Abstract
Piwi proteins are required for germ cell proliferation, differentiation, and germ line stem cell maintenance. In normal tissues, human and mouse Piwil2 are primarily expressed in testis but widely expressed in tumors. However, the underlying mechanism remains largely unknown. In vertebrates, transforming growth factor (TGF)-β signaling plays an important role in patterning embryo and control of cell growth and differentiation. A previous study has shown a role for Zili, a Piwil2 gene in zebrafish, in germ cells in zebrafish. Here we report that zili functions in patterning the early embryo and inhibits TGF-β signaling. Whole mount expression analysis shows that zili expresses not only in PGCs but also in axis. Ectopic expression of zili causes fusion of the eyes and reduction of mesodermal marker genes expression, suggesting that zili functions to inhibit Nodal signaling and mesoderm formation. Genetic interaction shows that zili inhibits Nodal and bone morphogenetic protein signaling. The results of protein interaction assays identify that Zili binds to Smad4 via its N-terminal domain and prevents the formation of Smad2/3/4 and Smad1/5/9/4 complexes to antagonize TGF-β signaling. This work shows that zili plays a role in early embryogenesis beyond germ line as a novel negative regulator of TGF-β signaling, extending the function of Piwi proteins in vertebrates.
Keywords: Cytokines/TGF-β, Development Differentiation, Gene Regulation, Smad/Transcription Factor, Zebra fish, Smad4, TGF-β, Zili, Piwil2
Introduction
The Piwi clade of Argonaute proteins is found in all animals examined so far, and genomes of multicellular animals encode multiple Piwi proteins (1). In Drosophila, the male and female germ lines express three proteins, including Piwi, Aubergine, and AGO3, and Piwi is required in germ cells, as well as in somatic niche cells, for regulation of cell division and maintenance of germ line stem cells (2). Mutations of piwi genes in mouse (miwi, mili, and miwi2) influence meiotic progression of developing sperm, but not so do the oogenesis (3–6). Mili and its human homolog (hili) are primarily expressed in testis among normal tissues but widely expressed in tumors. However, the underlying mechanism remains largely unknown (7, 8). In zebrafish, Piwil1 (Ziwi) colocalizes with vasa at the embryonic genital ridge and expresses at the gonad specifically in the adults (9), and the loss of Ziwi function results in a progressive loss of germ cells because of apoptosis during larval development (10). Furthermore, Houwing et al. (11) describe a function for Zili, another zebrafish Piwi protein gene, in transposon defense and germ cell differentiation, as well as a crucial function in meiosis.
TGF-β2 signal transduction has been shown to play a pivotal role in a wide variety of developmental events, ranging from the earliest steps in germ layer patterning of the pregastrula embryo to tissue healing, regeneration, and homeostasis in the adult (12). Smads are essential intracellular transducers for TGF-β signals (13–16). In response to TGF-β, receptor-activated Smads (R-Smads) are phosphorylated by type I receptors. Phosphorylated R-Smads form a complex with Smad4 and are transported into the nucleus, where Smads cooperate with specific DNA-binding transcription factors to regulate gene transcription in a context-dependent manner (13). Here, we present that zebrafish Piwi protein gene Piwil2 (Zili) suppresses TGF-β signaling by physically associating with Smad4 and preventing the formation of Smad2/3/4 and Smad1/5/9/4 complexes.
EXPERIMENTAL PROCEDURES
All of the animals were handled in strict accordance with good animal practices as defined by the National Zebrafish Resources of China and The Zebrafish Book (17), and all of the animal work was approved by the National Zebrafish Resources of China and West China Hospital.
Zebrafish Strain and Embryos
The zebrafish (Danio rerio) AB strain was provided by the National Zebrafish Resources of China and kept at 28.5 °C. The embryos were obtained by natural mating and cultured in embryo medium (17). Staging of the embryos was carried out according to Kimmel et al. (18).
Cloning of Zili mRNA
Based on homology analysis, hili and mili mRNA sequence were used to search the homologous zebrafish genomic and expressed sequence tag sequence. Four pairs of primers (85U and 1158L; 891U and 1953L; 1759U and 2760L; and 2627U and 3519L) were designed for amplifying the coding region of zili (supplemental Table S1). Subsequently, recombinant PCR was used for gaining the complete coding region. 5′- and 3′-rapid amplification of cDNA ends were performed using the SMARTTM rapid amplification of cDNA ends cDNA amplification kit (Clontech) with designed specific primers (3345U for 3′ and 182L for 5′ end; supplemental Table S1). The whole zili mRNA sequence was obtained by assembling the sequences acquired as described above.
Zili Antibody and Reverse Transcription-PCR to Detect Zili Expression
Zili antibody was raised in rabbits with the synthetic peptide MDPKRPTFPSPPGVI+C published by Houwing et al. (11). Primer sequences for amplifying the 470-bp β-actin were designed according to Kaslin et al. (19); the 373-bp fragment of zili was amplified by primers 2387U and 2760L (supplemental Table S1).
Constructs
The coding sequence of zili was cloned into the vector pcDNA3.1+ (Invitrogen) for capped mRNA synthesis. Myc/HA/FLAG tag coding sequence was added upstream of zili and Smads cDNA, respectively, and fused sequences were cloned into pcDNA3.1+ for capped mRNA synthesis and transfection. The full-length coding region of zili was also cloned into pEGFP-N1 for expression of Zili-GFP fusion protein to generate plasmid that was used for testing the effectiveness of zili-MO1. Interesting the fragments were cloned into pSPT18/19 (Roche Applied Science) for antisense RNA synthesis. TRIpure reagent, a gel extraction kit, and a high purity plasmid preparation kit were purchased from Bioteke Corporation (Beijing, China) for total RNA extraction, PCR product purification, and plasmid preparation.
Zebrafish Whole Mount Immunohistochemistry (IHC) and in Situ Hybridization
IHC was performed as described in the Zebrafish Book (17); in situ hybridization protocols were performed as described in the Zebrafish Book (17) and by Thisse and Thisse (20). For two-color whole mount RNA in situ hybridization between zili and vasa or nanos1, zili RNA probe was labeled by digoxigenin, and vasa or nanos1 was labeled by Fluorescein. Colocalization of vasa RNA and Zili protein was performed as described by Knaut et al. (21) and Ober and Schulte-Merker (22). vasa probe was labeled by digoxigenin.
Morpholino Oligonucleotides, in Vitro Synthesis of RNA, and Microinjection
Three zili morpholino antisense oligonucleotides, a translation blocker (zili-MO1, 5′-GAT CCA TTT CTT CCT CTG TTG CAC T-3′), a splice inhibitor (zili-MO2, 5′-TGG TGT TAC TGA ATG TCA CCT GAT C-3′), and 5-mis-pair control morpholino (zili-cMO, 5′-GAT gCA TTT gTT CgT CTc TTG gAC T-3′) were designed and synthesized by Gene-tools (Philomath, OR).
After lineage by appropriate restriction enzymes, capped mRNAs were synthesized using a mMESSAGE mMACHINE® kit (Ambion); antisense RNAs for in situ hybridization were synthesized using a digoxigenin RNA labeling kit (SP6/T7) (Roche Applied Science) and purified by MEGAclear (Ambion). Two nonoverlapping probes, localizing at positions 252–1293 and 2763–3626 bp, were designed for zili in situ hybridization detection.
Morpholinos or synthetic capped mRNAs were injected into single-cell embryos. The injection dose was an estimated amount received by a single embryo. For the RNA injection experiment, the control embryos were injected with GFP mRNA.
Cell Culture, Transfection, Immunostaining, Immunoprecipitation, Immunoblotting, and Reagents
Composite of capped mRNAs or mopholinos were injected into single-cell embryos, and 100 embryos at the 50% epiboly stage were used for immunostaining and immunoprecipitation analysis. HEK293 cells were grown in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. DNA transfection into HEK293 cells was performed using Lipofectamine 2000 (Invitrogen). The Universal protein extraction buffers were purchased from Bioteke Corporation (Beijing, China). Immunostaining, immunoprecipitation, and immunoblotting were performed as described in the product information. Antibodies against FLAG tag, HA tag, and Myc tag were purchased from Sigma-Aldrich and Abcam. Antibodies against Smad2/3/4 were purchased from Santa Cruz. Anti-phospho-Smad2 antibody was purchased from Cell Signaling Technology.
Two-hybrid Experiment
A bacterial two-hybrid experiment was performed as the described in the protocol provided by the exclusive Stratagene BacterioMatch® II two-hybrid system. Both zili and Smad4 were inserted into pBT and pTRG plasmid, respectively, by EcoRI and XhoI restriction.
RESULTS
Zili Expresses Not Only in PGCs but Also in Axis during Early Embryogenesis
The 3656-bp nucleotide sequence of zili mRNA has been deposited in the GenBankTM data base under accession number EF186090. For the functional regions analysis below, we analyze the zili mRNA sequence by bioinformatics protocols. As shown in supplemental Fig. S1, the predicted open reading frame from nucleotides 282 to 3422 is 3138 bp in length and encodes a polypeptide of 1046 amino acid residues. Similar to other Piwi protein members, Zili also has PAZ and Piwi conserved domains at position 483–569 and 741–1032, suggesting functional similarity and conservation.
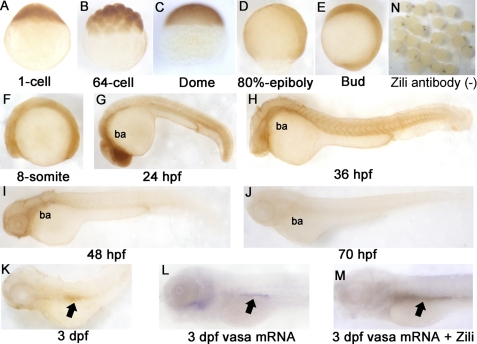
Later, Houwing et al. (11) submitted another 3389-bp sequence related to zili gene under accession number FJ168029. They analyzed Zili protein expression in primordial germ cells (PGCs) by an antibody raised against the N terminus of Zili, and found that Zili protein can be detected in PGCs from 3 days post-fertilization onwards. However, they did not detect the whole mount spatiotemporal expression pattern of Zili. Here, we also raised the antibody in rabbits according to the synthetic peptide MDPKRPTFPSPPGVI+C published by Houwing et al. (11) and analyzed the spatiotemporal expression pattern of Zili by using whole mount IHC. Consistent with Houwing et al., Zili can be detected in PGCs at 3 days post-fertilization (Fig. 1, K–M). However, expression of Zili also can be detected in axis before 3 days post-fertilization. The results show that the Zili protein appears in the one-cell stage and in all blastodermal cells until the dome stage (Fig. 1, A–C). At the onset of gastrulation, Zili expression starts to concentrate on the axis (Fig. 1D). When segmentation starts, Zili is expressed in axis ubiquitously (Fig. 1, E and F), and then branchial and pharyngeal arches gradually gain stronger expression (Fig. 1G). Interestingly, Zili expression on the axis begins to weaken after Prim-5 stage (24 hpf) (Fig. 1, H–K). At 3 days post-fertilization, Zili can be detected in PGCs easily but not in other parts of embryo.
FIGURE 1.
Spatiotemporal expression pattern of Zili protein in zebrafish embryos at indicated stages. Detection by whole mount IHC is shown. A–C, lateral views with the animal pole oriented at the top; D and E, lateral views with the anterior oriented toward the top; F–M, lateral views with the anterior oriented toward the left. ba, branchial and pharyngeal arches. PGCs are indicated by arrows in K–M. N, IHC without Zili antibody as negative control. dpf, days post-fertilization.
Next, expression of zili mRNA during zebrafish embryogenesis was examined by using whole mount in situ hybridization, and two nonoverlapping probes for zili, localizing at positions 252–1293 bp (probe 1) and 2763–3626 bp (probe 2), were used to demonstrate specificity and obtain identical spatially restricted expression patterns (20). The results of in situ hybridization show that spatiotemporal expression pattern of zili transcript is similar to Zili protein (supplemental Fig. S2). Differently, expression of zili transcript is detectable weakly in two clusters both laterally from the eight-somite stage onwards (supplemental Fig. S2, D–S). Double staining with zili and vasa or nanos1, PGCs markers, shows that the both lateral cluster domains of zili colocalize with the vasa or nanos1 expression domain, and the results detected by probe 2 are consistent with probe 1 (supplemental Fig. S3), suggesting the specificity and identical spatially restricted expression patterns of zili transcript. Additionally, we also performed reverse transcription-PCR to detect zili mRNA from the one-cell stage (supplemental Fig. S3M), and the amplified fragment was confirmed by sequencing. These expression patterns suggest that zili is maternal expressed and plays a role not only in PGCs development but also in early axis formation especially before 24 hpf.
Zili Functions to Inhibit Mesoderm Formation
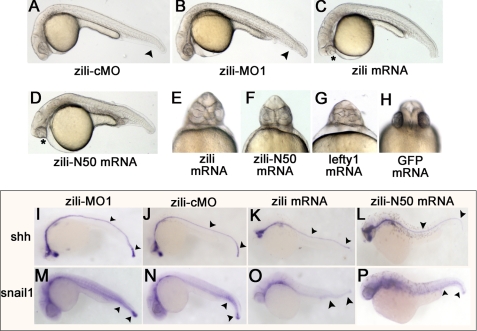
To study the function of zili, endogenous expression of zili was knocked down by injecting a translation blocker (zili-MO1) antisense morpholino, which was able to block production of the Zili-GFP fusion protein from a Zili-GFP fusion expression plasmid (supplemental Fig. S4, A–C). Embryos injected with 5 ng of zili-MO1 exhibited dorsalized phenotypes at 24 hpf, including loss of the ventral tail fin and a shorter tail (Fig. 2B), which were described by Mullins et al. (23). To test the specification of zili-MO1, zili mRNA was synthesized for rescuing the phenocopy mediated with zili-MO1. The results showed that 30 pg of zili mRNA can relieve the dorsalized phenotypes in 86% (n = 147) embryos (supplemental Fig. S4, E and J), suggesting that zili-MO1 could specifically block translation of zili mRNA. Furthermore, according to the recommendations of Eisen and Smith (24), a splice inhibitor (zili-MO2) was designed and synthesized against zili to identify the specific effects of zili-MO1 once again. Consistent with zili-MO1, injected embryos also exhibited dorsalized phenotypes (supplemental Figs. S4, F–J, and S5), suggesting that dorsalization is the specific phenotype caused by zili knockdown, and injection of 5 ng zili-MO1 was used in experiments below.
FIGURE 2.
Effects of zili knockdown and overexpression in zebrafish embryos. Lateral (A–D) and ventral (E–H) views of live embryos at 24 hpf are shown. A, embryos injected with 5 ng of zili-cMO as control did not show abnormal morphology. B, injection with 5 ng of zili-MO1 led to loss of the ventral tail fin and a shorter tail. The arrowheads in A and B show the tail fins and tails. C, injection with zili mRNA resulted in partial or complete fusion of the eyes (asterisk) and partial loss of the notochord and tail reduction. D, the phenotype injected with zili-N50 mRNA is similar to complete zili mRNA. E–H, fusion of eyes caused by zili overexpression (E) and zili-N50 mRNA (F) resembled the phenotype of lefty1 mRNA injection (G) or 30 pg of GFP mRNA injection as control (H). I–P, expression patterns of marker genes in embryos injected with 5 ng of zili-MO1 (I and M), 5 ng of zili-cMO (J and N), 30 pg of zili mRNA (K and O), or 50 pg of zili-N50 mRNA (L and P). The embryos at 24 hpf for shh and snail1 are shown in lateral views with the anterior oriented toward the left. The arrowheads in I–L show the floor plates and tail buds, and those in M–P show the caudal somites and tail buds.
To investigate the role of zili in embryonic development, we then injected zebrafish embryos with synthetic zili mRNA. Injection with 30 pg of zili mRNA caused partial or complete fusion of the eyes (Fig. 2, C and E) in 11% (n = 12) embryos at 24 hpf, resembling the lefty1 mRNA injection (Fig. 2G), which result from insufficient Nodal signaling of TGF-β pathway (25). Overexpression of zili also caused a partial loss of the notochord and tail reduction. Injection of zili mRNA also led to reduction of axial and lateral mesodermal marker shh and snail1 expression at 24 hpf (Fig. 2, I–K and M–O). These results suggest that zili functions to inhibit mesoderm formation.
50 Amino Acids at N Terminus of Zili (Zili-N50) Function Similar to Complete Zili Protein during Early Embryogenesis
Evidence above suggests that the lack of Zili protein lead to dorsalization of embryo in early development of zebrafish. However, Houwing et al. (11) reported that the screened zili mutant ziliG51STOP does not display any patterning defects, suggesting that 50 amino acids at N terminus of Zili protein may play a role to maintain the normal early embryogenesis. So we isolated the sequence encoding the 50 amino acids at the N terminus of Zili protein and transcribed to mRNA. Injection of 50 pg of Zili-N50 mRNA to the one-cell stage embryos also caused fusion of the eyes, partial loss of notochord, tail reduction, and reduction of shh and snail1 expression at 24 hpf (Fig. 2, D, F, L, and P). Moreover, we asked whether Zili-N50 could rescue the deficiency of complete Zili protein and maintain normal embryogenesis. Coinjection of Zili-N50 mRNA and zili-MO1 or 2 were performed, and phenocopy of dorsalization caused by zili-MOs can be recovered (supplemental Fig. S4, K–M). The results of the patterning defect caused by zili-N50 mRNA similar to complete zili mRNA and effects of zili-N50 mRNA to recover zili-MOs suggest that a role for zili-N50 is similar to complete Zili protein during early embryogenesis. This may provide some explanation as to why the mutant ziliG51STOP does not show any patterning defect during early embryogenesis.
Zili Inhibits the Activity of Nodal Factor sqt and cyc
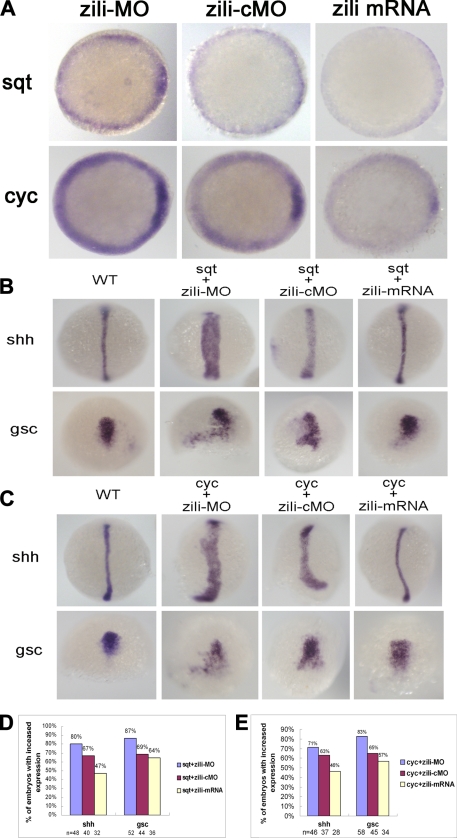
Because zili overexpression caused fusion of the eyes, suggesting inhibition of Nodal signaling of TGF-β pathway, we asked whether zili was involved in Nodal signals and mediated their activities. We analyzed Nodal signaling factor sqt and cyc expression in the zebrafish embryos in which activities of the zili were altered transiently. Injection with zili-MO induced sqt and cyc expression in 54% (n = 28) and 64% (n = 36) embryos and zili mRNA abolished these factors in 96% (n = 40) and 45% (n = 20) embryos at the 50% epiboly stage (Fig. 3A).
FIGURE 3.
Genetic interactions between zili and nodal signals. A, sqt and cyc expression in embryos injected with zili-MO or zili-cMO or zili mRNA. Embryos at the 50% epiboly stage are shown in animal pole views with the dorsal oriented toward the right. B and C, expression of the marker genes (indicated on the left). Dorsal views with the animal pole oriented toward the top for shh and gsc at bud and at the 60% epiboly stage are shown. D and E, statistical data for B and C, respectively. WT, wild type.
Subsequently, we tested the genetic interaction between zili and sqt/cyc that is essential for the formation of the axial mesoderm. Coinjection with 10 pg of zebrafish sqt mRNA and zili-cMO led to expanded expression of shh in 67% embryos (n = 40) at the bud stage. When the same amount of sqt mRNA and zili-MO were coinjected, the percentage of expanded shh increased to 80% (n = 48). In contrast, coinjection with sqt mRNA and 30 pg of zili mRNA caused the percentage of embryos with expanded shh expression decreasing to 47% (n = 32), and a similar case occurs when we examine the dorsal marker gsc (26) expression (Fig. 3, B and D). Furthermore, the genetic interaction between zili and cyc was consistent with zili and sqt. The percentage of expanded expression of shh and gsc decreased with the increased zili expression (Fig. 3, C and E). These results suggest that zili antagonizes the Nodal signal activity of the TGF-β pathway during the early development of zebrafish embryos.
Zili Suppresses Nodal Signaling by Physically Associating with Smad4 and Preventing the Formation of Smad2/3/4 Complex
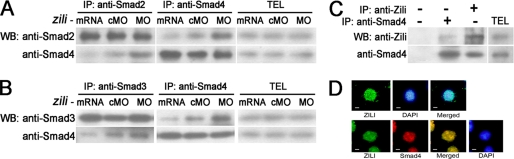
Nodal pathway activity is transduced by the R-Smads Smad2 and/or Smad3, together with the common mediator-Smad (co-Smad) Smad4 (16, 27). The Smad2/3 interacts with Smad4 to activate responsive gene expression. So we asked whether zili could inhibit the endogenous interaction between Smad2/3 and Smad4. Immunoprecipitation experiments in zebrafish embryos revealed that the amount of Smad2/3 coimmunoprecipitated with Smad4 decreased in the presence of the increasing amount of Zili (Fig. 4, A and B), although neither the Smad2/3 level nor the Smad4 level in total embryo lysate was altered. Next, to test the physical interaction between Zili and Smad2/3/4, we performed reciprocal immunoprecipitation of endogenous Zili and Smad2/3/4 in zebrafish embryos. The result indicated that Zili was coimmunoprecipitated with Smad4 (Fig. 4C) and not with Smad2/3 (data not shown). In addition, the receptor-mediated phosphorylation of Smad2 could not be altered by ectopic Zili expression (supplemental Fig. S6). Immunostaining in zebrafish embryo cells at the 50% epiboly stage revealed that Zili was located almost in the nuclei and overlapped well with Smad4 in the nucleus (Fig. 4D). Thus, Smad4 is a binding partner of Zili. These results suggest that Zili inhibits the interaction between Smad2/3 and Smad4 solely by competing with Smad2/3 for Smad4.
FIGURE 4.
Zili binds to Smad4 and inhibits the interaction of Smad4 with Smad2/3. A–D were performed in 50%-epiboly embryos. A and B, zili suppressed endogenous association of Smad2/3 with Smad4 in embryos. TEL indicates total embryos lysate, which is consistently used in C. C, endogenous interaction between Zili and Smad4. D, subcellular localization of FLAG-Zili and colocalization of FLAG-Zili (green) and HA-Smad4 (red) in embryo cells. 4′,6′-diamino-2-phenylindole (DAPI) was used to identify nuclei. Scale bars, 10 μm. WB, Western blot; IP, immunoprecipitation.
Zili Interacts with MH2 Domain of Smad4 via Its N-terminal Domain
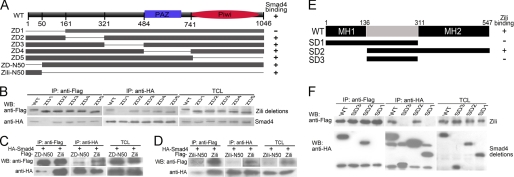
To further consolidate interaction between Zili and Smad4, we constructed a series of Zili protein deletion mutants to identify the functional domains of Zili that are required for interacting with Smad4. The Zili mutant ZD1, deletion of 161 amino acids at the N terminus of the Zili protein, failed to bind to Smad4, whereas other deletion mutants, including PAZ and Piwi conserved domain deletion, retained the ability to bind Smad4 (Fig. 5, A and B), suggesting that the 161 amino acids at N terminus of Zili protein is responsible for interaction with Smad4. This result and the discovery of Piwil2 in mouse (Mili) interacting with Tdrd1 by Wang et al. (28) suggest that the N terminus is an important functional region to Piwil2. Then we constructed Smad4 protein deletion mutants to identify the domain of Smad4 for binding to Zili. The results show that the MH2 domain of Smad4 is the binding site of Zili (Fig. 5, E and F), which is same site to which Smad2/3/1/5/9 bind (29). In addition, interaction between Zili and Smad4 was also validated by the two-hybrid experiment (supplemental Fig. S7).
FIGURE 5.
The specificity and domains of Zili-Smad4 interaction. A, schematic of different Zili deletion constructs. B–D, interactions between HA-Smad4 and different Zili deletion mutants with FLAG tag. E, schematic of different Smad4 deletion constructs. F, interactions between FLAG-Zili and different Smad4 deletion mutants with HA tag. The assays were performed in human HEK293 cells, TCL indicates total cell lysate, which is consistently used in Fig. 6. WB, Western blot; IP, immunoprecipitation; WT, wild type.
50-Amino Acid Region at N Terminus of Zili Protein Is Important for Binding to Smad4
According to the suggestion of mutant ziliG51STOP and the role for Zili-N50 in early embryogenesis described above, we asked whether the isolated 50 amino acids at the N terminus of the Zili protein could bind to Smad4. First, deletion of 50 amino acids at the N terminus of the Zili protein (ZD-N50) was constructed for the coimmunoprecipitation with Smad4. The result shows that the ability of Zili protein without the 50 amino acids at the N terminus to bind to Smad4 is obviously weaker than complete protein (Fig. 5C), suggesting that the 50-amino acid region at the N terminus of Zili is important for binding to Smad4. Then interaction of Smad4 with the isolated 50 amino acids (Zili-N50) was examined. As shown in Fig. 5D, Zili-N50 was coimmunoprecipitated with Smad4. Our results show that Zili functions in early embryogenesis, and the N terminus is the key region to this process. Houwing et al. (11) focus on the role for Zili in germ cell and Piwi-interacting RNAs, the well known function of Piwi protein depending on PAZ and PIWI domain (6). Taken together, our results and the well known function suggest that zili gene has multiple functional regions and possesses functional diversity, participating in different biological processes at different developmental stages.
Zili Also Suppresses BMP Signaling by Physically Associating with Smad4 and Preventing the Formation of Smad1/5/9/4 Complex
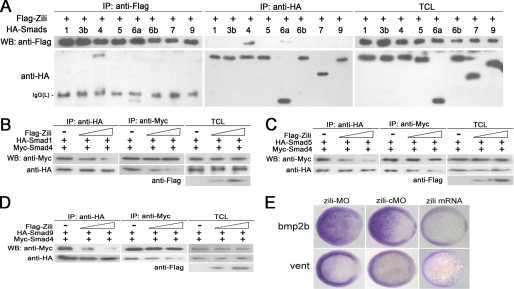
Because Smad4 is the co-Smad of TGF-β pathway and related to BMP signaling (12, 13, 30), we investigated the interaction between Zili and the rest of the Smads. In HEK293 cells, with Smad4 as a positive control, overexpressed Zili bound weakly to Smad6a, which is not the binding partner of Smad4; interaction with other Smads, including Smad1, 3b, 5, 6b, 7, and 9, was almost undetectable (Fig. 6A). This result suggests that Zili may compete with Smad1/5/9 for Smad4 to inhibit BMP signaling. Immunoprecipitation experiments showed that Zili overexpression inhibited the interaction of Smad4 with Smad1/5/9 in a dose-dependent manner (Fig. 6, B–D). These effects were confirmed by examination of the expression of the BMP ligand bmp2b and the known BMP transcriptional target vent (31) at the Shield stage (Fig. 6E).
FIGURE 6.
Zili prevents the formation of complex Smad1/5/9/4 and inhibits BMP signaling. A–D were performed in human HEK293 cells too. A, interactions of Zili with different zebrafish Smads. B–D, binding of Myc-Smad4 to HA-Smad1/5/9 was inhibited by coexpression of an increasing amount of FLAG-Zili (1 and 4 μg). E, bmp2b and vent expression in embryos injected with zili-MO or zili-cMO or zili mRNA. The embryos at the shield stage are shown in animal pole views with the dorsal oriented toward the right. Injection with zili-MO induced bmp2b and vent expression and zili mRNA abolished these factors in embryos. WB, Western blot; IP, immunoprecipitation; TCL, total cell lysate.
DISCUSSION
As described by Houwing et al. (11), the ziliG51STOP/G51STOP mutant is a good model for elucidating the role for Zili in Piwi-interacting RNAs control and germ cell differentiation. As shown by Houwing et al. in their Fig. 3B, by whole mount in situ hybridization of wild type and ziliG51STOP/G51STOP (zili−/− in the figure) gonads at 6 weeks post-fertilization (upper panel) and 7 weeks post-fertilization (lower panel), using an antisense probe for vasa, the authors observed a strong reduction in the amount of vasa-positive cells, which are completely lost at 8 weeks post-fertilization. Consistent with this result, with careful observation, supplemental Fig. S7A in Houwing et al. (11) paper shows that the brown staining reduces remarkably but is still clearly visible in the ziliG51STOP/G51STOP (zili−/− in the figure) gonads at 6 weeks of age compared with wild type siblings, suggesting the existence of protein that can be recognized by the Zili antibody used in the study. In brief, the remarkable reduction in the number of germ cells in zili mutant (ziliG51STOP/G51STOP) gonads at 6 weeks of age compared with wild type siblings (Fig. 3B in Houwing's paper) provides an explanation as to why the brown staining (indicating the existence of protein recognized by the Zili antibody) is reduced but visible in the mutant (ziliG51STOP/G51STOP) gonads at 6 weeks of age (supplemental Fig. S7A in Houwing's paper), and this explanation is evidenced by the significant reduction of Ziwi (red) protein in the mutant (ziliG51STOP/G51STOP) gonads at 6 weeks post-fertilization compared with wild type siblings (Fig. 3C in Houwing's paper). Considering that the mutant (G51STOP) identified by sequencing by Houwing et al. cannot encode entire Zili protein, the above results suggest the existence of a mutant Zili (G51STOP) protein that can also be recognized by the antibody raised against the N terminus of Zili (11) in the zili mutant (ziliG51STOP/G51STOP).
Morpholino antisense oligonucleotides function by binding to premessenger or messenger RNA, blocking access to the RNA. Diminution of the entire Zili protein induced by translation blocker (zili-MO1) and splice inhibitor (zili-MO2) morpholino used in this study was identified by the Zili antibody raised by the synthetic peptide published by Houwing et al. (supplemental Fig. S5B), and the specification of the zili-MOs is also confirmed (supplemental Fig. S4). Logically, the fact that the mutant (ziliG51STOP/G51STOP) screened by Houwing et al. does not display any patterning defect suggests that the 50 amino acids at the N terminus of the Zili protein may play a role in maintaining normal early embryogenesis. Our analysis of the 50 amino acids at the N terminus of the Zili protein may provide some explanation as to why the mutant ziliG51STOP keeps normal patterning during early embryogenesis.
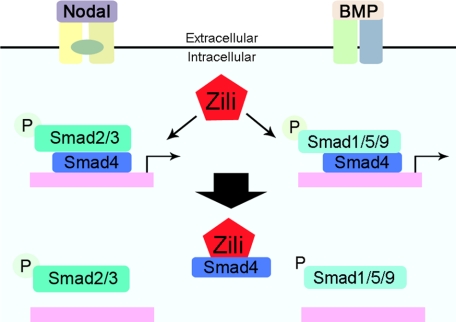
Stringent control of TGF-β signaling through the regulation of Smads is critical for normal embryo patterning. Smad4, co-Smad in TGF-β pathway, is involved in Nodal and BMP signaling and regulates genes related to different biological progress. Assembly of the Smad activator complex allows subsequent nuclear import of R-Smads. The inhibition of the interaction of Smad2/3 and Smad1/5/9 with Smad4 conversely serves as an important countermechanism for TGF-β signaling (32–34). Here we present several lines of evidence demonstrating that Zili inhibits the complex formation of phosphorylated R-Smads with Smad4, which provides some insights into how TGF-β signaling is terminated. First, zili inhibits the transcriptional activity of the Nodal and BMP ligands sqt, cyc, and bmp2b (Figs. 3A and 6E). Second, zili antagonizes the activity of Nodal and BMP signaling mediated with sqt, cyc, and bmp2b by detecting the marker genes shh, gsc, and vent (Figs. 3, B–E, and 6E). Third, overexpression of zili weakens the interaction of Smad4 with Smad2/3 and Smad1/5/9 (Figs. 4, A and B, and 6, B–D). Lastly, Zili binds to Smad4 and colocalizes in nucleus (Fig. 4, C and D). Because the receptor-mediated phosphorylation of Smad2 could not be affected by ectopic zili expression, Zili demolishes the complex of Smad2/3/4 and Smad1/5/9/4 solely by competing with Smad2/3 and Smad1/5/9 for Smad4 to inhibit the activity of TGF-β signaling (Fig. 7).
FIGURE 7.
A schematic model for antagonizing effects of zili on the TGF-β signaling. Zili precludes Smad4 from binding the Smad2/3 and Smad1/5/9.
To study the function of piwil2 gene in zebrafish (zili), we examined its expression patterns during early embryogenesis and got clues that zili played a role not only in PGC development but also in early axis formation. Ectopic expression of zili on zebrafish embryo development suggested that zili inhibited TGF-β signaling and mesoderm formation. Subsequently, the results of interaction between Zili and Smads proteins showed that Zili was a binding partner of Smad4 and prevented formation of the Smad2/3/4 and Smad1/5/9/4 complexes. Briefly, this work shows that zili functions to early embryogenesis beyond the germ line as a novel negative regulator of TGF-β signaling by interacting with Smad4, extending the function of Piwi proteins in vertebrates. Considering that inactivation of Smad4-mediated pathway could lead to cancer and mammal Piwil2 functioned in cancer and testis, our study might offer a new perspective to other biological approaches.
Supplementary Material
Acknowledgments
We thank Dr. Shaohua Yao for providing plasmids; Prof. Qiaqing Wu and Dr. Dongsheng Ren for providing BacterioMatch® II Two-Hybrid System XR Plasmid cDNA Library; and Dr. Hanshuo Yang and Fanxin Ma for technical support.
This work was supported by National Natural Science Foundation of China Grants 30770812 and 90919006 and National High-Tech Research and Development Program of China Grant 2008AA02Z102.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
- TGF
- transforming growth factor
- BMP
- bone morphogenetic protein
- R-Smad
- receptor-activated Smad
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- IHC
- immunohistochemistry
- PGC
- primordial germ cell
- hpf
- hours post-fertilization.
REFERENCES
- 1.Aravin A. A., Hannon G. J., Brennecke J. (2007) Science 318, 761–764 [DOI] [PubMed] [Google Scholar]
- 2.Cox D. N., Chao A., Baker J., Chang L., Qiao D., Lin H. (1998) Genes Dev. 12, 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng W., Lin H. (2002) Dev. Cell 2, 819–830 [DOI] [PubMed] [Google Scholar]
- 4.Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Lin H., Matsuda Y., Nakano T. (2004) Development 131, 839–849 [DOI] [PubMed] [Google Scholar]
- 5.Carmell M. A., Girard A., van de Kant H. J., Bourc'his D., Bestor T. H., de Rooij D. G., Hannon G. J. (2007) Dev. Cell 12, 503–514 [DOI] [PubMed] [Google Scholar]
- 6.Seto A. G., Kingston R. E., Lau N. C. (2007) Mol. Cell 26, 603–609 [DOI] [PubMed] [Google Scholar]
- 7.Lee J. H., Schütte D., Wulf G., Füzesi L., Radzun H. J., Schweyer S., Engel W., Nayernia K. (2006) Hum. Mol. Genet. 15, 201–211 [DOI] [PubMed] [Google Scholar]
- 8.Sugimoto K., Kage H., Aki N., Sano A., Kitagawa H., Nagase T., Yatomi Y., Ohishi N., Takai D. (2007) Biochem. Biophys. Res. Commun. 359, 497–502 [DOI] [PubMed] [Google Scholar]
- 9.Tan C. H., Lee T. C., Weeraratne S. D., Korzh V., Lim T. M., Gong Z. (2002) Gene Expr. Patterns 2, 257–260 [DOI] [PubMed] [Google Scholar]
- 10.Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D. V., Blaser H., Raz E., Moens C. B., Plasterk R. H., Hannon G. J., Draper B. W., Ketting R. F. (2007) Cell 129, 69–82 [DOI] [PubMed] [Google Scholar]
- 11.Houwing S., Berezikov E., Ketting R. F. (2008) EMBO J. 27, 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitman M., Raftery L. (2005) Development 132, 4205–4210 [DOI] [PubMed] [Google Scholar]
- 13.Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 14.ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 15.Tian T., Meng A. M. (2006) Cell Mol. Life Sci. 63, 672–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen M. M. (2007) Development 134, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 17.Westerfield M. (1993) The Zebrafish Book, University of Oregon Press, Eugene, OR [Google Scholar]
- 18.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 19.Kaslin J., Nystedt J. M., Ostergård M., Peitsaro N., Panula P. (2004) J. Neurosci. 24, 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thisse C., Thisse B. (2008) Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 21.Knaut H., Pelegri F., Bohmann K., Schwarz H., Nüsslein-Volhard C. (2000) J. Cell Biol. 149, 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ober E. A., Schulte-Merker S. (1999) Dev. Biol. 215, 167–181 [DOI] [PubMed] [Google Scholar]
- 23.Mullins M. C., Hammerschmidt M., Kane D. A., Odenthal J., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C. P., Jiang Y. J., Kelsh R. N., Nüsslein-Volhard C. (1996) Development 123, 81–93 [DOI] [PubMed] [Google Scholar]
- 24.Eisen J. S., Smith J. C. (2008) Development 135, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 25.Cheng S. K., Olale F., Brivanlou A. H., Schier A. F. (2004) PLoS Biol. 2, E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gritsman K., Zhang J., Cheng S., Heckscher E., Talbot W. S., Schier A. F. (1999) Cell 97, 121–132 [DOI] [PubMed] [Google Scholar]
- 27.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Saxe J. P., Tanaka T., Chuma S., Lin H. (2009) Curr. Biol. 19, 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attisano L., Wrana J. L. (2002) Science 296, 1646–1647 [DOI] [PubMed] [Google Scholar]
- 30.Kondo M. (2007) FEBS J. 274, 2960–2967 [DOI] [PubMed] [Google Scholar]
- 31.Ramel M. C., Lekven A. C. (2004) Development 131, 3991–4000 [DOI] [PubMed] [Google Scholar]
- 32.Inman G. J., Nicolás F. J., Hill C. S. (2002) Mol. Cell 10, 283–294 [DOI] [PubMed] [Google Scholar]
- 33.Xu L., Kang Y., Cöl S., Massagué J. (2002) Mol. Cell 10, 271–282 [DOI] [PubMed] [Google Scholar]
- 34.Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y. G., Meng A., Feng X. H. (2006) Cell 125, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.