.
Structural statistics
The notation of the NMR structures are as follows: <SA> are the final 90 simulated annealing structures, (SA)r is the restrained regularized mean structure of the IIAChb*(H89E)-IIBChb(C10S) complex. The number of experimental restraints used to calculate <SA> and (SA)r is the same.
| <SA> | (SA)r | ||
|---|---|---|---|
| Number of experimental NMR restraints | |||
| Intermolecular interproton distance restraints | 40 | ||
| IIBChb intramolecular interproton distance restraintsa | 78 | ||
| IIAChb* torsion angle restraints | 44 | ||
| IIBChb torsion angle restraints | 38 | ||
| 1DNH RDCs for IIBChb(C10S)b | 153 | ||
| 13Cα/13Cβ chemical shift restraints for IIBChb(C10S)c | 15 | ||
| R.m.s. deviation from interproton distance restraints (Å)d | 0.003 ± 0.003 | 0 | |
| R.m.s. deviation from torsion angle restraints (°)d | 0.35 ± 0.03 | 0.32 | |
| R.m.s. deviation from 13Cα/13Cβ shift restraints (ppm) | 0.60 ± 0.03 | 0.59 | |
| Measures of structural qualitye | |||
| Intermolecular repulsion energy (kcal·mol−1) | 3.8 ± 1.4 | 10.7 | |
| Intermolecular Lennard-Jones energy (kcal·mol−1) | −29.6 ± 6.3 | −31.0 | |
| Coordinate precision of the complex (Å)f | |||
| Backbone (N, Cα, C', O) atoms | 0.32 ± 0.11 | ||
| Interfacial sidechain heavy atomsg | 0.90 ± 0.07 | ||
a The intramolecular NOE-derived interproton distance restraints relate only to interfacial side chains and to the active site loop (residues 9–16) of IIBChb(C10S).
b The 1DNH RDCs were measured for free IIBChb(C10S) in two alignment media (10 mg/ml pf1 and 100 mm NaCl; and 17 mg/ml phage pf1 and 400 mm NaCl) (see Table 2). The active site loop (residues 9–16) is given complete torsional degrees of freedom, while the remainder of the backbone is treated as a rigid body comprising the x-ray coordinates of IIBChb(C10S) (PDB code 1IIB (9)); thus, the orientation of the alignment tensor is determined by the rigid portion of IIBChb(C10S), while the conformation of the active site loop is determined by the 1DNH RDCs within the loop. The RDC R-factors for the rigid portion of IIBChb(C10S) are given in Table 2, whereas those for the active site loop of the restrained regularized mean structure of the complex are provided in Table 4.
c The 13Cα and 13Cβ chemical shift restraints involve only the active site loop (residues 9–16) of IIBChb(C10S).
d None of the structures exhibit interproton distance violations >0.3 Å or torsion angle violations >5°.
e The intermolecular repulsion energy is given by the value of the quartic van der Waals repulsion term calculated with a force constant of 4 kcal·mol−1·Å−4 and a van der Waals radius scale factor of 0.78. The intermolecular Lennard-Jones van der Waals interaction energy is calculated using the CHARMM19/20 parameters and is not included in the target function used to calculate the structures.
f Defined as the average r.m.s. difference between the final 90 simulated annealing structures and the mean coordinates. The value quoted for the backbone (which excludes residues 75–84 of IIAChb that are disordered and have a coordinate precision of 1.9 Å) provides a measure of the precision with which the orientation and translation of the two proteins in the complex have been determined, and does not take into account the backbone accuracy of the NMR coordinates of IIAChb*(H89E) (26) and the x-ray coordinates of IIBChb(C10S) (9) used for conjoined rigid body/torsion angle dynamics docking. The accuracy of the restrained regularized mean coordinates of IIAChb*(H89E), excluding the loop residues 75–84 that were given backbone torsional degrees of freedom can be estimated from the coordinate precision (0.4 Å (26)) and the value of 8.7% for the cross-validated RDC R-factor for the NH bond vectors (this work), which suggests a coordinate accuracy comparable to a 1.5–2 Å resolution crystal structure (73, 74). The accuracy of the x-ray coordinates of IIBChb(C10S) (excluding the active site loop that was given backbone torsional degrees of freedom) is likely to be around 0.3 Å judging from the crystallographic resolution (1.8 Å) and R-factor (18.7 and 24.1% for Rfree) (9), as well as the RDC R-factors (this work, Table 2). The backbone precision for the active site loop of IIBChb(C10S), obtained from the ensemble of simulated annealing structures, is 0.55 Å. The percentage of residues in the most favored region of the Ramachandran map (81) is 95% for both IIAChb*(H89E) and IIBChb(C10S).
g The side chains of IIAChb*(H89E) given torsional degrees of freedom during simulated annealing are: residues 18, 19, 21, 22, 25, 26, 28, 29, 32–34, 36, 40, 41, 58, 59, 62, 65, 66, 69, 70, 72–74, 85–91, 93, 98, 101. (Note that a non-crystallographic symmetry restraint ensures that the conformation of equivalent residues for the three chains of IIAChb*(H89E) are identical.) The side chains of IIBChb(C10S) given torsional degrees of freedom are: residues 9–12, 14–22, 24, 37, 39, 40, 41–43, 46, 47, 59, 60, 62, 63, 81, 82, 84, 86, and 87.
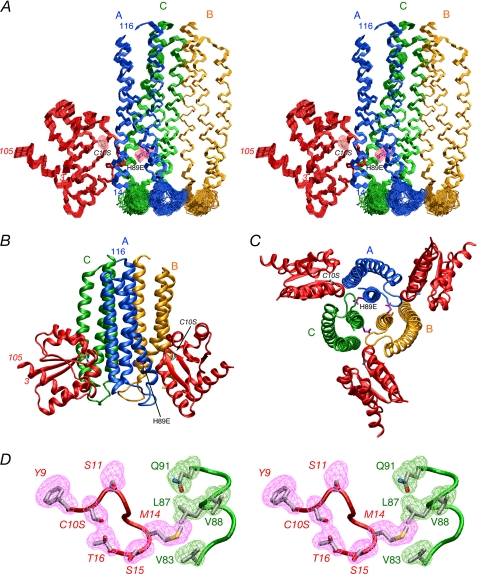
FIGURE 3.
Solution structure of the IIAChb*(H89E)- IIBChb(C10S) complex. A, stereoview of a superposition of the backbone (N, Cα, C) atoms of the final 90 simulated annealing structures with the A, B, and C subunits of the IIAChb*(H89E) symmetric trimer in blue, gold, and green, respectively, and IIBChb(C10S) in red. The active site residues, H89E and C10S are shown in purple and cyan, respectively, and the pink meshes represent the reweighted atomic density probability map (69) for these two residues (drawn at a value of 20% maximum). Only a single IIBChb molecule binding at the interface of the A and C subunits of IIAChb*(H89E) is shown; because IIAChb*(H89E) is a symmetric trimer there are three identical binding sites formed at the interfaces between the A and C subunit, the C and B subunits and the B and A subunits. B, ribbon diagram of the complex showing two IIBChb(C10S) molecules bound to the IIAChb*(H89E) trimer. C, ribbon diagram of the complex with a view orthogonal to that shown in B depicting three molecules of IIBChb(C10S) bound to the IIAChb*(H89E) trimer. The color coding in B and C is the same as that in A. D, stereoview showing a reweighted atomic probability density map (drawn at a value of 20% maximum and calculated from the final 90 simulated annealing structures) for some of the interfacial side chains displayed as purple and green meshes for IIBChb(C10S) and the C chain of IIAChb*(H89E), respectively. The backbones are shown as tubes color coded as in A; the side chains of the restrained regularized mean structure are color coded according to atom type (carbon, gray; oxygen, red; and nitrogen, blue). Residues of IIBChb(C10S) are labeled in italics.
TABLE 4.
RDC R-factors for IIBChb in the IIAChb*(H89E)-IIBChb(C10S) complex and corresponding phosphoryl transition state
| RDC R-factor (%)a |
||
|---|---|---|
| Complex | Transition state | |
| 1DNH RDCs measured on free IIBChb(C10S)b | ||
| 100 mm NaCl and 10 mg/ml pf1 | ||
| All data (73) | 8.1 | 16.7 |
| Active site loop (residues 9–16) (6)c | 7.8 | 105.4 |
| 400 mm NaCl and 17 mg/ml pf1 | ||
| All data (80) | 9.8 | 13.1 |
| Active site loop (residues 9–16) (6)c | 7.8 | 38.3 |
| 1DNH RDCs measured on free phospho-IIBChbb | ||
| 100 mm NaCl and 10 mg/ml pf1 | ||
| All data (66) | 44.1 | 15.1 |
| Active site loop (residues 9–16) (7)d | 86.9 | 2.8 |
| 200 mm NaCl and 15 mg/ml pf1 | ||
| All data (76) | 35.7 | 14.7 |
| Active site loop (residues 9–16) (7)d | 76.7 | 11.8 |
Overall Structure of the IIAChb*(H89E)-IIBChb(C10S) Complex
The overall structure of the complex is shown in Fig. 3 with one, two, and three molecules of IIBChb(C10S) bound.
IIAChb* is a symmetric trimer (26). Each subunit comprises a three-helix bundle (α1, residues 14–44; α2, residues 46–74, α3, residues 84–114) in an up-down topology with α2 antiparallel to α1 and α3. The trimer interface consists of a parallel coiled-coil formed by helix α3 of each subunit. The active site residue, His-89, is located in helix α3 and lies in a deep groove bounded by helix α1 of its own subunit and helices α2 and α3 of an adjacent subunit. In the representations shown in this report with subunits A, B, and C of IIAChb* colored in blue, gold, and green, respectively, there are three identical binding sites for IIBChb, located at the interface of subunits A and C, subunits C and B, and subunits B and A, with the first named subunit of each binding site bearing the active site His-89 (Fig. 3, B and C).
IIBChb is a mixed α/β protein comprising 5 helices (residues 12–30, 43–49, 63–71, 80–86, and 88–105), and 4 β-strands (residues 4–9, 34–39, 53–56 and 76–78) arranged in a -1x, -2x, -1x topology (9–11). The active site residue, Cys-10, is located in an exposed active site loop (residues 9–16) that forms a protrusion on the surface of the protein. The overall topology of IIBChb is similar to that of IIAMtl (7) although the percentage sequence identity is only 8%. In the NMR structure of phospho-IIBChb (11) but not the x-ray structure of IIBChb(C10S) (9), the active site loop has a conformation that is similar to that of IIBMtl (both unphosphorylated (7) and phosphorylated (8)) and the low molecular weight protein-tyrosine phosphatases (42), except that the residues located at i+1 and i+2 of the active site cysteine are replaced by only a single residue in IIBChb.
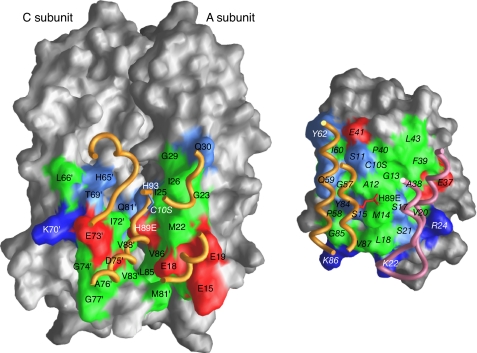
Because the three IIBChb binding sites on IIAChb* are identical, we will simply consider the interaction of IIBChb(C10S) with the A and C subunits of IIAChb*(H89E), where the contributing active site histidine (His-89) originates from the A subunit (Fig. 3A, 4 and 5). A total of 1836 Å2 is buried upon complexation, 927 Å2 originating from IIAChb*(H89E) and 909 from IIBChb(C10S). The interface accessible surface area contributed by the A and C subunits of IIAChb*(H89E) is approximately equal. The interface comprises 15 residues each from the A and C subunits of IIAChb*(H89E) and 29 residues from IIBChb(C10S); 4 residues of the latter interact simultaneously with the A and C subunits of IIAChb*(H89E) (Fig. 4C). The interface is ellipsoidal in shape with an eccentricity of ∼0.3 (where values of 0 and 1 indicate a perfect circle and a straight line, respectively), the gap volume index (ratio of gap volume to interface accessible surface area) is 3.3 Å, and the r.m.s. deviation of the interface atoms from a least-squares plane through these atoms is 3.7 Å (concave; Fig. 5, left panel) for IIAChb*(H89E) and 3.3 Å (convex; Fig. 5, right panel) for IIBChb(C10S), These values are typical of transient complexes (76, 77).
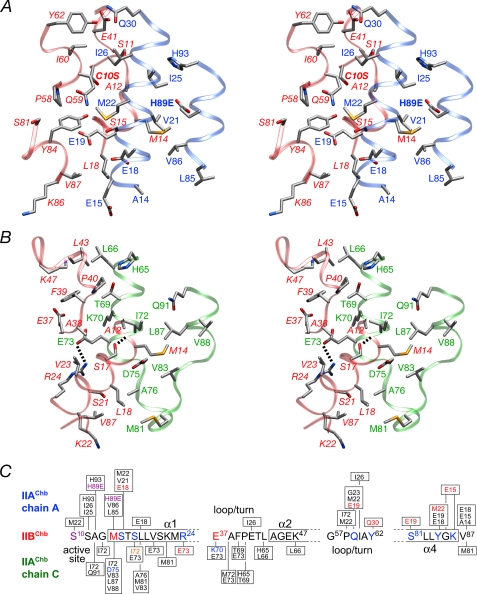
FIGURE 4.
The IIAChb*(H89E)-IIBChb(C10S) interface. A, stereoview of the interface between the A subunit of IIAChb*(H89E) and IIBChb(C10S) with the respective backbones shown as transparent blue and red ribbons, respectively. B, stereoview of the interface between the C subunit of IIAChb*(H89E) and IIBChb(C10S) with the respective backbones shown as transparent green and red ribbons, respectively. The dashed lines indicate intermolecular hydrogen bonds or salt bridges. The side chain atoms are colored according to atom type; carbon, gray; nitrogen, blue; oxygen, red. Residues of IIBChb(C10S) are labeled in italics. C, diagrammatic representation of the intermolecular contacts between the A and C subunits of IIAChb*(H89E) and IIBChb(C10S). Residues involved in side chain-side chain electrostatic interactions are colored in blue (donor) or red (acceptor). Ile-72 of the C subunit of IIAChb*(H89E) is colored in orange because its backbone carbonyl accepts a hydrogen bond from Ser-17 of IIBChb(C10S). The active site residues, H89E of IIAChb*(H89E), and C10S of IIBChb(C10S) are colored purple.
FIGURE 5.
Interaction surfaces for the IIAChb*(H89E)-IIBChb(C10S) complex. The left panel display the interaction surface (formed by the A and C subunits) on IIAChb*(H89E) for IIBChb(C10S); the right panel shows the interaction surface on IIBChb(C10S) for IIAChb*(H89E). The surfaces are color coded as follows: hydrophobic residues, green; uncharged residues bearing a polar functional group, cyan; negatively charged residues, red; positively charged residues, blue. Relevant portions of the backbone and active site residue of the interacting partner are displayed as tubes and bonds, respectively. Residues of IIBChb(C10S) are labeled in italics. Residues from the C subunit of IIBChb*(H89E) are indicated by an apostrophe after the residue number; in addition, the surfaces of the A and C subunits of IIAChb* that do not constitute the interaction surface are colored in dark gray and light gray, respectively.
Stereoviews depicting details of the interface are shown in Fig. 4, A and B, and a summary of the intermolecular contacts is provided in Fig. 4C. The active site loop, a 310 helix (residues 58–62) and helix α4 of IIBChb(C10S) are in contact with helices α1 and α3 of the A subunit of IIAChb*(H89E) (Fig. 4A). Helices α1 and α2, the C-terminal end of strand β2 and the subsequent 310 helix (residues 40–42) of IIBChb(C10S) contact helices α2 and α3 of the C subunit of IIAChb*(H89E), as well as some residues in the disordered loop connecting these two helices in IIAChb*(H89E) (Fig. 4B). The intermolecular interactions are largely hydrophobic with 50–60% of the interfacial residues being nonpolar. Met-14 of the active site loop of IIBChb(C10S) is involved in a very large number of intermolecular hydrophobic interactions with methyl group clusters of Val and Leu residues, specifically Val-21A and Val-86A of the A subunit and Val-83C, Leu-87C, and Val-88C of the C subunit of IIAChb*(H89E). Two other methionine residues, Met-22A and Met-81C of IIAChb*(H89E) are also involved in an array of intermolecular hydrophobic interactions, with C10S, Pro-58, Gln-59, and Tyr-84, and with Leu-18, Lys-22, and Val-87, respectively, of IIBChb(C10S). There are only two intermolecular salt bridge/hydrogen bonding interactions: between Arg-24 and Glu-73C, and between Ser-17 and the backbone carbonyl of Ile-72C (Fig. 4B). These are supplemented by 7 longer range electrostatic interactions, 5 involving the A chain and 2 the B chain of IIAChb*(H89E) (Fig. 4C). Of these, two involve interactions between charged side chains (Glu-15A and Lys-86, and Lys-70C and Glu-37). The remainder involve interactions either between polar groups or between polar and charged groups. An example of the former would include the interaction between the sulfur atom of Met-22A and the hydroxyl group of Tyr-84 (Fig. 4A). An example of the latter is the interaction between the carboxylate of Glu-19A and the hydroxyl group of Ser-81 and the amide group of Gln-59 (Fig. 4A).
As in previously solved complexes of the PTS (30, 31, 33–37), the center of each interface is largely hydrophobic, surrounded by a ring of polar and charged residues (Fig. 5). Of note is that the interaction surface of IIBChb contains both positively and negatively charged residues, similar to IIBGlc (31), IIBMtl (36) and IIBMan (35), but in contrast to HPr (33, 34, 37) where the charged residues on the interaction surface are entirely positive. This ensures that the IIB domains interact with their corresponding IIA domains, but not with enzyme I, despite the fact that the binding site on all the sugar-specific enzymes IIA is common to both HPr and IIB, and HPr uses the same binding surface to interact with both enzymes IIA and EI.
The Active Site and the Phosphoryl Transition State
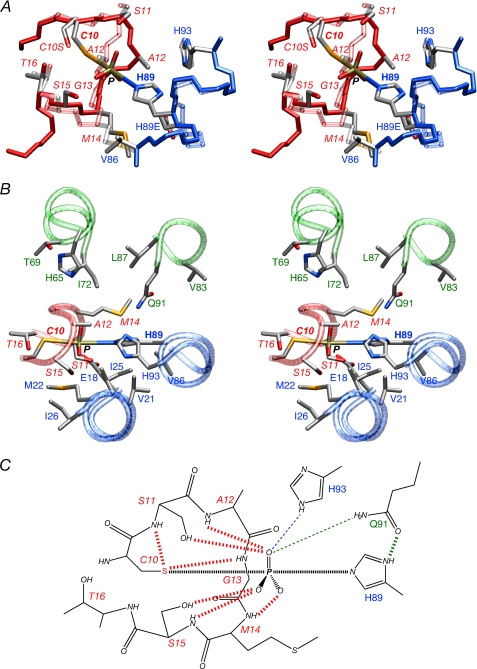
The conformation of the active site loop (residues 9–16) of IIBChb(C10S) in the complex is not compatible with the formation of a pentacoordinate phosphoryl transition state in which the phosphorus atom lies in the plane of the imidazole ring of His-89A and the P-Sγ-Cβ bond angle of Cys-10 lies between 90 and 130°. This is because the location of Ala-12 sterically occludes the formation of a phosphoryl transition state (Fig. 6A). The same is true of the x-ray structure of IIBChb(C10S) (9). Although there is a small displacement (1.3 Å) of the backbone of the active site loop of IIBChb(C10S) between the current structure (based on RDCs within the active site loop) and the crystal structure, the conformation of the loop in the two structures is actually very similar and the differences in backbone torsion angles are relatively small. In the conformation of the active site loop of phospho- IIBChb (11), however, the path of the polypeptide chain from residues 11–13 follows a mirror-like image relative to the unphosphorylated active site loop.
FIGURE 6.
The phosphoryl transition state of the IIAChb*-IIBChb complex. A, stereoview displaying the conformational change within the active site loop of IIBChb accompanying formation of a phosphoryl transition state. Superposition of the active site loop of the IIAChb*-P-IIBChb transition state (solid colors) and the IIAChb*(H89E)-IIBChb(C10S) complex (transparent colors) with the backbones of IIAChb* and IIBChb in red and blue, respectively (see text for further details). B, stereoview of the environment surrounding the His-89-P-Cys-10 transition state. The backbone is displayed as transparent tubes with IIBChb in red, and the A and C subunits of IIAChb* in blue and green, respectively. C, schematic of the interactions stabilizing the transition state. The thick dashed lines indicate hydrogen bonds, the thin dashed lines indicate electrostatic interactions that are too long to be classified as hydrogen bonds. It should be noted that only side chain torsion angle changes confined within a single rotamer would be required to permit hydrogen-bonding interactions from His-93A (Chain A) and Gln-91C (Chain C) of IIAChb* to the phosphoryl group in phospho-IIAChb*. Color coding: red, IIBChb; blue, A subunit of IIAChb*; green, C subunit of IIAChb*. Residues of IIBChb are labeled in italics.
To model the transition state, therefore, we kept the coordinates of IIAChb* and IIBChb within the complex fixed, with the exception of the backbone of the active site loop (residues 9–16) of IIBChb, the backbone immediately adjacent to the active site histidine (residues 87–91 of subunit A) of IIAChb*, and the interfacial side chains in close proximity to Cys-10 and His-89A, which were given torsional degrees of freedom and subjected to simulated annealing refinement on the basis of the experimental intermolecular NOE restraints and the RDCs collected on free phospho-IIBChb. In addition, restraints were included to define the geometry of the phosphoryl group relative to Cys-10 and His-89A in the transition state, as described for the phosphoryl transition state of the IIAMtl-IIBMtl complex (36). The results are shown in Fig. 6, A and B. The active site loop of IIBChb in the transition state adopts a very similar conformation to that seen in the previously determined NMR structure of phospho-IIBChb (11) with a backbone r.m.s. difference of 0.8 Å, and the RDCs within the active site loop are well satisfied (Table 4). The key conformational changes within the active site loop of IIBChb are the transition of the φ/ψ angles of Gly-13 from the right (∼ −60°/−40°) to left α-helical region (∼100°/30°) of the Ramachandran plot, a shift in the φ/ψ angles of Met-14 away from the α-helical region (from ∼ −50°/−50° to ∼ −110°/−70°), and a shift of the φ/ψ angles of Ala-12 from the β region (∼ −150°/70°) to the helical region (∼ −110°/−25°). The changes in the backbone of residues 87A-91A of IIAChb* are minimal with atomic r.m.s. displacements of less than 0.6 Å (Fig. 6A). It can be readily seen that as a consequence of the conformational change in the active site loop of IIBChb, Ala-12 no longer occludes the phosphoryl group and the path followed by the backbone readily permits the formation of a phosphoryl transition state His-89-P-Cys-10 intermediate.
Although the structure of phospho-IIAChb* has not been solved, the phosphorylated state is easily modeled from the structure of unphosphorylated IIAChb* (26): minimal changes in the χ1 and χ2 angles of His-89A are all that are required to permit the formation of hydrogen bonds from the side chain amide of Gln-91C and the Nϵ2-H proton of His-93A to the phosphoryl group. In the transition state, these distances are lengthened but bridging hydrogen bonds involving water molecules could clearly be formed (Fig. 6C). The phosphoryl group in the transition state is stabilized by an array of hydrogen bonds from the IIBChb active site loop, including the backbone amides of Ala-12, Met-14, and Ser-15, and the hydroxyl groups of Ser-11 and Ser-15; in addition, hydrogen bonds from the backbone amides of Ser-11 and Gly-13 to the sulfur atom to Cys-10 further stabilize the conformation (Fig. 6, B and C). Many of these interactions are maintained upon shortening of the S-P bond to form phospho-IIBChb (11). The interactions stabilizing the phosphoryl group in both the IIAChb*-P-IIBChb transition state and phospho-IIBChb (11) are very similar to those observed in phospho-IIBMtl (8) and the phosphorylated state of the low molecular weight protein-tyrosine phosphatases (42). Presumably, the reason that the active site loop of IIBChb undergoes a conformational change upon phosphorylation, whereas that of IIBMtl does not, resides in the fact that the single residue deletion within the active site loop of IIBChb, relative to IIBMtl, results in stereochemical strain that can only be overcome by numerous interactions between the protein backbone and the phosphoryl group.
Further examination of the IIAChb*-P-IIBChb transition state reveals that the phosphoryl group is buried within a largely hydrophobic environment provided by Ala-12 and Met-14 of IIBChb and Val-21A, Met-22A, Ile-25A, Ile-72C, and Leu-87C of IIAChb*. This configuration is common to all protein-protein complexes of the PTS solved to date (30, 31, 33–37), including the EIN-HPr complex that is common to all branches of the pathway, as well as the complexes involving sugar-specific components.
Concluding Remarks
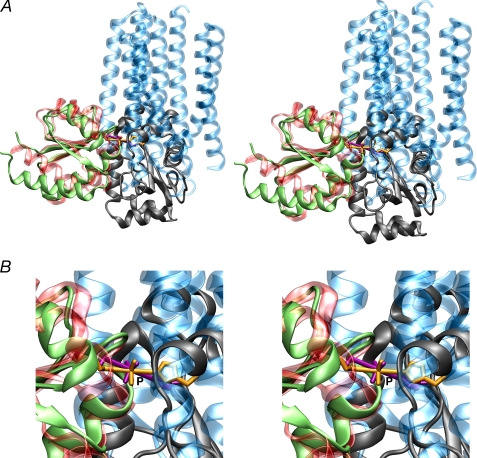
We have determined the solution structure of the IIAChb*(H89E)-IIBChb(C10S) complex, and shown that this structure is fully compatible with the formation of a phosphoryl transition state when the active site loop of IIBChb adopts the conformation found in phosphorylated IIBChb. As previously noted, the overall topology of IIBChb and IIBMtl are remarkably similar, despite 8% sequence identity, and the Cα atoms of 71 out of 106 residues can be superimposed with an atomic r.m.s. difference of 2 Å. Fig. 7 shows a comparison of the phosphoryl transition states of the two complexes with the coordinates of IIBChb and IIBMtl superimposed. The structures of the IIAChb* trimer and IIAMtl monomer bear no resemblance to one another, and with the exception of a single turn of helix that fortuitously overlaps, the structural elements making up the binding site are entirely different. Nevertheless, the position of the His-P-Cys phosphoryl transition state is remarkably similar. In addition, the distribution of hydrophobic, polar, and charged residues within the binding sites of IIAChb* and IIAMtl, and likewise IIBChb and IIBMtl, is broadly similar (compare Fig. 5 of this report with Fig. 6 of Ref. 36), although the interaction of IIAChb* with IIBChb involves a somewhat larger preponderance of hydrophobic residues than that between IIAMtl and IIBMtl, and the cleft in which the active site histidine of IIAChb* is located is both deeper and narrower than that for IIAMtl. Thus, one might argue that these two PTS complexes from distinct sugar branches of the pathway illustrate an example of convergent evolution of the surfaces of active sites (in terms of shape and distribution of residue type) generated by completely different underlying backbone structural elements.
FIGURE 7.
Comparison of the IIAChb*-IIBChb and IIAMtl-IIBMtl complexes. A, overall stereoview and, B, stereo close up of the His-P-Cys phosphoryl transition state, with IIBChb and IIBMtl superimposed. The backbone is displayed as a ribbon diagram and the His-P-Cys transition state as a stick diagram. IIAMtl and IIBMtl are shown in green and gray, respectively; IIAChb* and IIBChb are shown in transparent blue and red, respectively; and the His-P-Cys phosphoryl transition state is shown in gold for the IIAChb*-IIBChb complex and in purple for the IIAMtl-IIBMtl complex. The coordinates of the IIAMtl-IIBMtl complex are taken from Ref. 36 (PDB code 2FEW). The superposition of IIBChb and IIBMtl was carried out using the program O (79).
Acknowledgments
We thank Dan Garrett for software support and Chun Tang for many helpful discussions.
This work was supported, in whole or in part, by the intramural program of NIDDK, National Institutes of Health and the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G. M. C.).
The atomic coordinates and experimental NMR restraints (codes 2WWV and 2WY2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PTS
- phosphoenolpyruvate:sugar phosphotransferase system
- EI
- enzyme I
- HPr
- histidine-containing phosphocarrier protein
- Chb
- N,N′-diacetylchitobiose
- IIAChb
- IIBChb, and IICChb, A, B, and C domains, respectively, of the N,N′-diacetylchitobiose transporter IIChb
- IIAChb*
- double mutant of IIAChb comprising a 13-residue deletion at the N terminus and an Asp to Leu mutation at position 92 (wild-type numbering)
- NOE
- nuclear Overhauser effect
- HSQC
- heteronuclear single quantum coherence
- TROSY
- transverse relaxation optimized spectroscopy
- r.m.s.
- root mean square
- RDC
- residual dipolar coupling
- ppb
- parts per billion
- SA
- simulated annealing
- PDB
- Protein Data Bank.
REFERENCES
- 1.Deutscher J., Francke C., Postma P. W. (2006) Microbiol. Mol. Biol. Rev. 70, 939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundig W., Ghosh S., Roseman S. (1964) Proc. Natl. Acad. Sci. U.S.A. 52, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meadow N. D., Fox D. K., Roseman S. (1990) Annu. Rev. Biochem. 59, 497–542 [DOI] [PubMed] [Google Scholar]
- 4.Postma P. W., Lengeler J. W., Jacobson G. R. (1993) Microbiol. Rev. 57, 543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robillard G. T., Broos J. (1999) Biochim. Biophys. Acta 1422, 73–104 [DOI] [PubMed] [Google Scholar]
- 6.Siebold C., Flükiger K., Beutler R., Erni B. (2001) FEBS Lett. 504, 104–111 [DOI] [PubMed] [Google Scholar]
- 7.Legler P. M., Cai M., Peterkofsky A., Clore G. M. (2004) J. Biol. Chem. 279, 39115–39121 [DOI] [PubMed] [Google Scholar]
- 8.Suh J. Y., Tang C., Cai M., Clore G. M. (2005) J. Mol. Biol. 353, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 9.van Montfort R. L., Pijning T., Kalk K. H., Reizer J., Saier M. H., Jr., Thunnissen M. M., Robillard G. T., Dijkstra B. W. (1997) Structure 5, 217–225 [DOI] [PubMed] [Google Scholar]
- 10.Ab E., Schuurman-Wolters G., Reizer J., Saier M. H., Dijkstra K., Scheek R. M., Robillard G. T. (1997) Protein Sci. 6, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ab E., Schuurman-Wolters G. K., Nijlant D., Dijkstra K., Saier M. H., Robillard G. T., Scheek R. M. (2001) J. Mol. Biol. 308, 993–1009 [DOI] [PubMed] [Google Scholar]
- 12.Teplyakov A., Lim K., Zhu P. P., Kapadia G., Chen C. C., Schwartz J., Howard A., Reddy P. T., Peterkofsky A., Herzberg O. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16218–16223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberholzer A. E., Bumann M., Schneider P., Bächler C., Siebold C., Baumann U., Erni B. (2005) J. Mol. Biol. 346, 521–532 [DOI] [PubMed] [Google Scholar]
- 14.Oberholzer A. E., Schneider P., Siebold C., Baumann U., Erni B. (2009) J. Biol. Chem. 284, 33169–33176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao D. I., Silverton E., Seok Y. J., Lee B. R., Peterkofsky A., Davies D. R. (1996) Structure 4, 861–872 [DOI] [PubMed] [Google Scholar]
- 16.Márquez J., Reinelt S., Koch B., Engelmann R., Hengstenberg W., Scheffzek K. (2006) J. Biol. Chem. 281, 32508–32515 [DOI] [PubMed] [Google Scholar]
- 17.Herzberg O., Reddy P., Sutrina S., Saier M. H., Jr., Reizer J., Kapadia G. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia Z., Quail J. W., Waygood E. B., Delbaere L. T. (1993) J. Biol. Chem. 268, 22490–22501 [DOI] [PubMed] [Google Scholar]
- 19.Liao D. I., Kapadia G., Reddy P., Saier M. H., Jr., Reizer J., Herzberg O. (1991) Biochemistry 30, 9583–9594 [DOI] [PubMed] [Google Scholar]
- 20.Worthylake D., Meadow N. D., Roseman S., Liao D. I., Herzberg O., Remington S. J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10382–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sliz P., Engelmann R., Hengstenberg W., Pai E. F. (1997) Structure 5, 775–788 [DOI] [PubMed] [Google Scholar]
- 22.Nunn R. S., Marković-Housley Z., Génovésio-Taverne J. C., Flükiger K., Rizkallah P. J., Jansonius J. N., Schirmer T., Erni B. (1996) J. Mol. Biol. 259, 502–511 [DOI] [PubMed] [Google Scholar]
- 23.Schauder S., Nunn R. S., Lanz R., Erni B., Schirmer T. (1998) J. Mol. Biol. 276, 591–602 [DOI] [PubMed] [Google Scholar]
- 24.van Montfort R. L., Pijning T., Kalk K. H., Hangyi I., Kouwijzer M. L., Robillard G. T., Dijkstra B. W. (1998) Structure 6, 377–388 [DOI] [PubMed] [Google Scholar]
- 25.Orriss G. L., Erni B., Schirmer T. (2003) J. Mol. Biol. 327, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 26.Tang C., Williams D. C., Jr., Ghirlando R., Clore G. M. (2005) J. Biol. Chem. 280, 11770–11780 [DOI] [PubMed] [Google Scholar]
- 27.Kalbitzer H. R., Hengstenberg W. (1993) Eur. J. Biochem. 216, 205–214 [DOI] [PubMed] [Google Scholar]
- 28.Wittekind M., Rajagopal P., Branchini B. R., Reizer J., Saier M. H., Jr., Klevit R. E. (1992) Protein Sci. 1, 1363–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberstadt M., Grdadolnik S. G., Gemmecker G., Kessler H., Buhr A., Erni B. (1996) Biochemistry 35, 11286–11292 [DOI] [PubMed] [Google Scholar]
- 30.Hu J., Hu K., Williams D. C., Jr., Komlosh M. E., Cai M., Clore G. M. (2008) J. Biol. Chem. 283, 11024–11037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai M., Williams D. C., Jr., Wang G., Lee B. R., Peterkofsky A., Clore G. M. (2003) J. Biol. Chem. 278, 25191–25206 [DOI] [PubMed] [Google Scholar]
- 32.Garrett D. S., Seok Y. J., Liao D. I., Peterkofsky A., Gronenborn A. M., Clore G. M. (1997) Biochemistry 36, 2517–2530 [DOI] [PubMed] [Google Scholar]
- 33.Garrett D. S., Seok Y. J., Peterkofsky A., Gronenborn A. M., Clore G. M. (1999) Nat. Struct. Biol. 6, 166–173 [DOI] [PubMed] [Google Scholar]
- 34.Cornilescu G., Lee B. R., Cornilescu C. C., Wang G., Peterkofsky A., Clore G. M. (2002) J. Biol. Chem. 277, 42289–42298 [DOI] [PubMed] [Google Scholar]
- 35.Williams D. C., Jr., Cai M., Suh J. Y., Peterkofsky A., Clore G. M. (2005) J. Biol. Chem. 280, 20775–20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh J. Y., Cai M., Williams D. C., Jr., Clore G. M. (2006) J. Biol. Chem. 281, 8939–8949 [DOI] [PubMed] [Google Scholar]
- 37.Wang G., Louis J. M., Sondej M., Seok Y. J., Peterkofsky A., Clore G. M. (2000) EMBO J. 19, 5635–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keyhani N. O., Wang L. X., Lee Y. C., Roseman S. (2000) J. Biol. Chem. 275, 33084–33090 [DOI] [PubMed] [Google Scholar]
- 39.Keyhani N. O., Bacia K., Roseman S. (2000) J. Biol. Chem. 275, 33102–33109 [DOI] [PubMed] [Google Scholar]
- 40.Keyhani N. O., Boudker O., Roseman S. (2000) J. Biol. Chem. 275, 33091–33101 [DOI] [PubMed] [Google Scholar]
- 41.Keyhani N. O., Roseman S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14367–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M., Van Etten R. L., Stauffacher C. V. (1994) Biochemistry 33, 11097–11105 [DOI] [PubMed] [Google Scholar]
- 43.Goto N. K., Gardner K. H., Mueller G. A., Willis R. C., Kay L. E. (1999) J. Biomol. NMR 13, 369–374 [DOI] [PubMed] [Google Scholar]
- 44.Suh J. Y., Cai M., Clore G. M. (2008) J. Biol. Chem. 283, 18980–18989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 46.Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 47.Clore G. M., Gronenborn A. M. (1991) Science 252, 1390–1399 [DOI] [PubMed] [Google Scholar]
- 48.Bax A., Grzesiek S. (1993) Acc. Chem. Res. 26, 131–138 [Google Scholar]
- 49.Yang D. W., Kay L. E. (1999) J. Biomol. NMR 13, 3–10 [DOI] [PubMed] [Google Scholar]
- 50.Clore G. M., Starich M. R., Gronenborn A. M. (1998) J. Am. Chem. Soc. 120, 10571–10572 [Google Scholar]
- 51.Hansen M. R., Mueller L., Pardi A. (1998) Nat. Struct. Biol. 5, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 52.Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 53.Clore G. M., Gronenborn A. M. (1998) Trends Biotechnol. 16, 22–34 [DOI] [PubMed] [Google Scholar]
- 54.Cai M., Huang Y., Zheng R., Wei S. Q., Ghirlando R., Lee M. S., Craigie R., Gronenborn A. M., Clore G. M. (1998) Nat. Struct. Biol. 5, 903–909 [DOI] [PubMed] [Google Scholar]
- 55.Clore G. M., Gronenborn A. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5891–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clore G. M., Gronenborn A. M., Nilges M., Ryan C. A. (1987) Biochemistry 26, 8012–8023 [DOI] [PubMed] [Google Scholar]
- 57.Nilges M. (1993) Proteins 17, 297–309 [DOI] [PubMed] [Google Scholar]
- 58.Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwieters C. D., Clore G. M. (2001) J. Magn. Reson. 152, 288–302 [DOI] [PubMed] [Google Scholar]
- 60.Schwieters C. D., Kuszewski J., Clore G. M. (2006) Progr. NMR Spect. 48, 47–62 [Google Scholar]
- 61.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 62.Clore G. M., Gronenborn A. M., Tjandra N. (1998) J. Magn. Reson. 131, 159–162 [DOI] [PubMed] [Google Scholar]
- 63.Kuszewski J., Qin J., Gronenborn A. M., Clore G. M. (1995) J. Magn. Reson. B 106, 92–96 [DOI] [PubMed] [Google Scholar]
- 64.Nilges M., Gronenborn A. M., Brünger A. T., Clore G. M. (1988) Protein Eng. 2, 27–38 [DOI] [PubMed] [Google Scholar]
- 65.Clore G. M., Kuszewski J. (2002) J. Am. Chem. Soc. 124, 2866–2867 [DOI] [PubMed] [Google Scholar]
- 66.Schwieters C. D., Clore G. M. (2008) J. Phys. Chem. B 112, 6070–6073 [DOI] [PubMed] [Google Scholar]
- 67.Schwieters C. D., Clore G. M. (2001) J. Magn. Reson. 149, 239–244 [DOI] [PubMed] [Google Scholar]
- 68.Nicholls A., Sharp K. A., Honig B. (1991) Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 69.Schwieters C. D., Clore G. M. (2002) J. Biomol. NMR 23, 221–225 [DOI] [PubMed] [Google Scholar]
- 70.Clore G. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9021–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clore G. M., Garrett D. S. (1999) J. Am. Chem. Soc. 121, 9008–9012 [Google Scholar]
- 72.Bax A., Kontaxis G., Tjandra N. (2001) Methods Enzymol. 339, 127–174 [DOI] [PubMed] [Google Scholar]
- 73.Williams D. C., Jr., Cai M., Clore G. M. (2004) J. Biol. Chem. 279, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 74.Williams D. C., Jr., Lee J. Y., Cai M., Bewley C. A., Clore G. M. (2005) J. Biol. Chem. 280, 29269–29276 [DOI] [PubMed] [Google Scholar]
- 75.Ortega-Roldan J. L., Jensen M. R., Brutscher B., Azuaga A. I., Blackledge M., van Nuland N. A. (2009) Nucleic Acids Res. 37, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones S., Thornton J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13–208552589 [Google Scholar]
- 77.Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 78.Grzesiek S., Stahl S. J., Wingfield P. T., Bax A. (1996) Biochemistry 35, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 79.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 80.Sass H. J., Musco G., Stahl S. J., Wingfield P. T., Grzesiek S. (2001) J. Biomol. NMR 21, 275–280 [DOI] [PubMed] [Google Scholar]
- 81.Laskowski R. A., McArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]