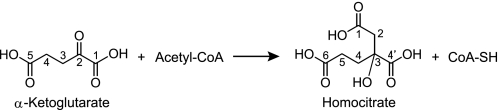
Abstract
Homocitrate synthase (HCS) catalyzes aldol-type condensation of acetyl coenzyme A (acetyl-CoA) and α-ketoglutarate (α-KG) to synthesize homocitrate (HC), which is the first and committed step in the lysine biosynthetic pathway through α-aminoadipate. As known in most enzymes catalyzing the first reactions in amino acid biosynthetic pathways, HCS is regulated via feedback inhibition by the end product, lysine. Here, we determined the crystal structures of HCS from Thermus thermophilus complexed with α-KG, HC, or lysine. In the HC complex, the C1-carboxyl group of HC, which is derived from acetyl-CoA, is hydrogen-bonded with His-292* from another subunit (indicated by the asterisk), indicating direct involvement of this residue in the catalytic mechanism of HCS. The crystal structure of HCS complexed with lysine showed that lysine is bound to the active site with rearrangement of amino acid residues in the substrate-binding site, which accounts for the competitive inhibition by lysine with α-KG. Comparison between the structures suggests that His-72, which is conserved in lysine-sensitive HCSs and binds the C5-carboxyl group of α-KG, serves as a switch for the conformational change. Replacement of His-72 by leucine made HCS resistant to lysine inhibition, demonstrating the regulatory role of this conserved residue.
Keywords: Allosteric regulation, Enzyme catalysis, Enzyme mechanisms, Enzyme structure, Protein structure
Introduction
Lysine is known to be biosynthesized by two completely different pathways. Fungi and yeast synthesize lysine through the α-aminoadipate pathway (1, 2), whereas most bacteria and plants synthesize amino acid through the diaminopimelate pathway (3, 4). Because higher animals cannot biosynthesize lysine, the enzymes of these pathways are attractive targets for designing antibiotics against pathogenic microorganisms (5, 6). We found that the extremely thermophilic bacterium, Thermus thermophilus, biosynthesizes lysine through the α-aminoadipate pathway (7). In biosynthesis, the enzymes involved in the former part, conversion of α-ketoglutarate (α-KG,3 2-oxopentanedioate) to α-aminoadipate, have amino acid sequence similarity to those of the tricarboxylic acid cycle and leucine biosynthesis as observed in fungi and yeast, whereas the enzymes involved in the latter part, α-aminoadipate to lysine biosynthesis, have amino acid sequence similarity to those involved in the conversion of glutamate to ornithine in arginine biosynthesis (7–14), although lysine biosynthesis differs from arginine biosynthesis in that the former involves proteinaceous amino group modification of biosynthetic intermediates instead of the acetylation used in the latter (15).
Amino acid biosynthesis is often controlled at levels of gene expression and enzyme activity, depending on nutrient availability in the environment. Homocitrate synthase (HCS; EC 2.3.3.14), responsible for the first reaction in lysine biosynthesis, transfers the acetyl group from acetyl-CoA to α-KG to yield homocitrate (HC; 2-hydroxybuthane-1,2,4-tricarboxylate) and CoA (Scheme 1). As commonly observed in enzymes responsible for the first reaction of amino acid biosynthesis, HCS from T. thermophilus (TtHCS) is feedback-inhibited by the end product, lysine (11). We have previously revealed that the promoter of the major lysine biosynthetic gene cluster, starting from the hcs gene of T. thermophilus, is controlled by lysine through leader peptide-mediated transcriptional attenuation (16).
SCHEME 1.
First reaction of lysine biosynthesis. HCS transfers the acetyl group from acetyl-CoA to α-KG to yield HC and CoA.
α-Isopropylmalate synthase (IPMS; EC 2.3.3.13) is the enzyme responsible for the first reaction of leucine biosynthesis. The enzyme catalyzes transfer of the acetyl group from acetyl-CoA to α-ketoisovalerate (α-KIV) to yield isopropylmalate in a manner similar to that of HCS (17). IPMS is inhibited by the end product, leucine (18). The crystal structures of IPMS from Mycobacterium tuberculosis (MtIPMS) were determined in α-KIV-bound and leucine-bound forms (19). The structure of α-KIV-bound MtIPMS reveals the active site organization of IPMS and the recognition mechanism of the hydrophobic isopropyl moiety of α-KIV. In the leucine-bound form, leucine is bound at the dimer interface formed between C-terminal regulatory domains, both of which are from different chains of the IPMS dimer. The leucine-binding site is located close to the site of mutation, giving resistance to a leucine analog (20), which was previously found in IPMS from Saccharomyces cerevisiae. These observations indicate that end product inhibition occurs through leucine binding to the C-terminal domain; however, the detailed allosteric regulatory mechanism remains to be elucidated, because no apparent conformational change around the active site is observed between leucine-bound and -unbound forms of MtIPMS. Recently, the crystal structure of the catalytic domain of citramalate synthase (CMS; EC 2.3.1.182) from Leptospira interrogans (LiCMS) complexed with acetyl-CoA and pyruvate was reported (21). CMS is another HCS paralogue, which catalyzes the first reaction of the isoleucine biosynthetic pathway, converting pyruvate and acetyl-CoA to citramalate and CoA. Interestingly, HCS is shorter than IPMS and CMS by about 150 amino acid residues, lacking the C-terminal portion corresponding to the regulatory domain for sensing allosteric inhibitors in these paralogues. This observation suggests that the regulatory mechanism of HCS is different from those of IPMS and CMS. Here, we report the crystal structures of TtHCS complexed with α-KG, HC, or lysine, which reveal the mechanism of substrate recognition and feedback inhibition.
EXPERIMENTAL PROCEDURES
Protein Preparation
We designed three expression systems for the hcs gene: (i) TtHCS without tag (TtHCSwt) used for crystallization of the lysine-bound complex; (ii) TtHCS with a C-terminal His6 tag (TtHCS-Chis) used for crystallization of α-KG and HC; and (iii) HCS with an N-terminal His8 tag (TtHCS-Nhis) for enzymatic assays of wild-type and mutant enzymes. To prepare the DNA fragment for TtHCSwt, the hcs gene from T. thermophilus was amplified by PCR using the oligonucleotides 5′-GGCGAATTCCATATGCGGGAGTGGAAGATT-3′ and 5′-GGCAAGCTTTTACGCCGTGATCCACTCCCG-3′. The amplified DNA fragment, designed to direct the production of full-length TtHCS (Met-1 to Ala-376), was cloned into the EcoRI/HindIII site of pBluescript II SK(+). To prepare the DNA fragment for TtHCS-Chis, similar PCR was performed using the oligonucleotides 5′-GGCGAATTCCATATGCGGGAGTGGAAGATT-3′ and 5′-GGCAAGCTTTTAGTGGTGGTGGTGGTGGTGCGCCGTGATCCACTCCCG-3′. The amplified DNA fragment, designed to direct the production of full-length TtHCS (Met-1 to Ala-376) with a His6 tag extension at the C-terminal end, was cloned into the EcoRI/HindIII site of pBluescript II SK(+). To prepare the DNA fragment of TtHCS-Nhis, the hcs gene was amplified by PCR using oligonucleotides 5′-CCCGAATTCATGCGGGAGTGGAAGATTATT-3′ and 5′-CCCGTCGACTTACGCCGTGATCCACTC-3′. The amplified fragment, which was designed to direct the production of full-length HCS with 24 additional residues containing a His8 tag extension and thrombin site at the N-terminal end, was cloned into the EcoRI/SalI site of pBluescript II SK(+). DNA fragments with the correct sequence were cloned into the NdeI/HindIII site of pET-26b(+) for TtHCSwt and TtHCS-Chis and into the EcoRI/SalI site of pHIS8 (22) for TtHCS-Nhis and introduced into Escherichia coli BL21-CodonPlus (DE3)-RIL cells. E. coli cells harboring pET26b(+)-TtHCSwt or pET26b(+)-TtHCS-Chis were grown in 2× YT medium (2 × YT medium: 1.6% tryptone, 1% yeast extract, and 0.5% sodium chloride) supplemented with kanamycin (50 μg ml−1) and chloramphenicol (30 μg ml−1) at 310 K. Cells harboring pHIS8-TtHCS-Nhis were grown in 2× YT medium containing kanamycin (50 μg ml−1) and chloramphenicol (30 μg ml−1) at 310 K. In each case, when optical density at 600 nm of the culture reached about 0.6, gene expression was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside, and the culture was continued for an additional 12 h at 298 K.
TtHCS-Chis was purified as follows. The cells were harvested, washed, and suspended in buffer A (20 mm Tris-HCl, pH 8.0, 150 mm NaCl) containing 10 mm α-KG and 10% (v/v) glycerol. Suspended cells were disrupted by sonication and centrifuged at 40,000 × g for 15 min. The supernatant was heated at 343 K for 20 min to denature proteins from E. coli. The heated solution was centrifuged at 40,000 × g for 15 min, and the supernatant was applied to a Ni2+-nitrilotriacetic acid column (Novagen, Darmstadt, Germany) equilibrated with buffer B (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10 mm α-KG, 10% glycerol) containing 25 mm imidazole. After washing with buffer B containing 25 mm imidazole, the proteins bound to the resin were eluted with buffer B containing 500 mm imidazole. Fractions containing TtHCS-Chis were pooled and concentrated to about 10 mg ml−1 using Vivaspin-20 centrifugal filtration with a 10 kDa cut-off (Vivascience, Goettingen, Germany). The concentrated sample was applied to a HiLoad 26/60 Superdex 75 gel filtration fast protein liquid chromatography column (GE Healthcare) equilibrated with buffer A supplemented with 10 mm α-KG, 5 mm MgSO4, and 10% glycerol and eluted at 1.25 ml min−1. Active fractions were collected. TtHCS-Chis thus purified had over 95% purity on SDS-PAGE. Selenomethionine (SeMet)-substituted TtHCS-Chis was produced in E. coli B834 (DE3) (F− ompT hsdSB (rB−mB−) gal dcm met (DE3)) (23). The cells were grown at 310 K for 20 h in 1.6 liters of SeMet core medium (Wako Pure Chemical Industries, Osaka, Japan) containing 16 g of glucose, 400 mg of MgSO4·7H2O, 6.7 mg of FeSO4·7H2O, 16 ml of vitamin growth supplement (Sigma), 50 μg ml−1 kanamycin, and 40 mg of SeMet (Wako Pure Chemical). The protein was purified using the protocol shown above. Native and SeMet-substituted proteins were concentrated to 10 mg ml−1 in buffer A supplemented with 10 mm α-KG, 5 mm MgSO4, and 10% glycerol and used for crystallization. The method of purifying TtHCS-Nhis was essentially the same as that of TtHCS-Chis except that 10 mm α-KG, 5 mm MgSO4, and 10% glycerol were not added in the purification steps. Protein samples to determine the crystal structure of the HCS·Mg2+·α-KG complex were prepared as follows. After purification by a Ni2+-nitrilotriacetic acid column, 4 mm EDTA was added to the sample and incubated at 277 K for 1 h. The sample was then dialyzed against buffer A for 2 h twice. The concentrated sample was applied to a HiLoad 26/60 Superdex 75 gel filtration fast protein liquid chromatography column equilibrated with buffer A containing 10 mm α-KG, 5 mm MgSO4, and 10% glycerol at a flow rate of 1.25 ml min−1. The protein fractions were pooled and concentrated to 10 mg ml−1.
TtHCSwt was purified as follows. The cells were harvested, washed, and suspended in buffer C (10 mm HEPES, pH 7.5, 1 mm EDTA). Suspended cells were disrupted by sonication and centrifuged at 40,000 × g for 15 min. The supernatant was heated at 343 K for 30 min to denature proteins from E. coli. The heated solution was centrifuged at 40,000 × g for 15 min. Ammonium sulfate was added at a final concentration of 50% to the supernatant. After centrifugation, the precipitant was suspended in buffer C containing 1.5 m ammonium sulfate and applied onto a HiLoad 26/60 phenyl-Sepharose column (GE Healthcare). Protein was eluted with a linear gradient from 1.5 to 0 m ammonium sulfate in 250 ml of buffer C at a flow rate of 5 ml min−1, and fractions containing TtHCSwt were pooled and concentrated to about 4 mg ml−1. The concentrated sample was applied to a Resource Q anion exchange column (GE Healthcare) equilibrated with buffer C and eluted with a linear gradient of 0–1 m ammonium sulfate in buffer C at a flow rate of 3 ml min−1. Fractions containing TtHCSwt were pooled and concentrated to about 10 mg ml−1 and applied to a HiLoad 26/60 Superdex 75 gel filtration fast protein liquid chromatography column equilibrated with buffer D (10 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm MgSO4) and eluted at 2.5 ml min−1. TtHCSwt had over 95% purity on SDS-PAGE. Protein was concentrated to 10 mg ml−1 in buffer D and used for crystallization.
Crystallization
Crystallization conditions of the α-KG complex were screened by the hanging drop vapor diffusion method using Crystal Screen kits (Hampton Research). Drops of 2 μl consisting of 1 μl of reservoir solution and 1 μl of TtHCS-Chis solution of 10 mg ml−1 supplemented with 5 mm MgSO4 and 10 mm α-KG were equilibrated against 500 μl of reservoir solution at 293 K. In addition, a similar TtHCS-Chis solution supplemented with 5 mm MgSO4, 10 mm α-KG, and 0.2 mm acetyl-CoA was used for crystallization of the HC complex. A few crystals were obtained from both droplets using solution 32 (2.0 m ammonium sulfate) of Crystal Screen I over a few days. Crystals of 0.1 × 0.2 × 0.3 mm, formed in 2.0 m ammonium sulfate, were used for x-ray diffraction. We also obtained several crystals of similar shape and size for SeMet-containing TtHCS-Chis under the same conditions. Crystals of the Mg2+·α-KG complex were obtained from a droplet with a reservoir solution of 0.1 m MES-NaOH, pH 6.0, and 1.6 m ammonium sulfate. Although we also attempted to crystallize TtHCS-Nhis, no crystals suitable for crystallographic analysis were obtained.
Crystallization conditions of TtHCSwt complexed with lysine were screened by the hanging drop vapor diffusion method using Crystal Screen kits. Drops of 2 μl consisting of 1 μl of reservoir solution and 1 μl of TtHCSwt solution of 10 mg ml−1 added to 10 mm lysine and 5 mm MgSO4 were equilibrated against 500 μl of reservoir solution at 293 K. A crystal obtained from a droplet using solution 25 (0.1 m MES-NaOH, pH 6.5, 0.01 m CoCl26H2O, and 1.8 m ammonium sulfate) of Crystal Screen 2 was used for crystallographic analysis.
Data Collection, Structure Determination, and Refinement
Prior to data collection, crystals were briefly soaked in reservoir solution supplemented with 20% (v/v) glycerol as a cryoprotectant, flash-cooled in a nitrogen gas stream at 95 K, and stored in liquid nitrogen. Diffraction data of the α-KG and HC complexes were collected at the BL-5A station of the Photon Factory, High Energy Accelerator Research Organization (KEK) (Tsukuba, Japan). Diffraction data of the lysine complex were collected at the NW12 station of the Photon Factory. Diffraction images were indexed, integrated, and scaled using the HKL-2000 program suite (24). SeMet multiple wavelength anomalous diffraction data were also collected at the BL-5A station of the Photon Factory. Details of data collection statistics are summarized in Table 1. Crystals of SeMet-labeled TtHCS-Chis contain one monomer per asymmetric unit. SeMet multiple wavelength anomalous diffraction data were analyzed using SOLVE (25). All six selenium atoms were identified from the six possible selenium atoms in the primary sequence. Automated model building was performed by cycling with RESOLVE (26). The data of native TtHCS-Chis crystals collected at a wavelength of 1.0 Å were used for subsequent molecular replacement and crystallographic refinement. Molecular replacement was performed with MOLREP (27) in the CCP4 program suite (28) using the program Refmac 5.2 (29), and model correction in the electron density map was performed with Coot (30). Data collection, refinement statistics, and the results of Ramachandran plots produced by the program PROCHECK (31) are summarized in Table 1. Although Ile-279 falls into the disallowed region in each complex, excellent map fitting ensures unambiguous model building. The figures were prepared using PyMOL (available on the World Wide Web).
TABLE 1.
X-ray data collection and refinement statistics
| SeMet-HCS |
Complexed with |
||||||
|---|---|---|---|---|---|---|---|
| Peak | Edge | Remote | Cu2+·α-KG | Cu2+·HC | Mg2+·α-KG | Co2+·lysine | |
| Data collection | |||||||
| Wavelength (Å) | 0.97945 | 0.97934 | 0.96421 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P6322 | P6322 | P6322 | P6322 | P6322 | P6322 | C2221 |
| Cell dimensions | |||||||
| a (Å) | 135.6 | 135.7 | 135.8 | 135.6 | 134.7 | 135.5 | 80.3 |
| b (Å) | 96.0 | ||||||
| c (Å) | 127.5 | 127.6 | 127.7 | 127.1 | 127.3 | 126.6 | 77.1 |
| Resolution (Å)a | 2.15 (2.23-2.15) | 2.15 (2.23-2.15) | 2.15 (2.23-2.15) | 1.80 (1.86-1.80) | 1.96 (2.03-1.96) | 2.15 (2.23-2.15) | 1.80 (1.85-1.80) |
| Total reflections | 812,613 | 822,773 | 825,517 | 1,362,109 | 1,035,025 | 793,789 | 204,867 |
| Unique reflections | 38,146 | 38,273 | 38,366 | 64,069 | 48,641 | 37,714 | 26,532 |
| Completeness (%)a | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (100.0) | 99.6 (99.7) | 99.9 (100.0) | 99.7 (99.6) |
| Average I/σ (I)a | 48.5 (11.1) | 46.7 (9.6) | 43.3 (7.4) | 30.1 (3.9) | 53.7 (3.5) | 40.8 (4.2) | 35.1 (4.1) |
| Rmerge (%)ab | 6.3 (27.6) | 6.3 (31.1) | 6.7 (38.3) | 7.0 (59.3) | 7.3 (49.5) | 11.2 (54.6) | 6.1 (39.7) |
| Phasing | |||||||
| No. of Se sites | 6 | ||||||
| Figure of meritc | 0.39 (0.67)c | ||||||
| Refinement | |||||||
| Resolution (Å) | 30.66-1.80 | 44.36-1.96 | 31.5-2.15 | 35.6-1.80 | |||
| R-factor/Rfree (%) | 19.9/22.4 | 19.4/22.2 | 20.5/23.2 | 17.1/21.3 | |||
| No. of protein atoms | 2,447 | 2,447 | 2,502 | 2,745 | |||
| No. of α-KG | 1 | 1 | |||||
| No. of HC | 1 | ||||||
| No. of Lys | 1 | ||||||
| No. of metal ion | 1 | 1 | 1 | 1 | |||
| No. of water molecules | 423 | 370 | 234 | 185 | |||
| Average B-factor (Å2) | |||||||
| Protein | 19.3 | 22.5 | 27.3 | 16.5 | |||
| α-KG | 28.3 | 56.0 | |||||
| HC | 26.5 | ||||||
| Lys | 20.4 | ||||||
| Metal ion | 21.5 | 26.5 | 37.3 | 16.4 | |||
| Water | 55.5 | 42.0 | 48.9 | 26.3 | |||
| Root mean square deviation from ideal values | |||||||
| Bond length (Å) | 0.013 | 0.013 | 0.014 | 0.012 | |||
| Bond angles (degrees) | 1.4 | 1.6 | 1.4 | 1.3 | |||
| Ramachandran plot (%) | |||||||
| Favored | 90.8 | 89.4 | 90.9 | 91.4 | |||
| Additional allowed | 8.1 | 9.2 | 7.7 | 7.6 | |||
| Generously allowed | 0.7 | 1.1 | 1.1 | 0.7 | |||
| Disallowed | 0.4 | 0.4 | 0.4 | 0.3 | |||
a Values in parentheses are for the highest resolution shell.
b Rmerge = Σ|Ii − 〈I〉|/Σ〈I〉.
c Figure of merit was calculated with the program SOLVE.
Preparation of Mutant Enzymes
The H72L mutation was introduced into the HCS-Nhis gene on pBluescript II SK(+) vector by a QuikChange kit (Stratagene) using oligonucleotide primers 5′-AAGGTGGTGACCCTCATCCAGTGCCGC-3′ and 5′-GCGGCACTGGATGAGGGTCACCACCTT-3′. The mutated gene with the desired mutation was introduced into pHIS8 as described for constructing pHIS8-TtHCS-Nhis. Protein expression and purification of the H72L mutant enzyme were performed by the method described above.
Enzyme Assay
TtHCS activity was measured at 323 K using 2,6-dichlorophenolindophenol by monitoring the decrease in absorbance at 600 nm (32). To determine the Km of wild-type TtHCS-Nhis for acetyl-CoA, the reaction mixture contained 2–400 μm acetyl-CoA, 200 μm α-KG, 5 mm MgSO4 in 1 ml of 100 mm HEPES, pH 7.5. To determine the Km of wild-type TtHCS-Nhis for α-KG, the reaction mixture contained 50 μm acetyl-CoA, 2–50 μm α-KG, 5 mm MgSO4 in 1 ml of 100 mm HEPES, pH 7.5. The reaction was initiated by the addition of 30 μg of enzyme to the reaction mixture. To determine the Km of α-KG and acetyl-CoA of the H72L mutant, the concentrations of α-KG and acetyl-CoA varied in the range of 10–500 and 5–100 μm, respectively. Inhibition by lysine was analyzed in a reaction mixture containing 50 μm acetyl-CoA, 2–100 μm α-KG, 0–100 μm lysine, 5 mm MgSO4 in 1 ml of 100 mm HEPES, pH 7.5, for wild-type TtHCS and in a reaction mixture containing 200 μm acetyl-CoA, 100–2000 μm α-KG, and 0–2000 μm lysine for TtHCS-H72L-Nhis.
To examine the effects of metal ions on the enzymatic activity of TtHCS, we first removed the bound metal ions co-purified with the enzyme by the following procedure. The purified protein was first dialyzed six times for 30 min against 20 mm Tris-HCl, pH 8.0, containing 10 mm EDTA to remove the divalent metal ions and then dialyzed for 30 min against 20 mm Tris-HCl, pH 8.0, eight times. The sample was used for the activity assay in the presence of various metal ions, as described above.
RESULTS AND DISCUSSION
Overall Structure of HCS
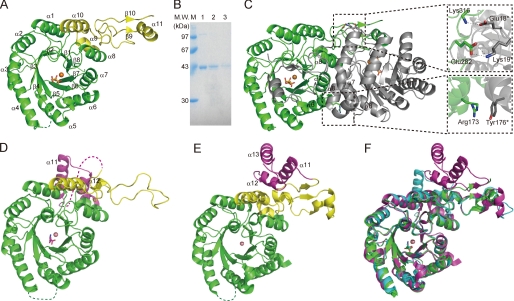
The crystal structure of TtHCS complexed with α-KG was determined by the multiple wavelength anomalous diffraction method and subsequent molecular replacement and refined to 1.80 Å (Fig. 1). This complex contains the electron density corresponding to α-KG in the catalytic pocket. In addition, in this complex, there was strong electron density near α-KG. TtHCS prepared without EDTA treatment binds divalent metal ions co-purified with TtHCS from the E. coli expression system, as described below. Although Mg2+ ion was added to the crystallization buffer and a substantial portion of initially bound metal ion was expected to be replaced by Mg2+ ion, which is biologically relevant for TtHCS, the electron density of the metal ion was too strong to be identified as Mg2+ ion. We also prepared TtHCS crystals in buffer containing α-KG, acetyl-CoA, and Mg2+ ions and determined the crystal structure at 1.96 Å resolution. The complex has electron density that matches an HC molecule, which is the product of the HCS reaction, in the catalytic pocket. In this complex, there was additional strong electron density similar to that of α-KG. The strong electron density observed in both complexes probably corresponds to that of mixtures of several metal ions, Mg2+, Cu2+, Zn2+, and Fe2+, contained in the crystallization buffer. Because Cu2+ contains more electrons than Mg2+, although Cu2+ was not the most abundant metal ion in the crystallization buffer, the electron density of the metal ion was well assigned as Cu2+ with a B-factor of 21.5 Å2, which is comparable with the average B-factor of protein in structural refinement (Table 1). Therefore, we tentatively named the complexes TtHCS·Cu2+·α-KG and TtHCS·Cu2+·HC for substrate-bound and product-bound complexes, respectively, for convenience. We also determined the crystal structure of another TtHCS complex containing α-KG and Mg2+ at 2.15 Å resolution, which was prepared using TtHCS by EDTA treatment, as described under “Experimental Procedures.” This complex was named TtHCS·Mg2+·α-KG, and its electron density for metal ion matched Mg2+ very well. For each crystal structure, an asymmetric unit contained a monomer of TtHCS, sharing a very similar structure with the root mean square deviation below 0.2 Å. We here describe the overall structure of the TtHCS·Cu2+·α-KG complex with the highest crystallographic resolution. TtHCS is composed of a (β/α)8 TIM barrel domain (from Met-1 to Gln-245), which is named after triose-phosphate isomerase, and C-terminal small domain I (from Pro-246 to Ala-320), forming an α9-α10-β9-α11-β10 fold (Fig. 1A). In the TIM barrel domain, the electron density of the loop region (Ser-98 to His-105) between β4 and α4 was not observed, indicating that this region is disordered in the crystal. Although some electron density was observed for the C-terminal small domain II (from Ser-321 to Ala-376), which is expected to form a three-helix bundle by homology with small domain II in the linker domain of MtIPMS and by secondary structure prediction with the PSIPRED server (33), we could not build a model of this region due to very weak density. When crystals of TtHCS·Cu2+·α-KG and TtHCS·Cu2+·HC were dissolved and subjected to SDS-PAGE, protein bands corresponding to the original size of full-length TtHCS were observed (Fig. 1B); therefore, the weak electron density suggests that this small domain has high mobility in the crystal.
FIGURE 1.
Structures of TtHCS. A, monomer structure of the TtHCS·Cu2+·α-KG complex. The (β/α)8 TIM barrel domain and C-terminal small domain I are in green and yellow, respectively. Metal Cu2+ is shown as orange spheres and α-KG is shown as orange sticks. The disordered region (Ser-98 to His-105) is shown as a dotted line. B, SDS-PAGE of TtHCS proteins dissolved from crystals. A single crystal was dissolved in 20 mm Tris-HCl, pH 8.0, and applied to 15% SDS-PAGE. Lane 1, freshly purified TtHCS-Chis; lane 2, TtHCS·Cu2+·α-KG dissolved from a crystal; lane 3, TtHCS·Cu2+·HC dissolved from a crystal. C, dimer structure of TtHCS·Cu2+·α-KG complex. Bound Cu2+ ion is shown as orange spheres, and α-KG is shown as sticks. One monomer is shown in green, and the adjacent subunit is shown in gray. Intersubunit hydrogen bonds are shown in enlarged insets. D, monomer structure of the TtHCS·Co2+·Lys complex. The (β/α)8 TIM barrel domain, C-terminal small domain I, and C-terminal small domain II are shown in green, yellow, and purple, respectively. Metal Co2+ is shown as pink spheres, and bound lysine is shown as magenta sticks. The disordered region (Leu-317 to Leu-336) is shown as a dotted line. E, monomer structure of the SpHCS·Zn2+complex. Domain colors are the same as in D. Metal Zn2+ is shown as pink spheres. F, superposition of the structure of the TtHCS·Cu2+·α-KG complex (green) with those of the TtHCS·Co2+·Lys complex (cyan) and the SpHCS·Zn2+ complex (magenta). α-KG and Lys are shown as green and cyan sticks, respectively.
In these crystal structures of TtHCS, two TtHCS molecules related by 2-fold crystallographic symmetry form a dimer (Fig. 1C). The surface area (3,508 Å2) buried by dimerization is large (21.6%) compared with the total surface area of the monomer (16,240 Å2), indicating that the structural unit of TtHCS is a dimer. Three helices, α6, α7, and α8, mainly contribute to the stabilization of dimer formation. The α8 helix interacts with β7-α7 and β6-α6 loops of the TIM barrel domain from another subunit, and the α7 helix interacts with the α7 helix in the TIM barrel domain from another subunit. Arg-173 on the α6 helix from one subunit interacts with Tyr-176* on the α6 helix from another subunit by π-cation interaction (asterisks denote residues from another subunit). The C-terminal small domain I of one monomer extends to cover the active site of another monomer. Glu-282 in the α10-β9 loop and Lys-316 in the α11-β10 loop from C-terminal small domain I form ion pairs with Lys-19* and Glu-18* in the β1-α1 loop of the TIM barrel domain from another subunit, respectively (Fig. 1C). These ion bonds between chains might be a factor elevating thermotolerance over 70 °C of TtHCS.
A DALI search (33) was performed to find proteins with a similar structure. HCS from Schizosaccharomyces pombe (SpHCS), whose crystal structure was determined most recently (34), showed the highest similarity (Z score = 42.9, root mean square deviation = 1.9 Å, Protein Data Bank code 3IVS). The TIM barrel domain structure of TtHCS is also similar to those of LiCMS and MtIPMS with root mean square deviation of 1.9 and 2.0 Å for 267 and 202 Cα atom pairs, respectively.
The crystal structure of TtHCS complexed with lysine was determined at 1.80 Å resolution by molecular replacement using the TtHCS·Cu2+·α-KG structure as the template. The structure contains both the TIM barrel domain and C-terminal small domains I and II (Fig. 1D). Small domain II is predicted to form a three-helix bundle structure. Although the structure lacks residues 317–336 in small domain II, putatively forming the first α-helix of the three-helix bundle, another portion, Gly-337 to Ala-376, forms two α-helices, as expected, covering the active site on another TIM barrel domain. The overall structure of the TIM barrel domain of the lysine-bound complex is similar to those of other complexes determined in this study. In C-terminal small domain I, significant conformational change is observed. In the TtHCS·Cu2+·α-KG complex, C-terminal small domain I forms an α9-α10-β9-α11-β10 fold structure, whereas these secondary structures were broken in the lysine complex (Fig. 1, A and D). In this complex, the electron density of the metal ion at the position equivalent to those of Cu2+ and Mg2+ was at the center of the TIM barrel domain in other complexes (Fig. 2D). The electron density of the metal ion was well assigned as Co2+ with a B-factor of 16.4 Å2, which is comparable with the average B-factor (16.5 Å2) of protein in structural refinement. We assume that Co2+ was incorporated into the active site because the crystallization buffer contained Co2+. The lysine-bound complex was named TtHCS·Co2+·Lys complex.
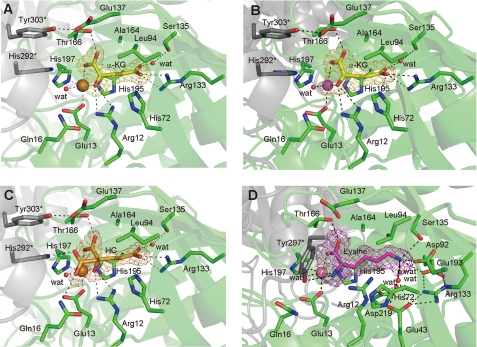
FIGURE 2.
α-KG·HC·lysine-binding residues of TtHCS. A, TtHCS·Cu2+·α-KG complex; B, TtHCS·Mg2+·α-KG complex; C, TtHCS·Cu2+·HC complex; D, TtHCS·Co2+·Lys complex. Residues interacting with ligands are shown as sticks. Bound Cu2+, Mg2+, and Co2+ ions are shown as orange, purple, and pink spheres, respectively. α-KG, HC, and Lys are shown as yellow, orange, and purple sticks, respectively. Water molecules are shown as red spheres. Hydrogen bonds are shown as dashed lines. Fo − Fc omit electron density maps for α-KG, HC, and Lys are shown as yellow, orange, and purple mesh, respectively, and contoured at 3.0 σ, 3.0 σ, 3.6 σ, and 3.5 σ, for A–D, respectively. Residues from the adjacent subunit are indicated by asterisks.
Bound Metal Ion
In TtHCS·Cu2+·α-KG and TtHCS·Cu2+·HC complexes, the electron density of the metal ion was at the center of the TIM barrel domain. To identify the bound metal ion, we performed ICP analysis of the purified enzymes. The results showed that purified protein contains Cu2+, Fe2+, and Zn2+ at 0.25, 0.12, and 0.04 mol/mol, respectively. These metal ions could be removed to the background level by treatment of the purified enzyme with EDTA. We analyzed metal ion dependence for the reaction of TtHCS after EDTA treatment. In the absence of metal ions, TtHCS exhibited only negligible activity. On the other hand, the addition of Mg2+, Mn2+, and Co2+ to the reaction buffer significantly restored TtHCS activity (Table 2). Thus, we assume that Mg2+ or Mn2+ ion is a biologically relevant divalent cation. Both metal ions (Cu2+ and Mg2+) are bound to TtHCS with six ligands in similar octahedral geometry (Fig. 2, A and B), coordinated by Oϵ1 of Glu-13 (2.2 and 2.4 Å bond length for HCS·Cu2+·α-KG and HCS·Mg2+·α-KG complexes, respectively), Nϵ2 of His-197 (2.1 and 2.4 Å), C1-carboxyl group (2.3 and 2.4 Å) and C2-oxo group (2.0 and 1.9 Å) of α-KG in the equatorial position, and Nϵ2 of His-195 (2.2 and 2.3 Å) and a water molecule (2.3 and 2.1 Å) in the axial position. A metal atom is also bound in octahedral geometry in the HCS·Cu2+·HC complex, which is mostly similar to that of the HCS·Cu2+·α-KG complex (Fig. 2, A and C).
TABLE 2.
Effect of metal ion on enzymatic activity
| Additive | Specific activity |
|---|---|
| milliunits/mg protein | |
| None | 0.0 ± 0.0 |
| MgSO4 | 41.5 ± 1.4 |
| MnCl2 | 39.2 ± 3.8 |
| CoCl2 | 33.3 ± 1.9 |
| CaCl2 | 7.5 ± 1.6 |
| NiCl2 | 3.6 ± 0.6 |
| ZnSO4 | 1.3 ± 0.2 |
| CuCl2 | 0.3 ± 0.0 |
| FeSO4 | 0.8 ± 0.2 |
In the TtHCS·Co2+·Lys complex, Co2+ was bound with a coordination similar to those of Cu2+ and Mg2+ in other complexes. Co2+ is coordinated by the α-carboxyl group (2.2 Å) and α-amino group (2.1 Å) of bound lysine in place of the C1-carboxyl group and the C2-keto group of α-KG in other complexes, respectively.
Determinants of the Substrate Specificity of TtHCS by Comparison of Structures between TtHCS and Homologous Enzymes
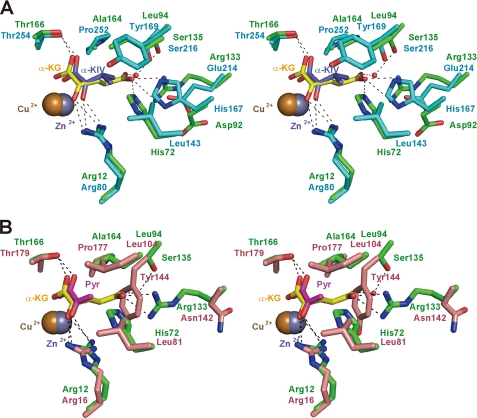
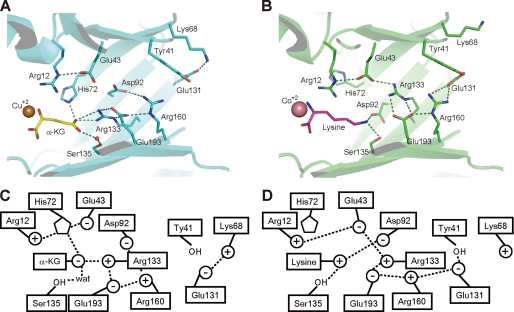
Although TtHCS has low sequence similarity to MtIPMS (22%) and LiCMS (25%), the residues forming the active site are present at similar positions in the three-dimensional structures of these enzymes, sharing similar conformations (Fig. 3). This observation indicates that these enzymes catalyze the aldol-type condensation reaction by a common mechanism. Whereas α-KG, the substrate for HCS, contains the carboxyl group at the C5 position, α-KIV and pyruvate, the substrates for IPMS and CMS, respectively, have hydrophobic moieties. According to the characteristics of the substrates, the mechanism to recognize the distal portion of the substrate differs among TtHCS, MtIPMS, and LiCMS. MtIPMS recognizes the isopropyl chain of α-KIV by the hydrophobic wall formed by Leu-143, His-167, Tyr-169, and Pro-252 (Fig. 3A). LiCMS recognizes the C3 methyl group of pyruvate by the more projected hydrophobic wall formed by Leu-81, Leu-104, Tyr-144, and Pro-177 (Fig. 3B). In the TtHCS·Cu2+·α-KG complex, α-KG is bound to the enzyme by specific interactions with several residues (Figs. 2A and 3, A and B). The C1-carboxyl group forms a hydrogen bond with Thr-166. The C2-keto group is recognized by the guanidium group of Arg-12. The hydrophobic portion, C3-C4 atoms, of the substrate makes van der Waals contacts with the side chains of Ala-164 and Leu-94. The C5-carboxyl group is stabilized by hydrogen bonds with Nϵ2 (2.7 Å) of His-72 and with Nη1 (2.8 Å) and Nη2 (3.3 Å) of Arg-133. Furthermore, the C5-carboxyl group makes a water molecule-mediated hydrogen bond with Oγ of Ser-135 (distances between the C5-carboxyl group and water and between water and Oγ of Ser-135 are 2.6 and 3.3 Å, respectively). Thus, the well designed recognition of α-KG assures that TtHCS recognizes α-KG with high affinity (11).
FIGURE 3.
Superposition of substrate-binding sites of TtHCS and related enzymes. A, TtHCS·Cu2+·α-KG complex (green) and MtIPMS·Zn2+·α-KIV complex (cyan); B, TtHCS·Cu2+·α-KG complex (green) and LiCMS·Zn2+·pyruvate (Pyr) complex (pink). Bound Cu2+ and Zn2+ ions are shown as orange and gray spheres, respectively. α-KG, α-KIV, and pyruvate are shown as yellow, dark blue, and magenta sticks, respectively. Hydrogen bonds between ligands and proteins are shown as dashed lines.
Catalytic Mechanism of TtHCS
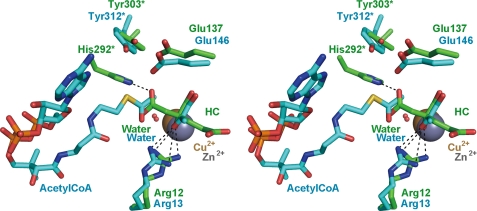
In the HC complex, HC, which is probably synthesized from α-KG and acetyl-CoA added to crystallization droplets by the HCS reaction, is bound to the active site of TtHCS in a manner similar to that of α-KG in the TtHCS·Cu2+·α-KG complex (Fig. 2C). The C1-carboxyl group, which is transferred from acetyl-CoA, forms hydrogen bonds with Nϵ of Gln-16 and Nϵ2 of His-292* from small domain I of another subunit. Interestingly, when the active sites are superimposed between TtHCS·Cu2+·HC and LiCMS·Zn2+·pyruvate·acetyl-CoA complexes, the C1-carboxyl group of HC is located close to the distal methyl group of acetyl-CoA (Fig. 4). The catalytic mechanism of HCS has been analyzed for HCS from S. cerevisiae (35). After α-KG and acetyl-CoA are bound, the catalytic reaction proceeds as follows. A general base abstracts a proton from the reactive methyl group of acetyl-CoA for enolization of the acetyl group, and a general acid stabilizes the enolated form of acetyl-CoA, which nucleophilically attacks the C2 atom of α-KG to form homocitryl-CoA. In the final step, a metal-bound water hydrolyzes homocitryl-CoA to yield homocitrate and CoA, aided by the general acid acting as a base. Through site-directed mutagenesis of HCS from S. cerevisiae, Qian et al. (35) proposed that the catalytic dyad of Glu-155 and His-309* corresponding to Glu-137 and His-292* in TtHCS, respectively, abstracts a proton from the methyl group of acetyl-CoA, where His-309* is responsible for deprotonation of the methyl group of acetyl-CoA, and Glu-155 stabilizes the protonated form of His-309* by electrostatic interaction. The conserved Arg-12 acts as a general acid in the catalytic mechanism. The present TtHCS·Cu2+·HC structure indicates that His-292*, which interacts with the C1-carboxyl group of HC, is the most probable candidate to aid the hydrolysis of homocitryl-CoA by acting as a base. The putative location of the C1-carboxyl group of HC close to the reactive methyl group of acetyl-CoA may support the hypothesis that His-292* also acts as a general base for proton abstraction from the reactive methyl group of acetyl-CoA. The crystal structure of TtHCS complexed with HC for the first time revealed direct structural evidence for the involvement of His-292* in the catalytic mechanism in this protein family. In the structures determined in this study, Glu-137 resides somewhat apart from His-292* (4.6 Å from Oϵ2 of Glu-137 to Nϵ2 of His-292*). Glu-137 forms a hydrogen bond with Tyr-303*. Because Tyr-303* stabilizes His-292* orientation by π-π stacking, we assume that Glu-137 plays a role in the catalytic mechanism indirectly by placing the active site residue His-292* in the correct orientation in TtHCS.
FIGURE 4.
Superposition of catalytic site between TtHCS·Cu2+·HC complex and LiCMS·Zn2+·pyruvate·acetyl- CoA complex. The TtHCS·Cu2+·HC complex and the LiCMS·Zn2+·pyruvate·acetyl-CoA complex are shown as green and cyan sticks, respectively. Hydrogen bonds between ligand and proteins are shown as dashed lines. His-302* of LiCMS, corresponding to His-292* of TtHCS, is not seen due to disorder of the regions containing this residue in the LiCMS structure.
Most recently, the crystal structure of SpHCS in apo form and complexes with α-KG were determined (34). The structures of SpHCS are very similar to those determined in this study. However, His-321*, corresponding to His-292* in TtHCS, is located 13 Å apart from Glu-167 (Glu-137 in TtHCS). On this point, authors argued the involvement of this residue in the acid-base mechanism by constructing a mutant of this residue and speculated the possible conformational change by acetyl-CoA binding. In this study, the TtHCS structure for the first time provided evidence of an interaction between His-292* and the acetyl moiety. Therefore, it is likely that the structures determined in this study are the highly catalytically relevant form of HCS.
Acetyl-CoA Recognition
The crystal structure of LiCMS was determined as a complex with acetyl-CoA and pyruvate (38). Acetyl-CoA is bound to LiCMS in a U-shaped conformation. The acetyl group of acetyl-CoA is positioned near pyruvate with the carbonyl oxygen of the acetyl group hydrogen-bonded with the side chain Nη2 of Arg-16 and the methyl group sandwiched between the side chain of Oϵ2 of Glu-146 and the C2 atom of pyruvate. Arg-16 and Glu-146 are conserved as Arg-12 and Glu-137 in TtHCS. Thus, it can be expected that TtHCS recognizes acetyl-CoA in a similar manner. In LiCMS, the cysteamine moiety of acetyl-CoA interacts with Gln-20 and Glu-239, and the carbonyl group of the β-alanine moiety of acetyl-CoA is recognized by Ser-51. These residues are conserved as Gln-16, Glu-227, and Thr-46 in TtHCS (supplemental Fig. 1A). The dimethyl group of pantheteinyl moiety is stabilized by hydrophobic interaction with Val-54 and Ala-299* in LiCMS, which are conserved as Ala-49 and Ala-289* in TtHCS. Gly-300* at the N-terminal helix dipole of α11 makes contact with the pyrophosphate group of acetyl-CoA. The residue is also conserved in TtHCS as Gly-290*. In contrast to the strong conservation of amino acid residues for recognition of the diphosphopanthetheinyl moiety of acetyl-CoA, residues recognizing the adenine ring and 3′-phosphoribose are not conserved in TtHCS. In LiCMS, the adenine moiety of acetyl-CoA is stabilized through π-π stacking interaction with Phe-83 of β3. The corresponding residue is replaced by Leu-77 in TtHCS. Arg-53, which electrostatically recognizes the 3′-phosphate group of the adenine ribose in LiCMS, is replaced by Val-48 in TtHCS. In addition, comparison of the protein surface around the acetyl-CoA binding site shows that the cavity recognizing the adenosine moiety has a different shape in TtHCS (supplemental Fig. 1, B and C). These observations may suggest that adenine and 3′-phosphoribose moieties are recognized in a different way in TtHCS.
Mechanism of Feedback Inhibition by Lysine
Despite the difference in the structure between lysine and α-KG, in the TtHCS·Co2+·lysine complex a lysine molecule occupies the active site in a manner similar to that of α-KG in the TtHCS·Cu2+·α-KG complex. The α-carboxyl and α-amino groups of bound lysine form hydrogen bonds with Thr-166 and Tyr-297*, respectively. The aliphatic side chain of lysine is stabilized by van der Waals contact with Ala-164 and Leu-94. The ϵ-amino group of bound lysine is stabilized by electrostatic interaction with Asp-92 and Glu-193 and by a hydrogen bond with Ser-135. The ϵ-amino group of bound lysine is further stabilized by Glu-43 and Asp-219 via bridging water molecules. Lysine binding to the active center is accompanied by displacement of a set of amino acid residues surrounding the C5-carboxyl group of α-KG (Figs. 2, A and D, and 5). The most significant change is found in the side chain rotation of Arg-133, Asp-92, and His-72 (Fig. 5). Arg-133, which forms an ion pair with α-KG in the TtHCS·Cu2+·α-KG complex, moves away from the active site while maintaining the electrostatic interaction with Glu-193 and forms a new ion pair with Glu-43. Asp-92, which recognizes Arg-160 in the TtHCS·Cu2+·α-KG complex, moves into the active site and interacts with bound lysine. Upon lysine binding, His-72, which forms a hydrogen bond with α-KG, changes its side chain orientation to stack with Arg-12. In the inhibitory complex, Arg-12, which also interacts with Glu-43, makes a van der Waals contact with the side chain of bound lysine. Conformational changes are also found in the opposite entrance side of the TIM barrel structure. Glu-131, which recognizes Lys-68 in the TtHCS·Cu2+·α-KG complex, rotates and forms a hydrogen bond with Tyr-41 and electrostatic interaction with Arg-160. These residues in the TIM barrel are all well conserved among HCSs (supplemental Fig. 2), suggesting that concerted conformational change upon binding the substrate/inhibitor is a common mechanism of inhibition in HCS.
FIGURE 5.
Comparison of recognition by TtHCS between α-KG and lysine. A, recognition of C5-carboxyl group of α-KG; B, recognition of ϵ-amino group of Lys; C and D, schematic drawing of recognition of α-KG and Lys, respectively.
To investigate the conformational change upon the transition between active and inhibitory forms, we compared overall structures between TtHCS·Cu2+·α-KG, TtHCS·Co2+·Lys, and SpHCS·Zn2+ complexes as an active form, an inhibitory form, and an apo form, respectively. The comparison suggests that the TIM barrel domain does not show gross conformational change (Fig. 1, A, E, and F). In addition, the structure of the C-terminal small domain I of the TtHCS·Cu2+·α-KG complex is similar to that of the corresponding domain of SpHCS. In the TtHCS·Co2+·lysine complex, two of three α-helices in small domain II are seen. When this structure is compared with that of SpHCS in the closed form, α11 and α12 of small domain II, which correspond to α12 and α13 in SpHCS, are displaced in the lysine complex. In the SpHCS·Zn2+ complex, Asp-394 on α13 electrostatically interacts with Arg-397 on the α13 helix from another subunit. In the TtHCS·Co2+·Lys complex, the interactions are lost by gross displacement; α11 helix is displaced by 44 Å with 308° rotation. On the other hand, His-292* and Tyr-303*, which stacks with His-292* to keep it in the correct orientation, move 13 Å away from the active site, and instead Tyr-297* occupies the vacant space to form a hydrogen bond with bound lysine in the TtHCS·Co2+·Lys complex (Fig. 2D). His-321*, which corresponds to His-292* in TtHCS, is also found to be present 13 Å apart from the catalytic site in SpHCS. Therefore, the large displacement of His-292* found in the lysine-bound complex may not be restricted to feedback inhibition.
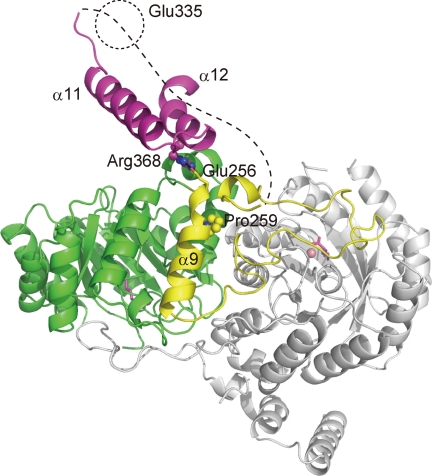
As described above, substantial differences in structure are found in the C-terminal domains. Feedback-resistant HCSs (Lys20p and Lys21p) from Saccharomyces cerevisiae (ScHCS), which share the same domain structure with TtHCS, were obtained by random mutagenesis (36). Arg-270 and Ser-386 are replaced with Lys and Phe, respectively, in ScHCSLys20p, whereas Gln-366 is replaced with Arg in ScHCSLys21p. Arg-270 and Ser-386 in ScHCSLys20p and Gln-366 in ScHCSLysS21p correspond to Pro-259, Arg-368, and Glu-335, respectively, in TtHCS. When these residues are mapped on the structure of the TtHCS·Co2+·Lys complex (Fig. 6), Arg-368 is on the loop between the α11 and α12 helices of the C-terminal small domain II, forming an ionic pair with Glu-256 on α9 from the small domain I, on which Pro-259 is located. Glu-335 could not be mapped on the determined structure because this residue is expected to be located on the first α-helix of the three-helix bundle, which was not modeled in this study. This residue is near Gly-337 at the N-terminal end of α11 of small domain II. Information on mutation mapping and the substantial displacement of the C-terminal domains upon lysine binding suggest that inhibitory complex formation is accompanied by rearrangement of the interaction between the TIM barrel domain and small domain II.
FIGURE 6.
Mapping of mutations conferring feedback resistance to ScHCSs on structure of TtHCS·Co2+·Lys complex. A dimer structure of the TtHCS·Co2+·Lys complex is shown. In a monomer, (β/α)8 TIM barrel domain, C-terminal small domain I, and C-terminal small domain II are shown in green, yellow, and purple, respectively. The adjacent subunit is in gray. Metal Co2+ is shown as pink spheres, and bound lysine is shown as magenta sticks.
In contrast to TtHCS, the feedback inhibitor, leucine, is bound at the regulatory domain in MtIPMS, where the regulatory domain is fused via the linker domain to the C terminus of the catalytic domain. The same mechanism is expected for CMS because the enzyme also possesses the C-terminal extension that could serve as a regulatory domain. Most recently, it was suggested that interdomain communication between the regulatory domain and catalytic domain is important for allosteric regulation in MtIPMS (37) and LiCMS (38), although it is unclear how the effector causes structural change at the catalytic site in both cases. Interdomain communication between the regulatory domain and catalytic domain might be important for allosteric regulation in this protein family.
Andi et al. (39) have reported the kinetic mechanism of feedback inhibition of ScHCS, showing that inhibition by lysine was competitive with α-KG, similar to TtHCS. Straightforward interpretation of the results, including ours, shows that lysine binds to the active site but not to the allosteric site. However, through further enzymatic characterization, they argued that the competition between lysine and α-KG arose from the two ligands binding with high affinity to two different conformations, with lysine stabilizing the less active form and α-KG stabilizing the more active form. In this study, the crystal structure of the inhibitory complex binding lysine definitively indicates that lysine is actually bound to the active site for inhibition, with local rearrangement of interacting residues. Currently, we cannot find any reasons for the contradictory interpretation, based on the crystal structure of the lysine complex. Considering the difference in sensitivity to lysine of the HCSs, sensitivity to lysine of TtHCS is higher than that of ScHCS by 1–2 orders of magnitude. We can therefore assume different inhibitory mechanisms between TtHCS and ScHCS, although both HCSs exhibit apparent competitive inhibition by lysine with α-KG, and, as described above, all amino acid residues responsible for lysine recognition are conserved in lysine-sensitive HCSs, including ScHCS.
Identification of His-72 as a Residue Involved in Feedback Inhibition by Lysine
Feedback inhibition by lysine in a competitive manner with α-KG (11, 39–41) and the present crystal structure of the TtHCS·Co2+·Lys complex indicate that lysine competes with α-KG for the overlapping sites in TtHCS. On the other hand, NifV, which is also the homocitrate synthase required to synthesize HC as the organic constituent of the Fe-Mo cofactor of nitrogenase in N2-fixing bacteria, is not regulated by lysine (42), although lysine-sensitive and -insensitive HCSs share amino acid sequence identity of about 25%. Comparison of the amino acid sequence between TtHCS and homologous enzymes indicates that His-72 is conserved only among lysine-sensitive HCSs, suggesting that this residue is involved in feedback inhibition by lysine in TtHCS; the residue is replaced with Trp in lysine-insensitive HCS (NifV) and hydrophobic residues in IPMS and CMS (supplemental Fig. 2). The crystal structures determined in this study reveal that His-72 is located in the substrate-binding pocket and interacts with the C5-carboxyl group of α-KG in the TtHCS·Cu2+·α-KG complex, whereas the same residue changes its side chain orientation to stabilize Arg-12 to stack bound lysine in the TtHCS·Co2+·Lys complex. These observations suggest that His-72 serves as a structural switch determining which form, active or inactive, should be taken, depending on the availability of α-KG or lysine. We therefore constructed mutant H72L and examined the kinetic properties, including sensitivity to lysine. The kinetic parameters of the H72L mutant are comparable with those of the wild-type enzyme, except that the Km value of α-KG is 32-fold larger than that of the wild-type enzyme (Table 3). The increased Km value of α-KG is expected because His-72 is also involved in binding the C5-carboxyl group of α-KG (Figs. 2 and 3). The wild-type enzyme is inhibited by lysine with a Ki value of 70.7 μm in a competitive manner with α-KG. In contrast, the H72L mutant exhibited a significantly increased Ki value, larger than 2,000 μm for lysine. These results indicate that His-72 functions not only as a residue recognizing the C5-carboxyl group of α-KG but also as a switch controlling the activity. The H72L mutation may sterically hinder Arg-12 from stacking the bound lysine.
TABLE 3.
Kinetic parameters of HCS and H72L mutant
| Enzyme |
Km |
kcat | Ki for lysine | |
|---|---|---|---|---|
| α-KG | Acetyl-CoA | |||
| μm | min−1 | μm | ||
| HCSa | 5.4 ± 0.7 | 10.9 ± 0.4b | 70.7 ± 6.1 | |
| 33 ± 5.3 | 41 ± 4 | |||
| H72La | 174 ± 13 | 14.9 ± 0.5b | >2000c | |
| 37 ± 4.2 | 31 ± 1 | |||
a Recombinant enzymes with N-terminal His8 tags were used.
b kcat for α-KG is lower than that for acetyl-CoA because HCS and the mutants are significantly subject to substrate inhibition by acetyl-CoA.
c The H72L mutant was significantly desensitized to feedback inhibition, and 50% inhibition was not achieved even in the presence of 2,000 μm lysine.
Supplementary Material
Acknowledgments
We are grateful to the staff of the Photon Factory for assistance with data collection. We thank Dr. Shinya Fushinobu for advice on structure determination.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and grants from the Nagase Science and Technology Foundation, from the Asahi Glass Foundation, and from the Charitable Trust Araki Medical and Biochemistry Memorial Research Promotion Fund. This work was performed with approval from the Photon Factory Program Advisory Committee (Proposals 2007G530 and 2009G540).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (codes 2ZTJ, 2ZTK, 2ZYF, and 3A9I) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- α-KG
- α-ketoglutarate
- α-KIV
- α-ketoisovalerate
- HCS
- homocitrate synthase
- HC
- homocitrate
- IPMS
- α-isopropylmalate synthase
- CMS
- citramalate synthase
- TtHCS
- T. thermophilus HCS
- MtIPMS
- M. tuberculosis IPMS
- LiCMS
- L. interrogans CMS
- SpHCS
- S. pombe HCS
- ScHCS
- S. cerevisiae HCS
- MES
- 2-morpholinoethanesulfonic acid
- SeMet
- selenomethionine.
REFERENCES
- 1.Broquist H. P. (1971) Methods Enzymol. 17, 112–129 [Google Scholar]
- 2.Vogel H. J. (1964) Am. Nat. 98, 435–446 [Google Scholar]
- 3.Chatterjee S. P., Singh B. K., Gilvarg C. (1994) Plant Mol. Biol. 26, 285–290 [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee J. K. (1985) Crit. Rev. Microbiol. 12, 131–151 [DOI] [PubMed] [Google Scholar]
- 5.Cox R. J. (1996) Nat. Prod. Rep. 13, 29–43 [DOI] [PubMed] [Google Scholar]
- 6.Xu H., Andi B., Qian J., West A. H., Cook P. F. (2006) Cell Biochem. Biophys. 46, 43–64 [DOI] [PubMed] [Google Scholar]
- 7.Kobashi N., Nishiyama M., Tanokura M. (1999) J. Bacteriol. 181, 1713–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida H., Nishiyama M., Kobashi N., Kosuge T., Hoshino T., Yamane H. (1999) Genome Res. 9, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 9.Miyazaki J., Kobashi N., Nishiyama M., Yamane H. (2001) J. Bacteriol. 183, 5067–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki J., Kobashi N., Fujii T., Nishiyama M., Yamane H. (2002) FEBS Lett. 512, 269–274 [DOI] [PubMed] [Google Scholar]
- 11.Wulandari A. P., Miyazaki J., Kobashi N., Nishiyama M., Hoshino T., Yamane H. (2002) FEBS Lett. 522, 35–40 [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki J., Kobashi N., Nishiyama M., Yamane H. (2003) J. Biol. Chem. 278, 1864–1871 [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki T., Miyazaki J., Yamane H., Nishiyama M. (2004) Microbiology 150, 2327–2334 [DOI] [PubMed] [Google Scholar]
- 14.Jia Y., Tomita T., Yamauchi K., Nishiyama M., Palmer D. R. (2006) Biochem. J. 396, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie A., Tomita T., Saiki A., Kono H., Taka H., Mineki R., Fujimura T., Nishiyama C., Kuzuyama T., Nishiyama M. (2009) Nat. Chem. Biol. 5, 673–679 [DOI] [PubMed] [Google Scholar]
- 16.Tsubouchi T., Mineki R., Taka H., Kaga N., Murayama K., Nishiyama C., Yamane H., Kuzuyama T., Nishiyama M. (2005) J. Biol. Chem. 280, 18511–18516 [DOI] [PubMed] [Google Scholar]
- 17.de Carvalho L. P., Blanchard J. S. (2006) Biochemistry 45, 8988–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Carvalho L. P., Argyrou A., Blanchard J. S. (2005) J. Am. Chem. Soc. 127, 10004–10005 [DOI] [PubMed] [Google Scholar]
- 19.Koon N., Squire C. J., Baker E. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalieri D., Casalone E., Bendoni B., Fia G., Polsinelli M., Barberio C. (1999) Mol. Gen. Genet. 261, 152–160 [DOI] [PubMed] [Google Scholar]
- 21.Ma J., Zhang P., Zhang Z., Zha M., Xu H., Zhao G., Ding J. (2008) Biochem. J. 415, 45–56 [DOI] [PubMed] [Google Scholar]
- 22.Jez J. M., Ferrer J. L., Bowman M. E., Dixon R. A., Noel J. P. (2000) Biochemistry 39, 890–902 [DOI] [PubMed] [Google Scholar]
- 23.Leahy D. J., Hendrickson W. A., Aukhil I., Erickson H. P. (1992) Science 258, 987–991 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1937–1940 [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 28.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 32.Hadler H. I., Erwin M. J. (1963) Biochemistry 2, 954–957 [DOI] [PubMed] [Google Scholar]
- 33.Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulfer S. L., Scott E. M., Couture J. F., Pillus L., Trievel R. C. (2009) J. Biol. Chem. 284, 35769–35780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian J., Khandogin J., West A. H., Cook P. F. (2008) Biochemistry 47, 6851–6858 [DOI] [PubMed] [Google Scholar]
- 36.Feller A., Ramos F., Piérard A., Dubois E. (1999) Eur. J. Biochem. 261, 163–170 [DOI] [PubMed] [Google Scholar]
- 37.de Carvalho L. P., Frantom P. A., Argyrou A., Blanchard J. S. (2009) Biochemistry 48, 1996–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P., Ma J., Zhang Z., Zha M., Xu H., Zhao G., Ding J. (2009) Biochem. J. 421, 133–143 [DOI] [PubMed] [Google Scholar]
- 39.Andi B., West A. H., Cook P. F. (2005) J. Biol. Chem. 280, 31624–31632 [DOI] [PubMed] [Google Scholar]
- 40.Luengo J. M., Revilla G., López M. J., Villanueva J. R., Martín J. F. (1980) J. Bacteriol. 144, 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andi B., Cook P. F. (2005) J. Biol. Chem. 280, 31633–31640 [DOI] [PubMed] [Google Scholar]
- 42.McLean P. A., Dixon R. A. (1981) Nature 292, 655–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.