Abstract
In trypanosomes, the predominant mechanisms of regulation of gene expression are post-transcriptional. The DEAD-box RNA helicase DHH1 was identified in a screen for gene products that are necessary for the instability of the GPI-PLC mRNA in insect-stage trypanosomes. Expression of an ATPase-deficient dhh1 mutant caused a rapid growth arrest associated with a decrease in polysomes, an increase in P-bodies and a slight decrease in average mRNA levels. However, the effect of dhh1 mutant expression on both turnover and translational repression of mRNAs was selective. Whereas there was little effect on the stability of constitutive mRNAs, the control of a large cohort of developmentally regulated mRNAs was reversed; many mRNAs normally downregulated in insect-stage trypanosomes were stabilized and many mRNAs normally upregulated decreased in level. One stabilised mRNA, ISG75, was characterised further. Despite the overall decrease in polysomes, the proportion of the ISG75 mRNA in polysomes was unchanged and the result was ISG75 protein accumulation. Our data show that specific mRNAs can escape DHH1-mediated translational repression. In trypanosomes, DHH1 has a selective role in determining the levels of developmentally regulated mRNAs.
Keywords: DHH1, Trypanosoma brucei, Microarray, Developmental gene regulation, Translation, ISG75
Introduction
The evolutionary divergence of trypanosomes from other model eukaryotes is reflected in several novel features in their biology. One example is gene expression; transcription by RNA polymerase II is constitutive, and monocistronic mRNAs arise through a coupled trans-splicing and polyadenylation reaction (LeBowitz et al., 1993; Matthews et al., 1994; Ullu et al., 1993). Consequently, most regulation of protein-coding gene expression has evolved to be dependent on post-transcriptional mechanisms (Clayton and Shapira, 2007; Haile and Papadopoulou, 2007), the exception being the regulated transcription by RNA polymerase I of a minority of loci that encode the superabundant cell-surface proteins (Gunzl et al., 2003; Kooter et al., 1987). Investigations into the regulation of genes transcribed by RNA polymerase II in trypanosomes have concentrated on the identification of factors that regulate mRNA levels and, in particular, half-life. The two developmental stages commonly used in the laboratory represent two successive life-cycle stages, the mammalian bloodstream form and the insect procyclic form, and work has concentrated on factors involved in the turnover of mRNAs that are differentially expressed between these two developmental forms. Several cis-elements necessary for correct mRNA levels and a smaller number of trans-acting factors regulating multiple mRNAs have been identified (e.g. Blattner and Clayton, 1995; Engstler and Boshart, 2004; Estevez, 2008; Furger et al., 1997; Hehl et al., 1994; Walrad et al., 2009).
The turnover of the majority of mRNAs is initiated by a shortening of the 3′ poly(A) tail (deadenylation) followed by one of two pathways of decay. The 3′-to-5′ exonucleolytic pathway is catalysed by the exosome (Dziembowski et al., 2007; Schaeffer et al., 2009) and is believed to be a major pathway in cultured mammalian cells (Mukherjee et al., 2002; Rodgers et al., 2002). The second pathway is initiated by hydrolysis of a phosphodiester bond in the 5′ cap (decapping) followed by 5′-to-3′ exonucleolytic degradation (Coller and Parker, 2004). In yeast, the majority of mRNA turnover, including that of both stable and unstable mRNAs, occurs through the 5′-to-3′ pathway (Cougot et al., 2004; LaGrandeur and Parker, 1999; Sheth and Parker, 2003). This pathway is also important for the turnover of some mRNAs in mammalian cells (Stoecklin et al., 2006)
The activities required for 5′-to-3′ mRNA decay are concentrated in cytoplasmic ribonucleoprotein processing bodies (P-bodies) that contain mRNA, the decapping (Dcp1 and Dcp2) and 5′-to-3′ hydrolytic enzymes (Xrn1) as well as proteins that enhance decapping — including Dhh1, Pat1, Edc1, Edc2 and Edc3 — and the Lsm1-7 complex (Balagopal and Parker, 2009; Franks and Lykke-Andersen, 2008; Parker and Sheth, 2007). In yeast, deadenylation of an mRNA results in the dissociation of translation initiation factors, and association with Lsm1-7 and the decapping enzyme (Tharun and Parker, 2001). The localisation of mRNAs to P-bodies seems to be one alternative to translation, and artificially altering the number of polysomes causes a rapid change in the number and/or size of P-bodies (reviewed in Balagopal and Parker, 2009; Franks and Lykke-Andersen, 2008; Parker and Sheth, 2007). Cycloheximide increases the number of polysomes and causes a reduction in P-body number and size, whereas puromycin releases mRNA from polysomes and increases the size and number of P-bodies in yeast (Sheth and Parker, 2003), mammals (Cougot et al., 2004) and trypanosomes (Holetz et al., 2007; Kramer et al., 2008). P-bodies respond to the amount of non-translated mRNA, and the movement of mRNA to and from P-bodies is also dynamic. Either degradation or mRNA storage can occur; subsequent recycling back to translation is particularly apparent after release from growth arrest (Bhattacharyya et al., 2006; Brengues et al., 2005). Furthermore, microscopically visible P-bodies are not required for mRNA turnover (Eulalio et al., 2007). It is likely that any cell contains a dynamic population of P-bodies of different sizes and those visualised by fluorescence microscopy are at the upper end of the size range (Franks and Lykke-Andersen, 2008).
How is the equilibrium between inclusion and exclusion from polysomes regulated? In yeast, the DEAD box RNA helicase Dhh1 (orthologous with Cgh-1 in Caenorhabditis elegans, Me31B in Drosophila, Xp54 in Xenopus and RCK in mammals) is an integral part of a molecular switch that removes an individual mRNA from translation. Dhh1 and Pat1 act in parallel pathways; Δdhh1 Δpat1 cells are unable to dissemble polysomes on glucose starvation (Coller and Parker, 2005) and overexpression of Dhh1 is sufficient to cause the vast majority of polysomes to disassemble (Coller and Parker, 2005). Because Dhh1 is concentrated in P-bodies, it is possible that the Dhh1 molecules that participate in the decision to exit translation accompany the mRNA from the cytoplasm to P-bodies.
The role of Dhh1 in removing mRNAs from translation is particularly apparent in oocytes and early embryos prior to the onset of zygotic transcription, because decapping does not occur in response to shortening of the poly(A) tail (Gillian-Daniel et al., 1998). Dhh1 is required for repression of translation of maternal mRNAs until the appropriate stage (Boag et al., 2008; Ladomery et al., 1997; Minshall et al., 2001; Nakamura et al., 2001; Navarro et al., 2001; Noble et al., 2008; Weston and Sommerville, 2006). In Xenopus oocytes, tethering of Dhh1 (Xp54) to a reporter mRNA was sufficient to repress translation if the mRNA was deadenylated and the Dhh1 was catalytically (ATPase) active (Minshall et al., 2009). The function of Dhh1 extends beyond the decision to remove an mRNA from polysomes: it enhances decapping and is present in complexes containing components of the degradation pathway (Coller et al., 2001; Fischer and Weis, 2002).
The 5′-to-3′ pathway for mRNA degradation in trypanosomes has been partially characterised. Orthologues of the deadenylase complexes are present (Schwede et al., 2008) and P-bodies have been defined by the presence of DHH1, SCD6 (orthologous with RAP55/LSm14 in vertebrates, Car-1 in C. elegans and Trailer hitch in Drosophila), and XRNA (the orthologue of Xrn1) (Holetz et al., 2007; Kramer et al., 2008; Li et al., 2006). In contrast to yeast and metazoa, components of the decapping enzyme (Dcp1 and Dcp2) as well as many enhancers of decapping (Lsm1, Edc3 and Pat1) are not readily identifiable in the trypanosome genome. The presence of XRNA in P-bodies is consistent with a role in mRNA degradation but the absence of the decapping enzyme and many accessory factors suggests an alternative pathway for initiation of mRNA degradation.
Here, the function of DHH1 in mRNA turnover in insect stage (procyclic) Trypanosoma brucei has been investigated. Two phenotypes have been characterised; the first produced by ~twofold overexpression of wild-type DHH1 and the second by expression of an ATPase-inactive dhh1 mutant at levels less than the endogenous DHH1. The two phenotypes were qualitatively similar but the expression of the dhh1 mutant caused quantitatively greater changes. Expression caused a decrease in polysomes, an increase in the number of visible P-bodies and a rapid growth arrest consistent with a repression of translation. There was a selective effect on mRNA levels: most were not greatly affected, whereas developmentally regulated mRNAs showed altered behaviour. A group of mRNAs normally downregulated in insect-relative to mammalian-form trypanosomes was stabilised. By contrast, the group of mRNAs that declined most rapidly was enriched in mRNAs normally upregulated in insect-relative to mammalian-form trypanosomes. The data provide evidence for selective action of DHH1 on developmentally regulated mRNAs in trypanosomes.
Results
DHH1 knockdown causes a stabilisation of the unstable GPI-PLC mRNA in procyclic trypanosomes
GPI-PLC mRNA is 50-fold less abundant in insect procyclic trypanosomes than in mammalian bloodstream forms. The major contributor to this differential expression is the instability of the mRNA in procyclic cells, in which it has a half-life of 3 minutes (Webb et al., 2005). Using tetracycline-inducible RNAi (LaCount et al., 2000), a candidate-based screen was performed to identify genes necessary for the low levels of GPI-PLC mRNA in procyclic cells. One gene identified in this screen encoded the DEAD box RNA helicase DHH1; upon induction of RNAi there was a small but distinct increase in the steady-state levels of GPI-PLC mRNA (Fig. 1A). The knockdown caused a cessation of proliferation between 24 and 48 hours after induction and, after 48 hours, DHH1 was reduced to 5-10% of normal levels (Fig. 1B,C and data not shown). Attempts were made to generate a DHH1−/− cell line by targeted gene deletion. Whereas heterozygote DHH1+/− cell lines were readily obtained, several attempts to produce DHH1−/− cells failed, supporting the finding that DHH1 is essential in trypanosomes (Schwede et al., 2008). DHH1−/− cells would have been used to investigate any mRNA phenotype. The DHH1 RNAi cell lines did not present a readily accessible alternative, because DHH1 protein persisted for several cell cycles after induction, a timescale that made any analysis of the primary phenotype difficult owing to the problem to distinguish it from secondary effects caused by the growth arrest.
Fig. 1.
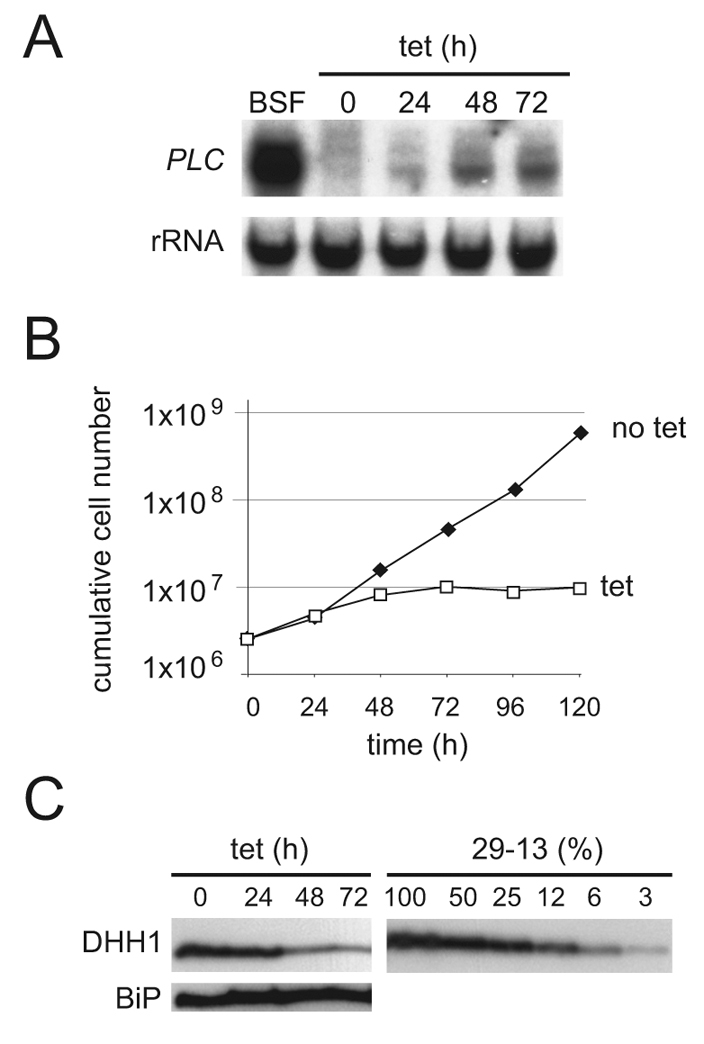
RNAi knockdown of DHH1 stabilises the GPI-PLC mRNA in procyclic cells. (A) GPI-PLC mRNA, (B) cell proliferation and (C) DHH1 protein were followed over a time course after induction of RNAi. For DHH1 protein, quantification was done by comparison with a titration of cell equivalents of the parental cell line T. brucei Lister 427 pLEW29:pLEW13 (Wirtz et al., 1999). One representative experiment of three is shown.
Overexpression of DHH1 and expression of dhh1 mutants causes a growth phenotype
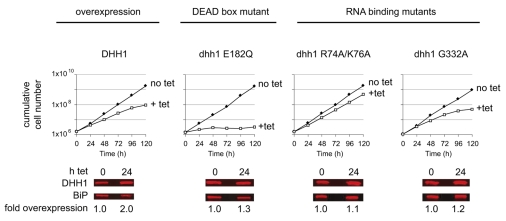
T. brucei DHH1 has 69% identity to yeast Dhh1 and the residues involved in catalytic activity and RNA binding are conserved (supplementary material Fig. S1). Wild-type and mutant forms of DHH1 were expressed from integrated tetracycline-inducible transgenes to investigate any role of DHH1 in mRNA turnover. Three dhh1 mutants were used: dhh1 E182Q carries a mutation in the DEAD-box motif, preventing ATP hydrolysis; two further mutants, dhh1 R74A/K76A and dhh1 G332A, were made based on equivalent mutations in yeast Dhh1 that greatly reduced RNA binding and were dominant for growth arrest and stabilisation of the unstable EDC1 mRNA (Cheng et al., 2005).
Wild-type and mutant forms of DHH1 were expressed in DHH1+/+ procyclic cells and, following induction, growth and DHH1 protein were monitored over a time course. The expression of dhh1 E182Q caused a rapid cessation of proliferation, whereas expression of wild-type DHH1, dhh1 R74A/K76A or dhh1 G332A resulted in a reduced growth rate (Fig. 2). None of the transgenes caused more than twofold overexpression after 24 hours, compared with wild type (Fig. 2). DHH1 is an abundant protein with an estimated ~400,000 molecules per cell (supplementary material Fig. S2), which is in several-fold excess over the estimated ~50,000 total mRNA molecules per cell (Haanstra et al., 2008). The wild-type level of DHH1 seems to be close to the maximum threshold for optimal growth, because DHH1+/− cell lines grew at the same rate as DHH1+/+ cells (data not shown), whereas twofold overexpression caused a decrease in growth rate (Fig. 2). The growth analysis was repeated with two further clones for each transgene: all three clones for each transgene produced similar growth phenotypes (supplementary material Fig. S3).
Fig. 2.
Growth phenotype after expression of DHH1 wild type and mutants. Cell proliferation after induction of DHH1, dhh1 E182Q, dhh1 R74A/K76A and dhh1 G332 transgenes. The increase in expression of DHH1 was determined by quantitative western blotting.
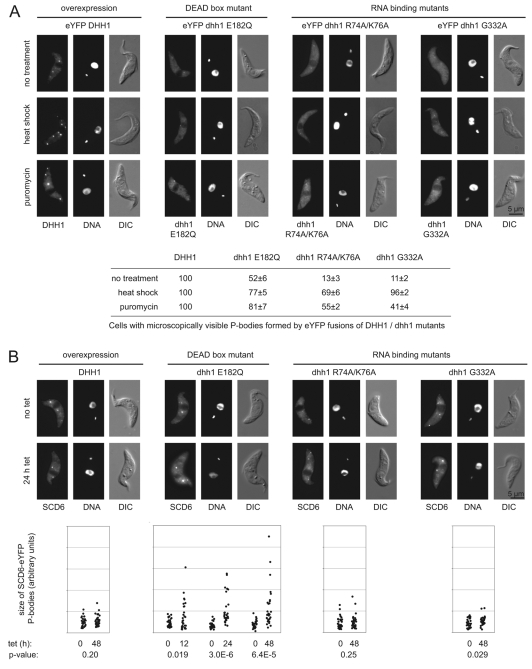
All three dhh1 mutants have impaired localisation to P-bodies and dhh1 E182Q causes an increase in P-bodies
eYFP-DHH1 is fully functional (Kramer et al., 2008), and the subcellular distribution of DHH1 wild type and mutants was determined using the cell lines expressing eYFP fusions. Both the dhh1 E182Q and the RNA-binding mutants showed reduced localisation to P-bodies when compared with the wild type. Wild-type eYFP-DHH1 localised into microscopically visible P-bodies in 100% of cells, whereas dhh1 E182Q, dhh1 R74A/K76A and dhh1 G332A did so in only 52%, 13% and 11% of cells, respectively (Fig. 3A and supplementary material Fig. S4). An increase in the number of P-bodies caused by either puromycin or heat shock (Kramer et al., 2008) slightly increased the localisation of the dhh1 mutants into P-bodies, but cells without visible localisation of mutant dhh1 to P-bodies were still common (Fig. 3A). These data indicate that RNA binding and ATPase activity are necessary for efficient localisation of DHH1 to P-bodies.
Fig. 3.
Subcellular localisation of DHH1 wild type and mutants. (A) Images of representative cells expressing eYFP-DHH1 wild type and mutants after 12 hours induction and the effect of heat shock and puromycin on the subcellular distribution. The percentage of cells (n=150) with microscopically visible P-bodies in each case is shown below. (B) Effect of expression of DHH1 wild type and mutants on subcellular localisation of SCD6-eYFP, an independent P-body marker. Representative images are shown 0 and 24 hours after induction. Quantitation of P-body size is shown below; 30 cells per time point were measured and P-values of a Student's t-test comparing uninduced and induced cells are shown.
The reduced localisation of the eYFP-dhh1 mutant proteins into P-bodies could be either an intrinsic feature of the mutant proteins or caused by a general reduction in P-bodies as a consequence of the expression of the mutants. These two possibilities were distinguished by expressing the DHH1 transgenes, without an eYFP tag, in a cell line expressing SCD6-eYFP, an independent P-body marker (Kramer et al., 2008). There was no significant change in P-body size or number following the expression of DHH1, dhh1 R76A/K74A or dhh1 G332A (Fig. 3B). dhh1-E182Q expression resulted in increased P-body size at 12 hours after induction without any decrease in number (Fig. 3B).
Expression of dhh1 E182Q results in a release of mRNA from polysomes
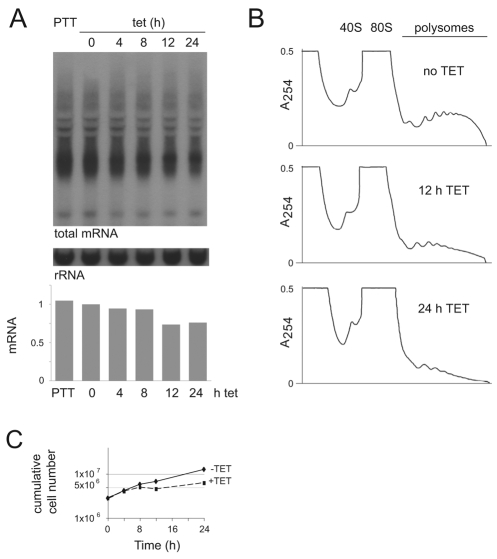
Expression of dhh1 E182Q produced a rapid phenotype, and all further experiments were concentrated on characterising this phenotype. The increase in size of P-bodies after dhh1-E182Q expression suggested a release of mRNAs from polysomes. Total mRNA levels and polysome profiles were determined over a time course after dhh1 E182Q induction (Fig. 4). Cycloheximide pretreatment of the cultures was not used (Coller and Parker, 2005) because it caused a clearly visible dissociation of P-bodies within a few minutes. The omission of cycloheximide did reduce the yield of polysomes, and subsequent experiments contained an assumption that the omission of cycloheximide had no selective effect on individual mRNAs (supplementary material Fig. S5).
Fig. 4.
Changes in total mRNA and polysomes after dhh1-E182Q induction. (A) Total mRNA was measured after induction of dhh1 E182Q by probing northern blots for the spliced leader present on all mRNAs. Values were normalised using the 0-hour time point. (B) Polysome analysis after induction of dhh1-E182Q expression. (C) Cell proliferation after induction of dhh1 E182Q to determine the point of arrest.
Total mRNA was measured by hybridising northern blots with an oligonucleotide complementary to the spliced leader present on all nuclear-encoded mRNAs. There was a gradual decrease in total mRNA to ~75% after 24 hours of dhh1-E182Q induction (Fig. 4A). By contrast, there was a marked reduction in polysomes after 12 hours of dhh1-E182Q induction and, by 24 hours, polysomes were several-fold lower than starting levels (Fig. 4B). Together, the data provide evidence that mRNAs are diverted from polysomes into P-bodies after dhh1-E182Q expression, although it remains unresolved whether the change is a direct effect of dhh1-E182Q expression or is caused by the growth arrest (Fig. 4C).
dhh1-E182Q expression results in increased levels of a subset of mRNAs
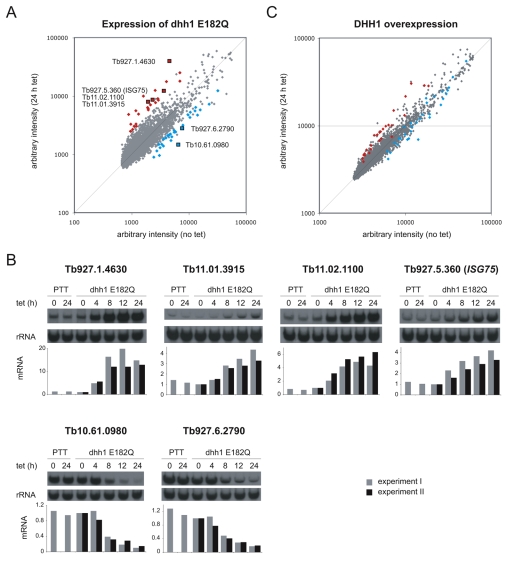
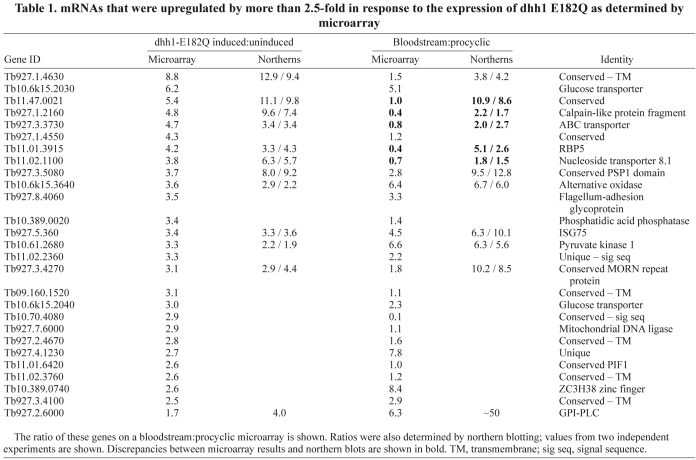
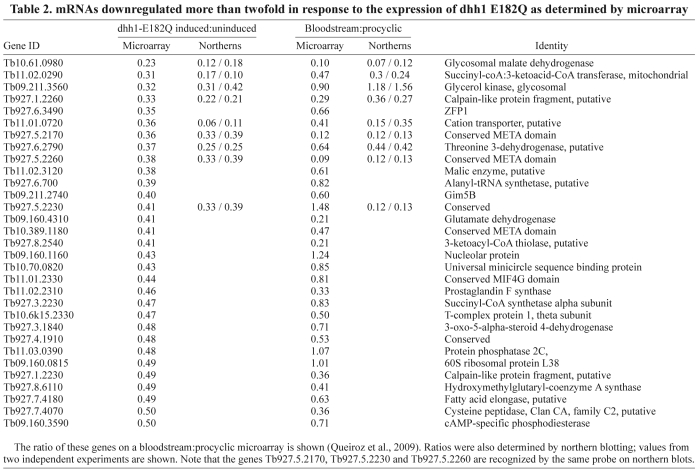
A microarray experiment comparing uninduced and 24-hour-induced cells was performed to determine the effects of dhh1-E182Q expression on individual mRNAs (ArrayExpress accession number: E-MEXP-2026). The data were normalised and filtered for quality and reproducibility, and pseudogenes were removed. The final data set contained 8276 genes, which is 79% of the putative genes present in the genome. Data normalisation was based on the standard assumption that the majority of mRNAs remained unchanged. However, induction for 24 hours caused a reduction of ~25% in total mRNA (Fig. 4A), and so an mRNA with a ratio of 1.0 (induced:uninduced) is actually reduced to ~75%. Taking this into account, arbitrary cut-offs were chosen: upregulated mRNAs had a ratio of >2.5 and downregulated mRNAs had a ratio of <0.5 (Fig. 5A). The lists of upregulated and downregulated mRNAs were then refined to remove those encoded by: (1) genes located in VSG sub-telomeric arrays, (2) genes related to variant surface glycoprotein or procyclin expression sites associated genes and (3) open reading frames that were unlikely to be real genes. The first two criteria were designed to eliminate incompletely characterised large gene families possibly transcribed by RNA polymerase I and under transcriptional control. Using these cut-offs, the vast majority of genes present in the final array data set were found to be unaltered, 26 mRNAs (0.4%) were upregulated, with the highest ratio being 8.8 (Table 1), and 31 genes were downregulated, with the lowest ratio being 0.23 (Table 2). Twelve of the upregulated and seven of the downregulated genes identified in the microarray experiment were validated by northern blotting. In all cases, the up- or downregulation was confirmed (Tables 1 and 2, and supplementary material Fig. S6A). Upregulation of one of the mRNAs, ISG75, was also confirmed using two further independent cell lines (supplementary material Fig. S6B).
Fig. 5.
Changes in mRNA after induction of expression of dhh1-E182Q or DHH1 transgenes. (A) Microarray dataset shown as a plot of the ratio of average intensities for each spot before and after dhh1-E182Q induction (the vast majority of genes are represented by a single spot). The most upregulated and most downregulated mRNAs (filtered as described in the text, see Tables 1 and 2) are coloured in red and blue, respectively. (B) The expression of four upregulated and two downregulated mRNAs, marked by black squares in A, was measured over a time course after induction of expression of dhh1 E182Q. Data from two independent experiments are shown. RNA from the parental cell line (Lister 427 PTT) with and without tetracycline is shown as a control. The data were normalised using the 0-hour time point. (C) Microarray dataset shown as plot of the ratio of average intensities for each spot before and after DHH1 transgene induction. For comparison, the mRNAs most up- and downregulated by the expression of dhh1 E182Q are coloured in red and blue, respectively.
Table 1.
mRNAs that were upregulated by more than 2.5-fold in response to the expression of dhh1 E182Q as determined by microarray
Table 2.
mRNAs downregulated more than twofold in response to the expression of dhh1 E182Q as determined by microarray
The microarray experiment used mRNA prepared 24 hours after induction, when cell proliferation had ceased (Fig. 4C). To determine whether the changes in mRNAs identified in the microarray experiment occurred prior to growth arrest, the expression of four upregulated and two downregulated mRNAs was measured over a time course after induction (Fig. 5B). For all upregulated mRNAs, the changes in mRNA levels were apparent after 4 hours, before growth arrest, which was first apparent at 8 hours (Fig. 4C). The selective and rapid onset of the increase in mRNA levels provides evidence that it is a direct result of dhh1-E182Q expression rather than a secondary effect of growth arrest. For the two downregulated mRNAs, the changes were significant after 8 hours but less obvious after 4 hours, and it is possible that the decrease is a secondary effect.
A second microarray experiment was performed to compare cells expressing wild-type DHH1 as a tetracycline-inducible transgene before and 24 hours after induction (ArrayExpress submission accession number: E-MEXP-2025; Fig. 5C). In this experiment, DHH1 expression was increased 2.0-fold, compared with 1.3-fold for the dhh1-E182Q microarray experiment. Wild-type DHH1 overexpression had little effect on total mRNA levels (supplementary material Fig. S7A), and there was only a small decrease in polysomes by 24 hours (supplementary material Fig. S7B). Alterations in individual mRNAs after expression of DHH1 were also smaller, as can be seen by comparing the deviation from the diagonals in Fig. 5A and 5C. Using the same filtering criteria as previously, there was a single upregulated mRNA, Tb927.1.4630, with a ratio of 2.6: the same mRNA that was most increased by dhh1-E182Q expression. The values obtained from the microarray were validated using northern blotting for ISG75 mRNA, which showed a 1.3- to 1.5-fold increase in three separate clones (supplementary material Fig. S7C). Both microarray experiments identified changes 24 hours after transgene induction. The validation by northern blotting indicated that mRNAs that changed rapidly after induction were correctly identified but it is likely that the microarray data identified some mRNAs that changed as a result of secondary effects.
The mRNAs most increased by DHH1 overexpression were the same as those most increased by dhh1-E182Q expression, and those most decreased by dhh1-E182Q expression were also decreased by DHH1 overexpression (Fig. 5C and supplementary material Tables S1 and S2). These data indicate that similar sets of mRNAs were affected in the two experiments, suggesting that the effect of dhh1-E182 expression was a stronger version of the mRNA phenotype caused by wild-type DHH1 overexpression.
dhh1-E182Q expression reverses both positive and negative developmental regulation of multiple mRNAs
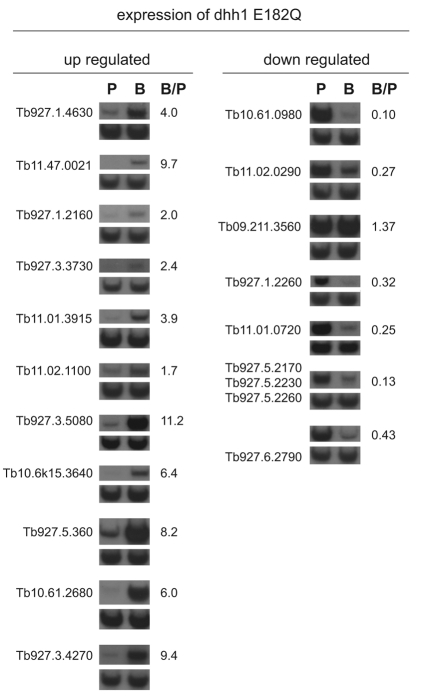
DHH1 was identified in a screen for genes necessary for mRNA instability that underlies developmentally regulated gene expression. A set of the mRNAs identified in the dhh1-E182Q microarray experiment was tested for developmentally regulated expression by northern blotting of mRNA from wild-type mammalian bloodstream and insect procyclic trypanosomes. Eleven mRNAs most increased by dhh1 E182Q were tested, and all eleven were >1.8 more abundant in bloodstream forms (Fig. 6 and Table 1) than insects forms. Seven mRNAs that decreased on dhh1-E182Q expression were also tested: six were expressed at levels between 2- and 14-fold higher in procyclic than in bloodstream forms, and one, the glycosomal glycerol kinase, was expressed equally in both developmental forms (Fig. 6 and Table 2). Thus, dhh1-E182Q expression in procyclic forms reversed the developmental regulation of a subset of mRNAs; the level of normally downregulated mRNAs increased and that of normally upregulated mRNAs decreased. It has been estimated that around 5% of mRNAs are differentially expressed between bloodstream and procyclic forms (Brems et al., 2005), so expression of dhh1 E182Q has a highly selective effect on developmentally regulated mRNAs.
Fig. 6.
Developmental regulation of mRNAs identified in the dhh1-E182Q microarray experiment. Quantification of relative mRNA expression in bloodstream and procyclic forms using northern blots with RNA from procyclic (‘P’) and bloodstream form cells (‘B’). Note that Tb927.5.2170, Tb927.5.2230 and Tb927.5.2260 are 99% identical and are recognised by the same probe. Data of two independent experiments are shown (one representative blot and average value of the quantification).
The most obvious functional connection linking the dhh1-E182Q-affected mRNAs is developmental regulation. Beyond that, it is difficult to be certain of a link, because many were of unknown function. However, mRNAs encoding proteins with predicted or known signal sequences and/or transmembrane helices were over-represented (54% compared with 21% in the array), a feature that was still apparent (46%) when the arbitrary cut-off was reduced from >2.5 to >2.0, producing an extended dataset of 67 genes (supplementary material Table S3). Of the 26 mRNAs most decreased by dhh1-E182Q expression, many encoded enzymes of central metabolism; procyclic forms utilize oxidative phosphorylation, whereas bloodstream forms do not.
Many attempts were made to obtain a bloodstream-form cell line carrying an inducible dhh1-E182Q transgene. Although many cell lines were isolated after selection, no cell line was recovered that uniformly expressed dhh1 E182Q. It is likely that there was selection against the transgene even with the very low levels of expression that occur in the absence of tetracycline.
The changes in mRNA levels correlate with changes in mRNA stability
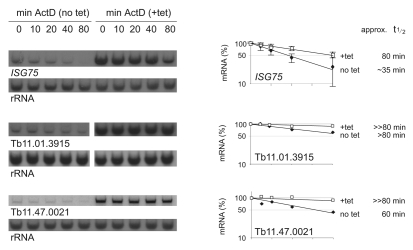
The changes in steady-state levels of individual mRNAs on expression of dhh1 E182Q could have resulted from a change in the rate of transcription, and/or post-transcriptional mechanisms, including a change in the rates of maturation, export to the cytoplasm and half-life in the cytoplasm. Three mRNAs whose levels were increased by dhh1-E182Q expression were selected and mRNA half-lives were estimated before and after dhh1-E182Q induction for 12 hours by measuring mRNA decay after addition of actinomycin D over a period of 80 minutes (Fig. 7). All three mRNAs had increased half-lives after dhh1-E182Q induction. For the ISG75 mRNA, there was an increase from 35 to 80 minutes, the other mRNAs (Tb11.01.3915 and Tb11.47.0021) had too long half-lives to allow quantification of the increase with confidence. The data provide evidence that an increase in mRNA stability contributes to the increase in mRNA levels on induction of dhh1-E182Q expression.
Fig. 7.
Increase in mRNA stability following expression of dhh1 E182Q. RNA half-lives were estimated by quantification over a time course after actinomycin-D addition using cells before or 12 hours after dhh1-E182Q induction. Average data of three (ISG75) or two (Tb11.01.3915 and Tb11.47.0021) independent experiments are shown. Standard deviations are indicated as error bars (ISG75).
The half-lives of two mRNAs (Tb927.6.2790 and Tb11.02.0290) whose levels were decreased by dhh1-E182Q expression were assayed in the same way. It was not possible to obtain reliable estimates of changes in half-life as they greatly exceeded the timescale of the experiment (supplementary material Fig. S8).
The increase in the level of ISG75 mRNA results in an increase in ISG75 protein
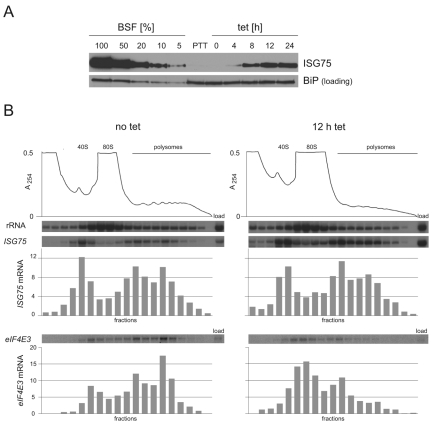
The most reliable measurement for change in mRNA half-life was obtained for ISG75 and this was investigated further. ISG75 is a plasma-membrane protein of unknown function expressed at higher levels in bloodstream than in procyclic forms (Ziegelbauer et al., 1992; Ziegelbauer and Overath, 1992). After 4 hours of dhh1-E182Q induction, ISG75 protein became detectable and then continued to accumulate (Fig. 8A). The increase in ISG75 protein could have resulted from simply the increased ISG75 mRNA or could have been caused by an increased fraction of the ISG75 mRNA being in polysomes, which, in turn, might contribute to the increased half-life. The distribution of ISG75 mRNA along a sucrose-gradient polysome fractionation was determined before and after induction of dhh1-E182Q expression in two independent experiments (Fig. 8B and supplementary material Fig. S9). In both cases, the proportion of ISG75 mRNA in the polysome fraction was similar before and after dhh1-E182Q induction, whereas the proportion of a control mRNA (eIF4E3) in the polysome fraction decreased (Fig. 8B). These data demonstrate that translation of ISG75 mRNA continues during dhh1-E182Q expression with a similar proportion of ISG75 mRNA in polysomes.
Fig. 8.
ISG75 protein and mRNA after dhh1-E182Q induction. (A) ISG75 protein expression was determined by western blotting over a time course after dhh1-E182Q induction. A titration of bloodstream-form (BSF) cell equivalents was used to estimate expression levels. BiP expression is shown as a loading control (Bangs et al., 1993). (B) Distribution of ISG75 mRNA on a polysome analysis before and 12 hours after dhh1-E182Q induction analysed by northern blotting. The distributions shown are relative with the total set at 100%. eIF4E3 mRNA was used as a control.
The amount of ISG75 protein per cell increased over 24 hours of induction but did not reach the levels present in bloodstream-form cells (Fig. 8A). There were two contributory factors to the lower levels of ISG75 protein: the increased ISG75 mRNA after dhh1-E182Q expression was still less than present in bloodstream forms (Table 1) and the ISG75 protein had a shorter half-life in procyclic cells (supplementary material Fig. S10).
dhh1-E182Q expression alters many developmentally regulated mRNAs
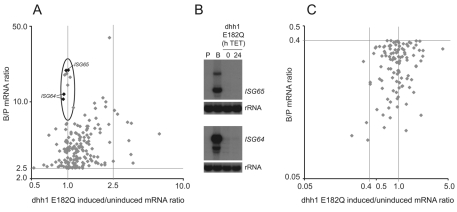
To determine whether the effect of dhh1-E182Q expression was generally applicable to developmentally regulated genes, the dhh1-E182Q microarray results were compared with a microarray experiment that compared bloodstream and procyclic mRNAs (Queiroz et al., 2009) (Array Express acc. no. E-MEXP-2028). Some of the values in the microarray experiment for differential expression between bloodstream and procyclic forms were significantly different to those obtained by northern blotting (no differential expression by microarray, clear differential expression by northern blot) (Fig. 6 and Table 1). The likely explanation for this discrepancy is that the microarray (JCVI PFGRC T. brucei 37K v2) has a single oligonucleotide for most open reading frames and is thus prone to occasional error. This precluded a robust global analysis, so a simpler comparison of the two microarray experiments was performed. The list of genes differentially expressed between the two life stages was filtered using the same criteria as for the dhh1-E182Q experiment and the two lists compared (Fig. 9). A more stringent definition of developmentally regulated genes was used [bloodstream:procyclic (B:P)>2.5 or <0.4] because this provided a sufficient number of data points. The majority (135/171) of mRNAs that were upregulated in bloodstream forms (B:P>2.5) were also increased (induced:uninduced>1) in procyclic forms on expression of dhh1 E182Q, suggesting that the majority are regulated through a pathway involving DHH1 (supplementary material Table S4). However, there was a group of mRNAs that were upregulated in bloodstream forms that was unaffected by dhh1-E182Q expression (circled in Fig. 9A), suggesting a second pathway for developmental regulation. This group included ISG64 and ISG65 mRNAs, and the observation was confirmed by northern blot (Fig. 9B). A similar analysis of mRNAs upregulated in procyclic relative to bloodstream forms (B:P<0.4) showed that a small majority (61/111) were also decreased by dhh1-E182Q expression in procyclic forms (induced:uninduced<1) (supplementary material Table S5 and Fig. 9C). This weaker association might indicate that many mRNAs upregulated in procyclic forms are not dependent on a DHH1 pathway for their increased expression.
Fig. 9.
Comparison of the dhh1-E182Q and the BSF:PCF microarray. Effect of dhh1-E182Q expression on (A) mRNAs developmentally upregulated in bloodstream forms (B:P >2.5), and (C) mRNAs developmentally downregulated in bloodstream forms (B:P <0.4). In both cases the B:P ratio is on the y-axis and the dhh1-E182Q-induced:uninduced value is on the x-axis. In A, the group of strongly developmentally regulated mRNAs unaffected by dhh1 E182Q are circled and two genes of this group have been analysed by northern blot (B).
Can any conclusions be made about mRNAs differentially expressed in other life-cycle stages? The BARP gene encodes the only known example of mRNAs that are upregulated specifically in epimastigotes, the proliferative form present in the insect salivary glands (Urwyler et al., 2007). BARP is encoded by a tandem array of 14 genes: in the microarray experiment, 12/14 increased on dhh1-E182Q expression (mean, 1.35; range, 1.1 to 1.8) and the remaining two were unaffected (1.0). This observation suggests that BARP mRNAs, as representatives of epimastigote mRNAs, might also be regulated by the DHH1 pathway. There was one mRNA (Tb09.211.3560, encoding glycerol kinase) that decreased on dhh1-E182Q expression but was not differentially expressed between bloodstream and procyclic forms. It is possible that this mRNA is differentially regulated in another life-cycle stage or that it is subject to another regulatory pathway.
These data provide evidence that a pathway involving DHH1 is used to downregulate many developmentally regulated mRNAs in bloodstream forms but that there is a second independent pathway to achieve the same end. The upregulation of some developmentally regulated mRNAs in procyclic forms is also dependent on a pathway including DHH1.
Discussion
There are three proliferative stages in the life cycle of African trypanosomes. Each occurs in a different host environment and has a unique pattern of gene expression, mainly achieved by post-transcriptional regulation. This regulation has been studied almost exclusively in the two experimentally tractable stages: the mammalian bloodstream form and insect midgut procyclic form. Typically, a developmentally regulated mRNA is 2- to 50-fold more abundant in one life-cycle stage than in the other. Here, the RNA helicase DHH1 was identified in an RNAi screen for factors necessary for the downregulation of one mRNA in procyclic forms and experiments concentrated on the phenotype produced by ectopic expression of dhh1 E182Q, an ATPase mutant that selective acted on the levels of developmentally regulated mRNAs.
The main findings are: (1) expression of dhh1 E182Q in procyclic forms at levels less than the endogenous wild-type DHH1 caused a cessation of proliferation between 4 and 8 hours after induction, a decrease in polysomes, and an increase in P-body size. (2) Induction of dhh1-E182Q expression did not greatly affect the levels of most mRNAs, which declined slowly to 75% of starting levels over 24 hours. (3) By contrast, a subset of mRNAs increased within 4 hours of induction and these were developmentally regulated mRNAs expressed at lower levels in procyclic than in bloodstream forms. (4) A second subset of mRNAs decreased on induction and these were mainly developmentally regulated mRNAs normally expressed at higher levels in procyclic than in bloodstream forms. (5) ISG75 mRNA, which increased during dhh1-E182Q expression, had an increased half-life, was translated, and ISG75 protein accumulated despite the overall decrease in polysomes. (6) The mRNAs altered by dhh1-E182Q expression were affected in a similar manner by twofold overexpression of wild-type DHH1.
These findings have implications for the regulation of gene expression in trypanosomes and identify an mRNA-turnover pathway dependent on DHH1 that is highly selective for many developmentally regulated mRNAs. In the wider context, the continued translation of ISG75 shows that the global repression of translation by DHH1 (Coller and Parker, 2005) can be modulated at the level of the individual mRNA.
DHH1 as a selective repressor of translation
Twofold overexpression of wild-type DHH1 produced a similar, but weaker, phenotype to dhh1-E182Q expression. This suggests that the dhh1-E182Q mutant might be mimicking excess DHH1 expression. In trypanosomes, dhh1 E182Q caused a decrease in polysomes combined with an increase in P-bodies within 12 hours. This phenotype is similar, but weaker, to that produced by overexpression of wild-type DHH1 in yeast (Coller and Parker, 2005). However, the experiments in yeast used a strong promoter on a multicopy plasmid and the degree of overexpression is likely to have been much higher than in the experiments here. The phenotype in trypanosomes suggests that overexpression of both wild-type and expression of dhh1 E182Q causes mRNA to exit polysomes. However, the action is selective, because ISG75 mRNA remained in polysomes during dhh1-E182Q expression, indicating that the action of DHH1 as a repressor of translation is regulated at the level of individual mRNA species, presumably by other trans-acting factors.
One protein that could possibly act as such a trans-acting factor has recently been identified in Trypanosoma cruzi (Dallagiovanna et al., 2007). The Pumilio-domain protein PUF6 binds to and colocalises with DHH1 in the epimastigote life-cycle stage from the insect but does not colocalise with DHH1 in the amastigote life-cycle stage from the mammalian host. Interestingly, six out of seven mRNAs that were identified as bound to PUF6 in epimastigotes were developmentally regulated. However, the T. brucei orthologues of six of these mRNAs were unaffected by the expression of dhh1 E182Q, and PUF1, the T. brucei orthologue of T. cruzi PUF6, is non-essential in procyclic cells (Luu et al., 2006).
dhh1 E182Q reverses developmental regulation of many mRNAs
A striking finding was the selective effect of dhh1 E182Q on the levels of developmentally regulated mRNAs; the differential expression of many mRNAs between the two life-cycle stages was effectively reversed. There was a corresponding increase in half-life in the three upregulated mRNAs investigated. This observation indicates that the ATPase activity of DHH1 is required for destabilisation of some developmentally regulated mRNAs. The decrease in some mRNAs that are normally upregulated in procyclic forms, following dhh1-E182Q expression, occurred coincidentally with growth arrest and it remains possible that the decrease was a secondary effect. The selective increases in mRNAs can only occur if the action of DHH1 is modulated at the level of the individual mRNA species, presumably by the unique complement of trans-acting factors bound to the mRNA.
Does dhh1-E182Q expression affect all developmentally regulated mRNAs? Two microarray experiments were compared: (1) mRNAs altered by dhh1-E182Q expression in procyclic forms, and (2) mRNAs differentially expressed between procyclic and bloodstream forms. The majority of the mRNAs downregulated in procyclic forms (B:P>1) increased on dhh1-E182Q expression, suggesting that the DHH1 pathway is a common route for this downregulation. Some other mRNAs normally downregulated in procyclic forms were unaffected, suggesting the presence of at least one other pathway independent of DHH1.
Many mRNAs normally upregulated in procyclic forms were unaffected by dhh1-E182Q expression. Presumably, developmental regulation of expression of these mRNAs is solely dependent on active destabilisation in the bloodstream-form stage; whether this process is DHH1 dependent is an important issue that we were unable to address, as bloodstream-form cell lines expressing a dhh1-E182Q transgene were not stable.
How does dhh1 E182Q reverse development regulation?
The ATPase activity of DHH1 is not necessary for unwinding short RNA helices but is needed for efficient recycling of DHH1 (Chen et al., 2008; Liu et al., 2008; Yang et al., 2007). There are between five and ten molecules of DHH1 per mRNA, an excess that is consistent with frequent cycles of remodelling mRNPs in the cytoplasm. The E182Q mutation inactivates the ATPase activity and probably increases the time any DHH1 molecule remains bound to an mRNA; a similar effect would be produced by overexpression of DHH1. For constitutive mRNAs, expression of dhh1 E182Q results in a movement out of polysomes, suggesting that DHH1-dependent remodelling is necessary to maintain the normal equilibrium. However, the majority of DHH1 is not associated with polysomes in T. cruzi (Holetz et al., 2007) or T. brucei (supplementary material Fig. S11); this might indicate that any interaction between DHH1 and a polysome is usually transient.
After mRNA release from polysomes, and despite the increase in P-bodies, there was no large increase in the rate of decay over 24 hours of dhh1-E182Q induction, indicating that removal from polysomes in itself is not sufficient to increase decay rates. These results contrast with the effect of heat shock, which also leads to a rapid decrease in polysomes but is accompanied by an increased rate of mRNA turnover resulting in a 50% decrease in total mRNA after 1-hour heat shock (Kramer et al., 2008).
ISG75 mRNA was stabilised by dhh1-E182Q expression, and the increase in mRNA resulted in increased ISG75 protein, indicating that the mRNA remained in translation. The rate of turnover of an mRNA is affected by the equilibrium between inclusion and exclusion from polysomes, and presence of an mRNA in polysomes protects it from the general mRNA-turnover pathways. For example, the stability of PGK1 mRNA was correlated to the rate of translation initiation in yeast (LaGrandeur and Parker, 1999). One possible explanation for the stabilisation was that ISG75 mRNA had increased access to polysomes and was consequently stabilised. However, the fraction of ISG75 mRNA in polysomes showed no significant increase on dhh1-E182Q expression. Thus, the stabilisation by dhh1 E182Q resulted from slowing of the pathway necessary for the reduced half-life in procyclic forms possibly linked to 5′-to-3′ degradation by XRNA (Li et al., 2006).
The mRNAs that decrease most rapidly on dhh1-E182Q expression are mainly developmentally regulated mRNAs that are normally increased in procyclic forms compared with bloodstream forms (B:P<0.4). The simplest explanation is that the decrease in these mRNAs is a secondary event to the decrease in polysomes; polysomes normally have a stabilising effect. If this is the case, differential regulation of translation in the two life-cycle stages contributes towards the developmental regulation of many mRNAs. There is a clear example of a cis-element that both destabilises and represses translation of the unstable EP procyclin mRNA in bloodstream-form trypanosomes (Schurch et al., 1997). However, the possibility that DHH1 has a direct role in stabilising these mRNAs cannot be ruled out.
Differences between trypanosomes and yeast
In yeast, Dhh1 functions both to determine whether an mRNA is removed from polysomes (Coller and Parker, 2005) and also interacts with components of the decapping pathway. In trypanosomes, DHH1 has a clear role in determining whether mRNAs remain in polysomes but the genome contains no obvious orthologues of several genes involved in decapping although activities are present (Milone et al., 2002). The failure to identify orthologues is based on sequence alone and the large evolutionary divergence between trypanosome (Excavata) and yeast and metazoa (both Opisthokonta) means that the finding is tentative. However, in yeast and metazoa, Lsm4 and Lsm5 are present both in P-bodies as part of the Lsm1-7 complex, and in the nucleus as part of Lsm2-8; in trypanosomes, both Lsm4 and Lsm5 are restricted to the nucleus (Tkacz et al., 2008) (S.K. and M.C., unpublished). In trypanosomes, any role of DHH1 in decapping awaits the identification of the decapping enzyme. However, the stabilisation of developmentally downregulated mRNAs provides evidence for such an interaction. In yeast, the absence of Dhh1 led to the accumulation of the normally short-lived EDC1 mRNA, whereas ectopic expression of two dhh1 DEAD-box mutants (D195A and E196A) did not lead to EDC1 accumulation (Cheng et al., 2005). This contrasts with the accumulation of unstable mRNAs in trypanosomes, suggesting differences in substrate selection for the DHH1 pathway.
In conclusion, the transcriptional control used for developmentally regulated gene expression in most eukaryotes is not available for RNAP-II-transcribed genes in trypanosomes and the results here provide evidence that a pathway including DHH1 has become central to the destabilisation of many developmentally regulated mRNAs in procyclic forms. The issues that now need to be addressed are first, the mechanism of the selective action of DHH1 and, second, the details of the mRNA-turnover pathway.
Materials and Methods
Trypanosomes
T. brucei Lister 427 procyclic cells or bloodstream forms expressing VSG MITat 1.5 were used. For inducible expression of transgenes, either T. brucei Lister 427 pLEW29:pLEW13 (Wirtz et al., 1999) or PTT (Philippe Bastin, Institute Pasteur Paris, France) were used. Procyclic cells were cultured in SDM-79 (Brun and Schonenberger, 1979) and bloodstream forms were grown in HMI-9 medium (Hirumi and Hirumi, 1994). Transgenic trypanosomes were generated using standard procedures (McCulloch et al., 2004). All experiments used logarithmically growing trypanosomes.
Cloning and transgenic cell lines
For RNAi knockdown of DHH1, the entire open reading frame of DHH1 was cloned into p2T7-177 (Wickstead et al., 2002). SCD6-eYFP was constitutively expressed from its endogenous locus (Kramer et al., 2008). The dhh1 DEAD-box mutant dhh1 E182Q (GAG to CAG), the dhh1 RNA-binding mutants dhh1 R74A/K76A (CGT to GCT, and AAA to GCG, respectively) and G332A (GGT to GCT), and the wild-type DHH1 protein were inducibly expressed from pLEW100 (Wirtz et al., 1999) in PTT (Philippe Bastin, Institute Pasteur Paris, France). Heat-shock treatment was done for 60 minutes at 41°C and puromycin treatment was done at 50 μg/ml for 30 minutes.
Quantitative RNA analysis by northern blotting
Northern blots were done as described (Kramer et al., 2008). Phosphoimager analysis was used for quantification and ribosomal RNA was used to adjust values for loading. Probes were full-length open reading frames with the exception of Tb927.3.3730 (4968 to 5538 of the ORF), Tb10.61.2680 (168 to 1497 of the ORF), Tb927.3.4270 (2125 to 2748 of the ORF), eIF4E3 (1 to 495 of the ORF). All probes recognised single bands. Total mRNA was measured using an oligonucleotide complementary to the spliced leader (Kramer et al., 2008).
Western blots
For western blots, standard procedures were used. Detection was either done by ECL or using the Odyssey Infrared Imaging System (LI-COR). For quantification, the Odyssey software was used (background method: the average of a three pixel width line at the top and bottom of each band was subtracted from each pixel).
Microscopic imaging
Cells were washed with SDM-79 without serum, fixed at a density of 1×107 cells/ml with 2.4% paraformaldehyde overnight, washed once in PBS and stained with Hoechst H33258. Fluorescence microscopy was carried out using a Zeiss Axioimager M1 microscope and a Plan-Apochromat 100/1.4 Oil DIC objective. Images were taken with the monochrome CCD camera AxioCam MR using AxioVision software (Zeiss).
P-body quantification
Fluorescent images of a randomly selected cell, focussed on the largest P-body, were taken and pixel intensities integrated over the area of the P-body using ImageJ (ImageJ, 1.40g, Wayne Rasband, National Institutes of Health, USA). Note that this method for quantifying P-body size is an approximation and does not provide absolute values for P-body sizes, since it neglects the third dimension and also underestimates the size of very big P-bodies due to saturation.
Polysome analysis
Polysome analysis was performed as previously described (Kramer et al., 2008). Cells were not treated with cycloheximide prior to, or during, the procedure. For RNA preparation, 800-μl fractions were collected and mixed with 800 μl phenol/chloroform 1:1 (v/v). After addition of 5 mM EDTA, 0.5% SDS (v/v) and 160 μl 20×SET (3 M NaCl, 20 mM EDTA, 0.6 M Tris-HCl pH 8) the samples were vortexed and centrifuged (5 minutes, 4°C, 16,000 g). The aqueous phase was collected and mixed with tRNA (20 μg/ml) and one volume isopropanol. After 15 minutes incubation at 4°C, RNA was precipitated by centrifugation (15 minutes, 4°C, 16,000 g), washed once in 70% ethanol, air-dried and resuspended in 20 μl H2O, of which 4 μl were used per northern blot lane.
Microarray
RNA for microarray experiments was prepared using the Qiagen RNAeasy Midi kit, including the DNAse digestion on the column. Hybridisations were done as described (Archer et al., 2008).
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust. Part of the microarray analysis was performed whilst M.C. held the Karush Fellowship at the MBL, Woods Hole. We thank Nancy Standart and Angela Schwede for critical reading of the manuscript, Jenny Reed for excellent technical assistance, Peter Overath for the ISG75 antibody, Jay Bangs for the BiP antibody, Steven Reed for the T. cruzi P0 antibody, Keith Gull for the BB2 antibody, Osvaldo de Melo for the PABP2 antibody and Philippe Bastin for the PTT cell line. The polysome experiments were performed in Richard Jackson's lab. We thank the Pathogen Functional Genomics Resource Centre (PFGRC), NIAID and the J. Craig Venter Institute (JCVI) for providing T. brucei microarrays. R.Q. holds a stipend from the Deutsche Akademische Austauschdienst. L.E. held a BBSRC PhD studentship. Deposited in PMC for immediate release.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/5/699/DC1
References
- Archer S., Queiroz R., Stewart M., Clayton C. (2008). Trypanosomes as a model to investigate mRNA decay pathways. Methods Enzymol. 448, 359-377 [DOI] [PubMed] [Google Scholar]
- Balagopal V., Parker R. (2009). Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21, 403-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs J. D., Uyetake L., Brickman M. J., Balber A. E., Boothroyd J. C. (1993). Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J. Cell Sci. 105, 1101-1113 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006). Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71, 513-521 [DOI] [PubMed] [Google Scholar]
- Blattner J., Clayton C. E. (1995). The 3′-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase-encoding genes mediate developmental regulation. Gene 162, 153-156 [DOI] [PubMed] [Google Scholar]
- Boag P. R., Atalay A., obida S., Reinke V., Blackwell T. K. (2008). Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brems S., Guilbride D. L., Gundlesdodjir-Planck D., Busold C., Luu V. D., Schanne M., Hoheisel J., Clayton C. (2005). The transcriptomes of Trypanosoma brucei Lister 427 and TREU927 bloodstream and procyclic trypomastigotes. Mol. Biochem. Parasitol. 139, 163-172 [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R., Schonenberger (1979). Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36, 289-292 [PubMed] [Google Scholar]
- Chen Y., Potratz J. P., Tijerina P., Del Campo M., Lambowitz A. M., Russell R. (2008). DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. USA 105, 20203-20208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Coller J., Parker R., Song H. (2005). Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA 11, 1258-1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C., Shapira M. (2007). Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156, 93-101 [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. (2004). Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73, 861-890 [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. (2005). General translational repression by activators of mRNA decapping. Cell 122, 875-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., Parker R. (2001). The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7, 1717-1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallagiovanna B., Correa A., Probst C. M., Holetz F., Smircich P., de Aguiar A. M., Mansur F., da Silva C. V., Mortara R. A., Garat B., et al. (2007). Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. J. Biol. Chem. 283, 8266-8273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Seraphin B. (2007). A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15-22 [DOI] [PubMed] [Google Scholar]
- Engstler M., Boshart M. (2004). Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 18, 2798-2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A. M. (2008). The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 36, 4573-4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970-3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Weis K. (2002). The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21, 2788-2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T. M., Lykke-Andersen J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32, 605-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A., Schurch N., Kurath U., Roditi I. (1997). Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 17, 4372-4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillian-Daniel D. L., Gray N. K., Astrom J., Barkoff A., Wickens M. (1998). Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol. Cell. Biol. 18, 6152-6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L. C., Chung H. M., Lee P. T., Lee M. G. (2003). RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2, 542-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanstra J. R., Stewart M., Luu V. D., van Tuijl A., Westerhoff H. V., Clayton C., Bakker B. M. (2008). Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J. Biol. Chem. 283, 2495-2507 [DOI] [PubMed] [Google Scholar]
- Haile S., Papadopoulou B. (2007). Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 10, 569-577 [DOI] [PubMed] [Google Scholar]
- Hehl A., Vassella E., Braun R., Roditi I. (1994). A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 91, 370-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H., Hirumi K. (1994). Axenic culture of African trypanosome bloodstream forms. Parasitol. Today 10, 80-84 [DOI] [PubMed] [Google Scholar]
- Holetz F. B., Correa A., Avila A. R., Nakamura C. V., Krieger M. A., Goldenberg S. (2007). Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356, 1062-1067 [DOI] [PubMed] [Google Scholar]
- Kooter J. M., Van der Spek H. J., Wagter R., d'Oliviera C. E., Van der Hoeven F., Johnson P. J., Borst P. (1987). The anatomy and transcription of a telomeric expression site for variant specific surface antigens in Trypanosoma brucei. Cell 51, 261-272 [DOI] [PubMed] [Google Scholar]
- Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J. D., Clayton C., Carrington M. (2008). Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 121, 3002-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Bruse S., Hill K. L., Donelson J. E. (2000). Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111, 67-76 [DOI] [PubMed] [Google Scholar]
- Ladomery M., Wade E., Sommerville J. (1997). Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 25, 965-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T., Parker R. (1999). The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA 5, 420-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz J. H., Smith H. Q., Rusche L., Beverley S. M. (1993). Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7, 996-1007 [DOI] [PubMed] [Google Scholar]
- Li C. H., Irmer H., Gudjonsdottir-Planck D., Freese S., Salm H., Haile S., Estevez A. M., Clayton C. (2006). Roles of a Trypanosoma brucei 5′->3′ exoribonuclease homolog in mRNA degradation. RNA 12, 2171-2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Putnam A., Jankowsky E. (2008). ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 105, 20209-20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu V. D., Brems S., Hoheisel J. D., Burchmore R., Guilbride D. L., Clayton C. (2006). Functional analysis of Trypanosoma brucei PUF1. Mol. Biochem. Parasitol. 150, 340-349 [DOI] [PubMed] [Google Scholar]
- Matthews K. R., Tschudi C., Ullu E. (1994). A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8, 491-501 [DOI] [PubMed] [Google Scholar]
- McCulloch R., Vassella E., Burton P., Boshart M., Barry J. D. (2004). Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262, 53-86 [DOI] [PubMed] [Google Scholar]
- Milone J., Wilusz J., Bellofatto V. (2002). Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 30, 4040-4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Thom G., Standart N. (2001). A conserved role of a DEAD box helicase in mRNA masking. RNA 7, 1728-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Kress M., Weil D., Standart N. (2009). Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell 20, 2464-2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Gao M., O'Connor J. P., Raijmakers R., Pruijn G., Lutz C. S., Wilusz J. (2002). The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21, 165-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Amikura R., Hanyu K., Kobayashi S. (2001). Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233-3242 [DOI] [PubMed] [Google Scholar]
- Navarro R. E., Shim E. Y., Kohara Y., Singson A., Blackwell T. K. (2001). cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128, 3221-3232 [DOI] [PubMed] [Google Scholar]
- Noble S. L., Allen B. L., Goh L. K., Nordick K., Evans T. C. (2008). Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559-572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646 [DOI] [PubMed] [Google Scholar]
- Queiroz R., Benz C., Fellenberg K., Hoheisel J. D., Clayton C. (2009). Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10, 495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers N. D., Wang Z., Kiledjian M. (2002). Regulated alpha-globin mRNA decay is a cytoplasmic event proceeding through 3′-to-5′ exosome-dependent decapping. RNA 8, 1526-1537 [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., Sanchez-Rotunno M., Arraiano C. M., van Hoof A. (2009). The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 16, 56-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch N., Furger A., Kurath U., Roditi I. (1997). Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 89, 109-121 [DOI] [PubMed] [Google Scholar]
- Schwede A., Ellis L., Luther J., Carrington M., Stoecklin G., Clayton C. (2008). A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36, 3374-3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P. (2006). ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7, 72-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Parker R. (2001). Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8, 1075-1083 [DOI] [PubMed] [Google Scholar]
- Tkacz I. D., Cohen S., Salmon-Divon M., Michaeli S. (2008). Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 160, 22-31 [DOI] [PubMed] [Google Scholar]
- Ullu E., Matthews K. R., Tschudi C. (1993). Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesised tubulin transcripts. Mol. Cell. Biol. 13, 720-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S., Studer E., Renggli C. K., Roditi I. (2007). A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol. Microbiol. 63, 218-228 [DOI] [PubMed] [Google Scholar]
- Walrad P., Paterou A., Acosta-Serrano A., Matthews K. R. (2009). Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog. 5, e1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb H., Burns R., Ellis L., Kimblin N., Carrington M. (2005). Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short lived protein factor. Nucleic Acids Res. 33, 1503-1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A., Sommerville J. (2006). Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 34, 3082-3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., Gull K. (2002). Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125, 211-216 [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89-101 [DOI] [PubMed] [Google Scholar]
- Yang Q., Del Campo M., Lambowitz A. M., Jankowsky E. (2007). DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell 28, 253-263 [DOI] [PubMed] [Google Scholar]
- Ziegelbauer K., Overath P. (1992). Identification of invariant surface glycoproteins in the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267, 10791-10796 [PubMed] [Google Scholar]
- Ziegelbauer K., Multhaup G., Overath P. (1992). Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267, 10797-10803 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.