Abstract
Hsp90 is an ATP-dependent molecular chaperone. The best characterized inhibitors of Hsp90 target its ATP binding pocket, causing nonselective degradation of Hsp90 client proteins. Here, we show that the small molecule celastrol inhibits the Hsp90 chaperoning machinery by inactivating the co-chaperone p23, resulting in a more selective destabilization of steroid receptors compared with kinase clients. Our in vitro and in vivo results demonstrate that celastrol disrupts p23 function by altering its three-dimensional structure, leading to rapid formation of amyloid-like fibrils. This study reveals a unique inhibition mechanism of p23 by a small molecule that could be exploited in the dissection of protein fibrillization processes as well as in the therapeutics of steroid receptor-dependent diseases.
Keywords: Amyloid, Cancer Therapy, Chaperone/Chaperonin, Nuclear Receptors, Protein Folding, Celastrol, Hsp90, Kinases, Fibrils, p23
Introduction
The combination of constitutive chaperones and heat- or other stress-induced proteins modulates the protein folding environment of the cell and links stress signaling processes with protein homeostasis (1). It is thus not surprising that a number of pathological conditions such as neurodegenerative and cardiovascular diseases as well as cancer are associated with an abundance of mutated and misfolded proteins that overwhelm or deregulate chaperone activity (2, 3). The ubiquitous molecular chaperone Hsp90 is a potential target for the treatment of cancer because of its central role in chaperoning client proteins whose dysregulations contribute to malignant phenotypes (3–5). Hsp90, together with co-chaperones such as p23 and Cdc37, mediates the proper folding of these client proteins. Most of the small molecule inhibitors of Hsp90 developed to date target the ATP-binding site of Hsp90 (6, 7). These inhibitors such as geldanamycin and its derivative 17-allylamino-17-demethoxygeldanamycin compete with ATP for binding to Hsp90 (8, 9), leading to the ubiquitination and proteasomal degradation of the client proteins (6, 10, 11).
Chemical genomic approaches identified celastrol as another compound that targets the Hsp90 chaperoning machinery (12). Celastrol does not compete with 17-allylamino-17-demethoxygeldanamycin for the ATP binding pocket of Hsp90, and the two drugs are mildly synergistic in inhibiting androgen receptor signaling (12). Celastrol induces the death of melanoma (13), prostate (14), and pancreatic cancer cells and inhibits growth of glioma xenografts (15) and prostate cancer growth in nude mice (14, 16). Recent work has also introduced celastrol as a proteostasis regulator to treat the loss-of-function lysosomal storage diseases through activation of the unfolded protein response (17). At the molecular level, celastrol induces a heat-shock response by activating the transcription factor Hsf1 (18) and antioxidant proteins in yeast and mammalian cells (19). Celastrol also modulates the inducible nitric-oxide synthase (20), topoisomerase II (21), NF-κB pathway (20, 22, 23), potassium channels (24), and the proteasome activity (16).
Because celastrol treatment of cells resulted in destabilization of several Hsp90 client proteins, initial reports concluded that celastrol targets Hsp90 directly (12, 14). Molecular docking and immunoprecipitation data later showed that celastrol binds the N-terminal domain of Hsp90, interfering with the Hsp90-Cdc37 interaction (12, 14). A more recent report using NMR spectroscopy and chemical and mutational analysis, however, clearly demonstrated that celastrol reacts with the cysteines of Cdc37, inducing Cdc37 aggregation and inhibiting Hsp90-Cdc37 interaction (25). It was suggested in a previous publication (14) that celastrol does not interfere with Hsp90 binding to p23, a finding that is at odds with data from another study (12) and our data in this report. Therefore, the mechanism by which celastrol inhibits the Hsp90 machinery remains inconclusive. To clarify the effect of celastrol on the Hsp90 machinery, we used a reconstituted chaperone/co-chaperone in vitro system that we developed for proper chaperoning of the progesterone receptor (PR),2a physiological client of Hsp90. This comprehensive assay uses five core components of the Hsp90 machinery, Hsp90, Hsp70, Hsp40, Hop, and p23, for chaperoning PR (26). Noticeably, PR chaperoning does not require the co-chaperone Cdc37 necessary for ChK1 kinase reconstitution (27, 28). For in vivo studies, we used HeLa cells expressing PRB and the human breast cancer cell line Hs578T.
We found that celastrol inhibits the Hsp90 chaperoning of PR in vitro. Treatment of cells with celastrol induces the degradation of PR at low doses and, in agreement with other reports, the degradation of several kinase clients of Hsp90 at higher doses or longer treatment. Celastrol affected Hsp90 and Hsp70 activity in our in vitro system. However, the co-chaperone p23 seems to be the most sensitive target of celastrol. In correlation with celastrol action, siRNA knockdown experiments targeting p23 show a greater sensitivity of steroid receptors compared with kinase clients of Hsp90 in HeLa and Hs578T cell lines. In the present study we demonstrated that interaction of celastrol with purified p23 alters the structure of this co-chaperone, causing it to polymerize into amyloid-like fibrils. In agreement with this finding, using immunocytochemistry and immunoelectron microscopy, we showed that p23 forms high molecular weight assemblies in cells treated with celastrol. These results reveal a novel inhibitory mechanism by which a small molecule can inactivate a protein by inducing its fibrillization.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human Hsp90β was expressed in Sf9 cells and purified as described previously. Hsp70, Ydj1, Hop, Cdc37, and p23 were expressed and purified as described previously (26, 27).
Progesterone Receptor Complex Assembly with Purified Proteins
Purified PR was adsorbed onto PR22 antibody-protein A-Sepharose and was assembled into complexes, as described previously (26), using ∼0.05 μm PR plus 1.4 μm Hsp70, 0.8 μm Hsp90 dimer, 0.2 μm Ydj1, 0.08 μm Hop, and 2.6 μm p23 in a reaction solution made of 20 mm Tris/HCl, pH 7.5, 5 mm MgCl2, 2 mm dithiothreitol, 0.01% Nonidet P-40, 50 mm KCl, and 5 mm ATP. After incubation for 30 min at 30 °C, 0.1 μm [3H]progesterone (American Radiolabeled Chemicals, Inc., St. Louis) was added for incubation in ice for 3 h. The complexes were then washed with reaction buffer and assessed for bound progesterone by liquid scintillation and for protein composition by SDS-PAGE.
Site-directed Mutagenesis
The QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce specific mutations in pET23a-hp23. All mutant clones were verified by DNA sequencing.
Cell Culture and Transcription Assays
HeLa cells that stably express PRB and chloramphenicol acetyltransferase (CAT) reporter gene under the control of two copies of the progesterone-response element (29) were grown in HEPES-buffered minimal essential media supplemented with 10% fetal bovine serum and nonessential amino acids. Transcriptional activity of PR was determined by the amount of CAT protein synthesized after stimulation of cells for 10 h with the progesterone analog R5020 (NENTM PerkinElmer Life Sciences). CAT protein (50 μg of cytosolic protein) was quantified by CAT enzyme-linked immunosorbent assay (Roche Applied Science).
Immunocytochemistry and Confocal Microscopy
Monolayer cells were grown on glass coverslips, fixed with formaldehyde, permeabilized with 0.1% Triton, and blocked with 10% goat serum and 5% glycerol in phosphate-buffered saline. A monoclonal primary antibody against PR (PR6) and goat anti-mouse secondary antibody labeled with Alexa Fluor 594 (Invitrogen) were used at a 1:1000 dilution. All incubations with antibodies were performed at room temperature for 1 h. Coverslips were washed and mounted on slides with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Cells were visualized and recorded using a 2LSM510 confocal laser scanning microscope (Carl Zeiss Inc., Oberkochen, Germany). To analyze the celastrol effect, HeLa PRB cells were grown on glass coverslips and treated with 5 μm DMSO, geldanamycin, or celastrol for 18 h. JJ3 antibody against p23 and anti-mouse secondary antibody labeled with Alexa Fluor 488 were used at a 1:500 dilution following the above protocol. Cells were imaged using a Zeiss Imager M1 microscope. Deconvolution of Z-stack images was done using an inverse filter algorithm with autolinear normalization.
Activity
ATPase activity assays were done as described previously (29) in the presence of geldanamycin, celastrol, or DMSO control.
siRNA Knockdown
HeLa PRB cells at 30–40% confluence in 6-well plates were transfected with 100 nm siRNA duplexes to p23 using DharmaFect 1 reagent according to the manufacturer's protocol (Dharmacon, Lafayette, CO). The nonspecific control VIII was used to assess the nonsequence-specific effects. Cells were harvested and analyzed at 90 h. Protein expression was assessed by Western blotting using 15 μg of protein lysate.
Cytosol Preparation, Immunoprecipitation, and Western Blotting
Cells were lysed in buffer A (10 mm Tris/HCl, pH 7.5, 50 mm KCl, 1 mm dithiothreitol) with protease inhibitors. After centrifugation (100,000 × g, 45 min), clarified lysates were incubated with protein A-Sepharose H90.10 (antibody to Hsp90) beads for 90 min in ice. The immunoprecipitates were washed five times with 1 ml of buffer A. Bound proteins were eluted with SDS sample buffer, resolved by 10% SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Proteins were then detected by Western blotting with antibodies against p23 (JJ3) or Hop (F5).
NMR Spectroscopy
All NMR data were collected at 25 °C with a Bruker Avance spectrometer operating at 1H frequency of 500 MHz. 200 μm samples of p23 dissolved in 10 mm Tris/HCl, pH 7.5, 50 mm KCl were used. Celastrol was dissolved in DMSO before addition to p23. Control experiments with DMSO alone were also recorded as DMSO causes small changes in p23 chemical shifts. Samples were probed for several days by acquiring 1H-15N heteronuclear single quantum coherence spectra.
Analytical Size Exclusion Chromatography
Samples of p23 (50 μm) in 10 mm Tris/HCl, pH 7.5, 50 mm KCl were treated with a 5-fold molar excess of celastrol dissolved in DMSO and with DMSO only for 2 h at 37 °C. A 50-μl aliquot of the mixture was passed through an analytical Superdex 200 size exclusion column.
Thioflavin-T Binding
Triplicate samples of p23 were treated with celastrol for 2–6 h at room temperature (25 °C). 5 mm thioflavin-T was added and then analyzed for enhancement of thioflavin-T fluorescence using a PTI-QM2001 fluorometer (Photon Technology International, Lawrenceville, NJ) at the excitation wavelength of 450 nm following emission from 470 to 500 nm (30).
Negative Staining and Electron Microscopy
Proteins were treated with celastrol in 10 mm Tris/HCl, pH 7.8, 50 mm KCl for 2–6 h at 25 °C. Unless specified otherwise, the molar ratio of protein/celastrol used in this study was about 1:2. Samples were spotted on a 300-mesh copper Formvar/carbon grid and air-dried. Grids were negatively stained with 4% uranyl acetate, washed, air-dried, and examined on either a JEOL 1200 EX or Philips Tecnai T12 transmission electron microscope. Immunoelectron microscopy analysis was done at the electron microscopy core facility of the Medical College of Georgia using JJ3 at 1:100 and Au7goat anti-mouse antibody labeled with gold following a standard protocol.
Reagents
Celastrol and dihydrocelastrol were purchased from Cayman Chemical, Ann Arbor, MI, and Gaia Chemical Corp., Gaylordsville, CT, respectively.
RESULTS
Steroid Receptors Are Particularly Sensitive to Celastrol
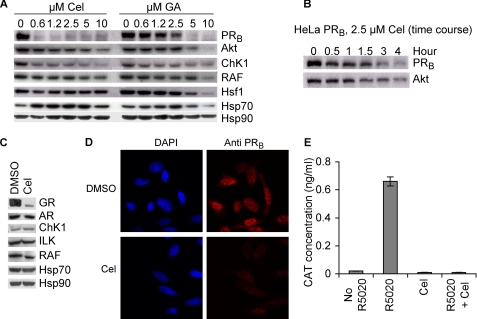
Our initial studies showed that, in agreement with what was previously reported for other kinases (12), celastrol induced a dose-dependent degradation of Akt, ChK1, and RAF and a modest induction of Hsp70 (Fig. 1A). Surprisingly, degradation of PR occurred at lower doses of celastrol compared with geldanamycin. Furthermore, time course experiments suggested that the loss of PR occurred more quickly compared with the loss of Akt (Fig. 1B). Treatment of the human breast cancer cell line Hs578T with 5 μm celastrol for 3 h also led to a more efficient degradation of the glucocorticoid receptor (GR) as compared with the kinases ChK1, ILK, and RAF (Fig. 1C). Interestingly, in this short treatment time, androgen receptor was less affected in the Hs578T cells (Fig. 1C). Immunohistochemical staining for PR in HeLa PRB cells treated with celastrol showed loss of this protein client (Fig. 1D), and gene reporter assays showed that the transcriptional activity of PR was lost after celastrol treatment (Fig. 1E). These data underscore a profound difference between the action of celastrol and other Hsp90 inhibitors. Indeed, the vast literature on inhibitors of Hsp90 that target the ATP binding pocket did not reveal any significant difference in destabilizing Hsp90 clients (i.e. steroid receptor and kinases). The two emerging distinctions between the Hsp90 chaperoning of steroid receptors versus kinases are the roles of the co-chaperones p23 and Cdc37. p23 seems to play no role in an in vitro assay for chaperoning ChK1 (27, 28), but it is essential in stabilizing PR- and GR-chaperoning complexes (26, 31–33). In contrast, Cdc37 is essential for chaperoning kinases (34), but it has no role in steroid receptor chaperoning (27). In this context, and in agreement with previously published data on celastrol inactivation of Cdc37 (14), our in vitro reconstitution of ChK1 using purified proteins (27) was inhibited by celastrol (data not shown). Therefore, because PR and GR were degraded faster than kinases in cells treated with celastrol, we focused our studies on PR, whose reconstitution in vitro does not require Cdc37.
FIGURE 1.
Celastrol destabilizes the Hsp90 client PR in cells. A, HeLa PRB cells were treated for 4 h with various amounts of celastrol (Cel) or geldanamycin (GA). Cell lysates were prepared and blotted for the presence of Hsp90, Hsp70, and the Hsp90 clients. B, HeLa PRB cells were treated with 2.5 μm celastrol for various periods of time, and cell lysates were blotted for Hsp90 clients PRB and Akt. C, Hs578T cells were treated for 4 h with 5 μm celastrol, and cytosols were prepared and blotted for the presence of Hsp90, Hsp70, and the Hsp90 clients. AR, androgen receptor. D, HeLa PRB cells were treated with 2.5 μm celastrol for 4 h, and PRB stability was visualized by immunohistochemistry. DAPI, 4′,6-diamidino-2-phenylindole. E, effect of celastrol on the transcriptional activity of PR was assessed by measuring the production of the reporter gene CAT under control of two copies of the progesterone-response element in HeLa PRB cells. Cells were pretreated with 2.5 μm celastrol for 2 h prior to adding the progesterone agonist R5020 (125 nm) for an additional 10 h. Experiment was done twice, and each sample was done in triplicate.
p23 Is the Co-chaperone Most Sensitive to Celastrol Treatment
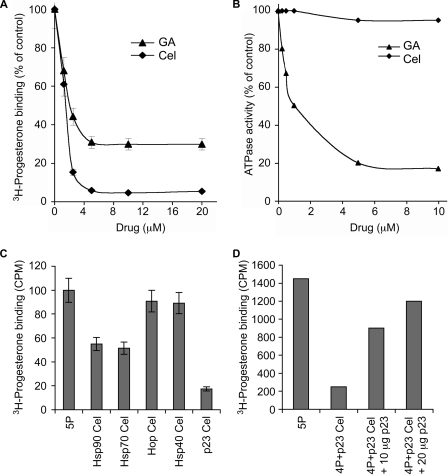
PR reconstitution in vitro using purified chaperones (i.e. Hsp90, Hsp70, Hop, Hsp40, and p23) (26) has been fundamental in furthering our understanding of how geldanamycin and related compounds inhibit Hsp90-dependent chaperoning (35). It is therefore a powerful system to dissect how celastrol inhibits the Hsp90 chaperoning machinery. As shown in Fig. 2A, celastrol inhibited PR chaperoning in vitro. When compared with geldanamycin, chaperoning of PR was more sensitive to inhibition by celastrol, which induced a more complete inhibition of the Hsp90 machinery. This celastrol effect was not through ATP binding and hydrolysis by Hsp90, as we determined that celastrol had no influence on the ATPase activity of Hsp90 (Fig. 2B).
FIGURE 2.
Celastrol inhibits in vitro chaperoning of PR by affecting p23 function. A, PR hormone binding activity was reconstituted using purified chaperones in the presence of various amounts of celastrol (Cel, diamonds) or geldanamycin (GA, triangles). Binding of [3H]progesterone is expressed as a percentage of the DMSO-treated control. The experiment was done twice, and each sample was done in duplicate. B, celastrol has no effect on the ATPase activity of Hsp90. ATPase activity of Hsp90, stimulated by the addition of the activator Aha1, was measured in the presence of various concentrations of celastrol (Cel, diamonds) or geldanamycin (GA, triangles). Activity is expressed as the amount of [32P]ATP converted to [32P]ADP relative to the untreated control. Data are representative of two experiments. C, 20 μm purified chaperones were individually treated with 2-fold molar excess of celastrol prior to being used to reconstitute PR hormone binding activity. Excess celastrol was removed by extensive buffer exchange, and the treated chaperone was used with the other four untreated chaperones in reconstitution of PR. 5P represents reconstitution of PR with p23, Hsp90, Hsp70, Hop, and Hsp40 (Ydj1). The experiment was repeated three times. Each sample was done in triplicate. D, untreated p23 compensates for the celastrol-inactivated p23. Purified p23 was treated with five time molar excess of celastrol, and excess of celastrol was removed as in C. This celastrol-treated p23 was used in PR reconstitution reaction in the presence of indicated amounts of untreated p23. 4P stands for the four proteins Hsp90, Hsp70, Hop, and Hsp40 (Ydj1).
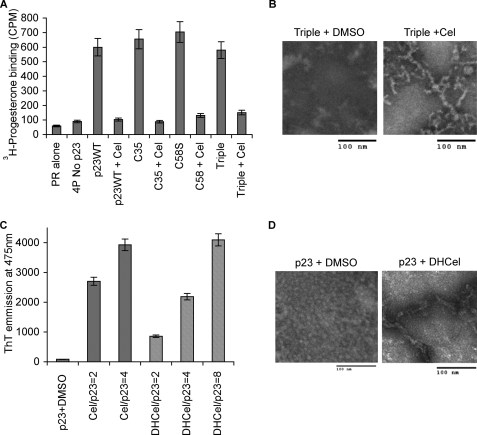
We then tested the effect of celastrol on individual chaperones used to reconstitute PR. Each chaperone was preincubated with celastrol. Free celastrol was then removed by extensive buffer exchange prior to using the celastrol-treated chaperone for PR reconstitution with the other untreated chaperones. The results presented in Fig. 2C show that the chaperone activities of Hsp90 and Hsp70 were reduced ∼50% by celastrol treatment, and Hop and Hsp40 (Ydj1 in budding yeast) were not affected. Surprisingly, treatment of p23 inhibited PR reconstitution activity by about 80%, an inhibition level similar to what is obtained when the reconstitution assay is performed without p23 (Fig. 7A) (26). Addition of untreated p23 to the reaction compensated for celastrol-inactivated p23 in a concentration-dependent manner (Fig. 2D). These data suggest that p23 is the chaperone most sensitive to celastrol.
FIGURE 7.
Celastrol induction of p23 fibrils in wild-type and mutant p23. A, ability of wild-type full-length p23 and its core domain p23C35 (residues 1–119) to chaperone PR was compared with that of p23 having the mutation C58S or the three mutations C40S, C58S, and C75S (Triple). 4P represents reconstitution of PR with Hsp90, Hsp70, Hop, and Hsp40 (Ydj1). Data represent three experiments in duplicate. Cel, celastrol. B, fibril formation upon celastrol treatment of p23 triple mutant (C40S, C58S, and C75S) was monitored by electron microscopy. C, ability of dihydrocelastrol to induce p23 fibrils was tested by thioflavin-T (ThT) binding using inhibitor-to-protein ratios of 2, 4, or 8 as indicated. Data represent three experiments. D, ability of dihydrocelastrol to induce p23 fibrils was monitored by electron microscopy.
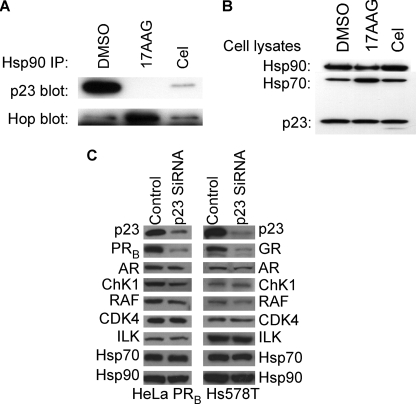
Additional experiments to test the effect of celastrol on p23 confirmed previous data (12) and showed that celastrol disrupted the p23-Hsp90 complexes in HeLa cells (Fig. 3A) but did not increase the cellular interaction of Hop with Hsp90 the way geldanamycin did (Fig. 3A) (12). Knockdown experiments with siRNA against p23 in HeLa PRB cells and in the Hs578T cells showed similar client destabilization profiles compared with celastrol treatment. Lack of p23 induced a dramatic destabilization of PR and GR, whereas the kinases ChK1, RAF, CDK4, and ILK were significantly less affected (Fig. 3C). Accordingly, as seen in celastrol treatment (Fig. 1C), androgen receptor is also less sensitive to the lack of p23 (Fig. 3C).
FIGURE 3.
Celastrol destabilizes cellular Hsp90-p23 complexes. A, HeLa cells were treated with 3 μm celastrol, 17-allylamino-17-demethoxygeldanamycin (17AAG), or DMSO control for 18 h. Cell lysates were prepared, and Hsp90 immunoprecipitates (IP) were blotted for p23 or Hop. B, assessment of total Hsp90, Hsp70, or p23 in HeLa cell lysates shown in A. C, HeLa PRB and Hs578T cells were treated with control or siRNA against p23 for 96 h. Cytosols were blotted for p23, steroid receptors (PR, androgen receptor (AR), and GR), kinases (ChK1, RAF, CDK4, and ILK), Hsp70, and Hsp90.
Celastrol Binds to p23 and Induces Rapid Formation of Amyloid-like Fibrils
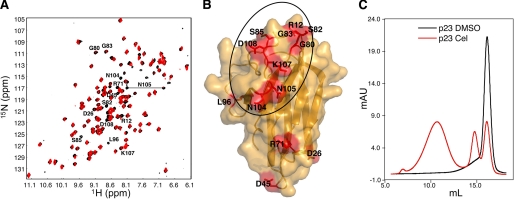
To determine how celastrol inhibits p23, we examined the 15N-labeled p23 core domain (residues 1–119) by NMR spectroscopy in the absence and presence of celastrol. Addition of a 5-fold molar excess of celastrol relative to p23 led to the selective weakening or complete disappearance of several signals in the 1H-15N heteronuclear single quantum coherence spectrum of p23 (Fig. 4A). Amino acids that showed changes (disappearance and/or weakening of signals) and for which NMR resonance assignments were available (36) were mapped on the surface of the p23 crystal structure (Fig. 4B) (37). The majority of affected residues were found to be in the vicinity of a hydrophobic region of p23 corresponding to the p23-Hsp90 interface (36, 38). Remarkably, prolonged treatment with celastrol led to the disappearance of all signals in the 1H-15N heteronuclear single quantum coherence spectrum of p23. This loss of signal suggested that celastrol had caused the protein to aggregate or to form high molecular weight species. The multimerization of p23 was confirmed by size exclusion chromatography. Celastrol-treated p23 eluted in the column void volume, unlike native p23, which has the expected retention time for a 23-kDa monomer (Fig. 4C).
FIGURE 4.
Celastrol affects the structure of p23. A, 1H-15N heteronuclear single quantum coherence spectrum of 15N-labeled p23 recorded immediately after addition of a 5-fold molar excess of celastrol dissolved in DMSO is shown in red, and the corresponding spectrum after addition of the same amount of DMSO only is shown in black. Assigned resonances with weaker signals or disappeared signals following addition of celastrol are labeled. Prolonged treatment with celastrol led to the disappearance of all signals in the spectrum of p23 (data not shown). B, molecular surface representation of p23 (residues 1–119). The side chains of residues labeled in A are shown in red. The p23 area involved in binding Hsp90 is circled. C, 25 μm purified p23 was treated with 5-fold molar excess of celastrol or DMSO for 2 h at 37 °C and analyzed by size exclusion chromatography using a Superdex 200 column. Cel, celastrol.
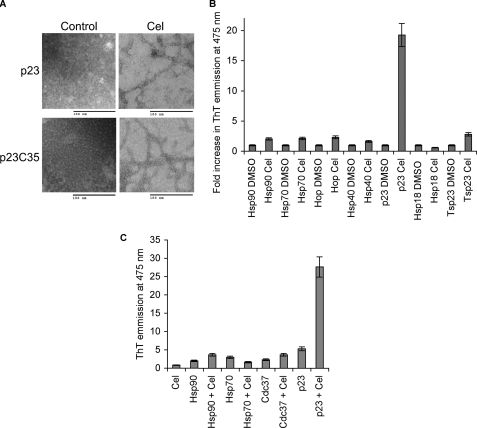
Electron microscopy of celastrol-treated p23 revealed the formation of amyloid-like fibrils not present in the untreated controls (Fig. 5A). These fibrils are distinct from amyloid fibrils as they occurred within 2–4 h under physiological conditions and were SDS-sensitive. Furthermore, the fibrils were not recognized by the antibodies A11 and OC that react with amyloid fibrils and amyloid oligomers (data not shown) (39). Fibril formation was also consistent with the pronounced enhancement of thioflavin-T fluorescence after treatment of p23 with celastrol (Fig. 5B). Thioflavin-T is a reagent known to fluoresce more intensely upon specific interaction with amyloid fibrils, but its binding mode is poorly understood (40). None of the other chaperones and co-chaperones used for in vitro reconstitution of PR (i.e. Hsp90, Hsp70, Hsp40, and Hop) were significantly affected by treatment with celastrol (Fig. 5B). Similarly, thioflavin-T binding by Tsp23, a member of the p23 family (41), and the small heat-shock protein Hsp18, whose β sheet-based fold is similar to that of p23 (37), were not induced by celastrol (Fig. 5B).
FIGURE 5.
Celastrol treatment causes p23 to form amyloid-like fibrils. A, 25 μm purified full-length p23 (p23WT) and its core domain p23C35 (residues 1–119) were treated with 2-fold molar excess of celastrol (Cel) and analyzed by electron microscopy. B, chaperone proteins Hsp90, Hsp70, Hop, Hsp40 (Ydj1), and Hsp18 along with p23 were treated with 2-fold molar excess of celastrol and analyzed for enhancement of thioflavin-T (ThT) fluorescence emission upon treatment with celastrol. The experiments were done two times, and all samples were tested in duplicate. C, Cdc37 treated with celastrol does not bind thioflavin-T. 25 μm Hsp90, Hsp70, Cdc37, and p23 were treated with 100 μm celastrol or DMSO control and incubated for 1 h at 37 °C and then 2 h at room temperature. Thioflavin-T binding was monitored using 96-well plates and the TECAN microplate reader at the excitation wavelength of 450 nm following emission at 475 nm.
Because celastrol was shown previously to trigger the aggregation of Cdc37, we tested whether celastrol could induce fibrillization of Cdc37. As shown in Fig. 5C, celastrol-treated Cdc37 does not bind thioflavin-T any more than the untreated protein. These data underscore the specific nature of this action of celastrol on p23.
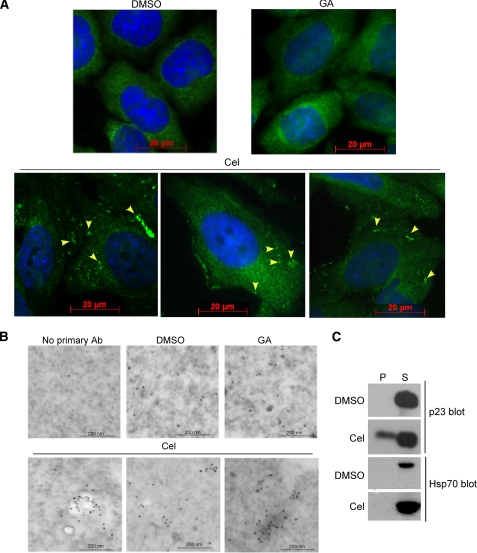
To test whether any change in p23 structure might be detectable in cells, HeLa PRB cells were treated with celastrol or geldanamycin and analyzed by immunocytochemistry and immunoelectron microscopy using a monoclonal antibody against p23. As seen in Fig. 6, A and B, although geldanamycin had no effect on p23 distribution, celastrol treatment induced a clustering of the co-chaperone. Biochemical analysis of cell fractions showed that p23 was partially excluded from the cytosolic/soluble fraction and appeared in the insoluble fraction, confirming that in celastrol-treated cells p23 forms a higher molecular weight species (Fig. 6C). This was not the case for Hsp70, indicating that celastrol did not cause gross unfolding of other cellular proteins.
FIGURE 6.
Celastrol induces oligomerization of p23 in HeLa cells. A, immunocytochemistry analysis of cells treated with DMSO, 5 μm geldanamycin (GA), or celastrol (Cel) using the monoclonal antibody (JJ3) against p23. Yellow arrowheads indicate p23 clusters that are absent in the DMSO controls or geldanamycin-treated cells. The nucleus is stained with 4′,6-diamidino-2-phenylindole (blue) Scale bars represent 20 μm. B, immunoelectron microscopy analysis of cells treated with DMSO, 5 μm geldanamycin, or celastrol using the monoclonal antibody (Ab) (JJ3) against p23 and a gold-labeled goat anti-mouse antibody. Compared with DMSO or geldanamycin treatment, celastrol causes clustering of p23 that might indicate its fibrillization. Scale bars represent 200 nm. C, celastrol causes the redistribution of p23 into the insoluble fraction of HeLa cells. HeLa cells were treated with 3 μm celastrol or DMSO for 4 h and were fractionated into soluble (S) and insoluble pellet (P) fractions. Proportional amounts of each fraction were loaded and analyzed by Western blot for p23 and Hsp70.
Fibril Induction by Celastrol and Its Reactivity with Cysteines
Recent studies suggest that celastrol irreversibly reacts with cysteine residues (13, 18, 19, 25). Our mass spectrometry analysis also showed that celastrol modified p23 on Cys-40, Cys-58, and Cys-75. These modifications in p23 were reversible by reducing agents such as β-mercaptoethanol or dithiothreitol (data not shown). To evaluate the potential contribution of celastrol-modified cysteines to disrupt p23 function, p23 variants with serine substitution at Cys-58 alone or in combination with the other cysteine-to-serine mutations were tested in PR reconstitution assays. These mutations had no effect on the activity of p23 as they failed to prevent celastrol from inhibiting the reconstitution of PR (Fig. 7A). Moreover, the triple cysteine-to-serine mutant retained its ability to form fibrillar species upon celastrol treatment (Fig. 7B), indicating that celastrol reaction with residues Cys-40, Cys-58, and Cys-75 of p23 is not what triggers the formation of amyloid-like fibrils. This interpretation was further supported by our observation that dihydrocelastrol, which lacks the quinone methide moiety and has no reactivity with cysteine residues, still induced p23 to form fibrils and bind thioflavin-T (Fig. 7, C and D). We also attempted to mutate all five cysteines of p23, including Cys-20 and Cys-76, to serines, but these cumulative mutations seem to destabilize the protein, leading to its precipitation during the purification process. Although a few small molecules have been reported to inhibit amyloid fibril formation (42), to our knowledge, this is the first example of a small molecule triggering rapid polymerization of a protein into amyloid-like fibrils at near-physiological conditions.
DISCUSSION
Celastrol elicits potent anticancer activities in cell and animal models (12, 13, 15, 16), and it has also been identified as a promising compound to treat neurodegenerative diseases associated with misfolded proteins (18). Although celastrol seems to have a pleiotropic activity, many of its actions have been linked to its inactivation of the Hsp90 chaperoning pathway (12). We found that cellular levels of PR and GR were more sensitive to celastrol treatment than were kinase clients of Hsp90. Our data indicate that levels of these steroid receptors were dramatically reduced within 3–5 h of celastrol treatment. Longer exposure of cells to celastrol (24 h) ultimately caused the degradation of androgen receptors and protein kinases (12, 14). Testing of celastrol-treated purified chaperones in vitro showed that the Hsp90 co-chaperone p23 is the most sensitive target of celastrol. Celastrol binding to p23 altered the co-chaperone structure by triggering its oligomerization into amyloid-like fibrils as demonstrated by changes in NMR spectra, elution profile in size exclusion chromatography, induced binding of thioflavin-T, and visualization of fibrillar structures by electron microscopy. Consistent with our in vitro finding, clustering of p23 was observed by immunocytochemistry and immunoelectron microscopy of celastrol-treated cells. The celastrol-induced oligomerization of p23 is rapid and occurs under physiological conditions. These fibrils are distinct from amyloid fibrils as they are SDS-sensitive, which suggests that their formation may be reversible. Furthermore, they neither react with the antibody OC that recognizes amyloid fibrils nor with the antibody A11 that recognizes pre-fibril oligomers (39).
Although amyloid fibril formation has been linked to numerous diseases, the differences noted between p23 fibrils and amyloid fibrils make us wonder if celastrol-mediated fibrillization of p23 contributes to the well documented beneficial action of celastrol in cells. Several reports have indicated that enhanced levels of cellular p23, by either overexpression or by disruption of Hsp90-p23 complexes, reduce the toxicity of misfolded proteins (43–45) and influence protein refolding, as shown for the cystic fibrosis transmembrane conductance regulator harboring the mutation ΔF508 linked to cystic fibrosis (46).
The formation of p23 fibrils may have a similar effect by stabilizing p23. Consistent with this possibility, p23 fibrils are still present in cells after 18 h of celastrol treatment and not cleared by proteasome or autophagy pathways. Celastrol could be used as a chemical template to design small molecule inhibitors that would more specifically target p23-dependent or Cdc37-dependent clients of Hsp90.
The importance of the quinone methide moiety (i.e. the group reacting with cysteines) in the cellular mechanism of action of celastrol is unclear. This group seems crucial to celastrol action on melanoma cellular assays because dihydrocelastrol, which lacks this moiety, fails to inhibit melanoma cell viability (13). However, dihydrocelastrol can induce a heat-shock response, although some compounds with a quinone methide group, such as brazilein and hematein, cannot (18).
Although celastrol inactivates Cdc37 by covalent modification of its cysteines (25), our data showed that dihydrocelastrol is capable of inducing p23 fibrils in vitro, albeit with less efficacy than celastrol (Fig. 7, C and D). Therefore, noncovalent binding of celastrol to p23 may be the trigger for its fibrillization. If this mechanism holds true, one could design celastrol-derived noncovalent binders of p23 that would selectively inhibit this protein. These celastrol derivatives would be helpful in better deciphering the mechanism of p23 fibrillization. This may also facilitate the design of drugs that would selectively target steroid receptor-dependent diseases through inactivation of the co-chaperone p23.
Acknowledgments
We thank Dr. David O. Toft for support and advice during the initial stage of this project and Bridget Stensgard and Elizabeth S. Bruinsma for their technical assistance. We are grateful to Dr. Sungjo Park for recording atomic force microscopy images of celastrol-induced p23 fibrils and to Dr. Charles G. Glabe for OC and A11 antibodies. We thank staff members of the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for their assistance in performing the mass spectrometry analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants GM071514 (to M. R.-A.) and CA109449 (to G. M.). This work was also supported by the Mayo Foundation (to M. R.-A.) and the American Heart Association Grant 06-30077N (to M. R.-A.).
- PR
- progesterone receptor
- GR
- glucocorticoid receptor
- CAT
- chloramphenicol acetyltransferase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 2.Latchman D. S. (2001) Cardiovasc. Res. 51, 637–646 [DOI] [PubMed] [Google Scholar]
- 3.Whitesell L., Lindquist S. L. (2005) Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 4.Chiosis G., Vilenchik M., Kim J., Solit D. (2004) Drug Discov. Today 9, 881–888 [DOI] [PubMed] [Google Scholar]
- 5.Tsutsumi S., Neckers L. (2007) Cancer Sci. 98, 1536–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taldone T., Gozman A., Maharaj R., Chiosis G. (2008) Curr. Opin. Pharmacol. 8, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taldone T., Sun W., Chiosis G. (2009) Bioorg. Med. Chem. 17, 2225–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbins C. E., Russo A. A., Schneider C., Rosen N., Hartl F. U., Pavletich N. P. (1997) Cell 89, 239–250 [DOI] [PubMed] [Google Scholar]
- 9.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1997) Cell 90, 65–75 [DOI] [PubMed] [Google Scholar]
- 10.Wandinger S. K., Richter K., Buchner J. (2008) J. Biol. Chem. 283, 18473–18477 [DOI] [PubMed] [Google Scholar]
- 11.Pearl L. H., Prodromou C., Workman P. (2008) Biochem. J. 410, 439–453 [DOI] [PubMed] [Google Scholar]
- 12.Hieronymus H., Lamb J., Ross K. N., Peng X. P., Clement C., Rodina A., Nieto M., Du J., Stegmaier K., Raj S. M., Maloney K. N., Clardy J., Hahn W. C., Chiosis G., Golub T. R. (2006) Cancer Cell 10, 321–330 [DOI] [PubMed] [Google Scholar]
- 13.Abbas S., Bhoumik A., Dahl R., Vasile S., Krajewski S., Cosford N. D., Ronai Z. A. (2007) Clin. Cancer Res. 13, 6769–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T., Hamza A., Cao X., Wang B., Yu S., Zhan C. G., Sun D. (2008) Mol. Cancer Ther. 7, 162–170 [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Zhou Y., Fan Y., Zhou D. (2008) Cancer Lett. 264, 101–106 [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Chen D., Cui Q. C., Yuan X., Dou Q. P. (2006) Cancer Res. 66, 4758–4765 [DOI] [PubMed] [Google Scholar]
- 17.Mu T. W., Fowler D. M., Kelly J. W. (2008) PLoS Biol. 6, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westerheide S. D., Bosman J. D., Mbadugha B. N., Kawahara T. L., Matsumoto G., Kim S., Gu W., Devlin J. P., Silverman R. B., Morimoto R. I. (2004) J. Biol. Chem. 279, 56053–56060 [DOI] [PubMed] [Google Scholar]
- 19.Trott A., West J. D., Klaiæ L., Westerheide S. D., Silverman R. B., Morimoto R. I., Morano K. A. (2008) Mol. Biol. Cell 19, 1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin H. Z., Hwang B. Y., Kim H. S., Lee J. H., Kim Y. H., Lee J. J. (2002) J. Nat. Prod. 65, 89–91 [DOI] [PubMed] [Google Scholar]
- 21.Nagase M., Oto J., Sugiyama S., Yube K., Takaishi Y., Sakato N. (2003) Biosci. Biotechnol. Biochem. 67, 1883–1887 [DOI] [PubMed] [Google Scholar]
- 22.Zhang D. H., Marconi A., Xu L. M., Yang C. X., Sun G. W., Feng X. L., Ling C. Q., Qin W. Z., Uzan G., d'Alessio P. (2006) J. Leukocyte Biol. 80, 309–319 [DOI] [PubMed] [Google Scholar]
- 23.Sethi G., Ahn K. S., Pandey M. K., Aggarwal B. B. (2007) Blood 109, 2727–2735 [DOI] [PubMed] [Google Scholar]
- 24.Sun H., Liu X., Xiong Q., Shikano S., Li M. (2006) J. Biol. Chem. 281, 5877–5884 [DOI] [PubMed] [Google Scholar]
- 25.Sreeramulu S., Gande S. L., Göbel M., Schwalbe H. (2009) Angew. Chem. Int. Ed. Engl. 48, 5853–5855 [DOI] [PubMed] [Google Scholar]
- 26.Kosano H., Stensgard B., Charlesworth M. C., McMahon N., Toft D. (1998) J. Biol. Chem. 273, 32973–32979 [DOI] [PubMed] [Google Scholar]
- 27.Arlander S. J., Felts S. J., Wagner J. M., Stensgard B., Toft D. O., Karnitz L. M. (2006) J. Biol. Chem. 281, 2989–2998 [DOI] [PubMed] [Google Scholar]
- 28.Felts S. J., Karnitz L. M., Toft D. O. (2007) Cell Stress Chaperones 12, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chadli A., Graham J. D., Abel M. G., Jackson T. A., Gordon D. F., Wood W. M., Felts S. J., Horwitz K. B., Toft D. (2006) Mol. Cell. Biol. 26, 1722–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grad I., McKee T. A., Ludwig S. M., Hoyle G. W., Ruiz P., Wurst W., Floss T., Miller C. A., 3rd, Picard D. (2006) Mol. Cell. Biol. 26, 8976–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J., Corbisier R., Stensgard B., Toft D. (1996) J. Steroid Biochem. Mol. Biol. 56, 31–37 [DOI] [PubMed] [Google Scholar]
- 33.Dittmar K. D., Demady D. R., Stancato L. F., Krishna P., Pratt W. B. (1997) J. Biol. Chem. 272, 21213–21220 [DOI] [PubMed] [Google Scholar]
- 34.Pearl L. H. (2005) Curr. Opin. Genet. Dev. 15, 55–61 [DOI] [PubMed] [Google Scholar]
- 35.Pratt W. B., Toft D. O. (2003) Exp. Biol. Med. 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Yamout M. A., Venkitakrishnan R. P., Preece N. E., Kroon G., Wright P. E., Dyson H. J. (2006) J. Biol. Chem. 281, 14457–14464 [DOI] [PubMed] [Google Scholar]
- 37.Weaver A. J., Sullivan W. P., Felts S. J., Owen B. A., Toft D. O. (2000) J. Biol. Chem. 275, 23045–23052 [DOI] [PubMed] [Google Scholar]
- 38.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krebs M. R., Bromley E. H., Donald A. M. (2005) J. Struct. Biol. 149, 30–37 [DOI] [PubMed] [Google Scholar]
- 41.Felts S. J., Toft D. O. (2003) Cell Stress Chaperones 8, 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacchettini J. C., Kelly J. W. (2002) Nat. Rev. Drug. Discov. 1, 267–275 [DOI] [PubMed] [Google Scholar]
- 43.Bose S., Weikl T., Bügl H., Buchner J. (1996) Science 274, 1715–1717 [DOI] [PubMed] [Google Scholar]
- 44.Freeman B. C., Toft D. O., Morimoto R. I. (1996) Science 274, 1718–1720 [DOI] [PubMed] [Google Scholar]
- 45.Weikl T., Abelmann K., Buchner J. (1999) J. Mol. Biol. 293, 685–691 [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]