Abstract
Centriole duplication is of crucial importance during both mitotic and male meiotic divisions, but it is currently not known whether this process is regulated differently during the two modes of division. In Caenorhabditis elegans, the kinase ZYG-1 plays an essential role in both mitotic and meiotic centriole duplication. We have found that the C-terminus of ZYG-1 is necessary and sufficient for targeting to centrosomes and is important for differentiating mitotic and meiotic centriole duplication. Small truncations of the C-terminus dramatically lower the level of ZYG-1 at mitotic centrosomes but have little effect on the level of ZYG-1 at meiotic centrosomes. Interestingly, truncation of ZYG-1 blocks centrosome duplication in the mitotic cycle but leads to centrosome amplification in the meiotic cycle. Meiotic centriole amplification appears to result from the overduplication of centrioles during meiosis I and leads to the formation of multipolar meiosis II spindles. The extra centrioles also disrupt spermatogenesis by inducing the formation of supernumerary fertilization-competent spermatids that contain abnormal numbers of chromosomes and centrioles. Our data reveal differences in the regulation of mitotic and meiotic centrosome duplication, particularly with regard to ZYG-1 activity, and reveal an important role for centrosomes in spermatid formation.
Keywords: Centrosome amplification, Meiosis, ZYG-1, C. elegans
Introduction
The bipolarity of the spindle is essential for proper chromosome segregation during mitosis and meiosis and can be achieved through at least two mechanisms. Microtubules can be nucleated and organized into a bipolar structure by chromatin and associated microtubule motor proteins. Alternatively, a pair of microtubule-organizing centers (MTOCs), known as centrosomes, can establish the two poles of the spindle thereby imparting bipolarity on the structure (Doxsey et al., 2005). The eggs of many species lack centrosomes and thus rely upon the chromatin-mediated pathway to assembly female meiotic spindles. By contrast, in mitotic and male meiotic cells, the centrosome-dependent pathway predominates and control of centrosome structure and number is of crucial importance.
To build a bipolar spindle, the centrosome-dependent pathway relies on the precise duplication of centrosomes (Tsou and Stearns, 2006a). At the onset of each cell cycle, cells possess a single centrosome that consists of a central pair of barrel-shaped centrioles surrounded by a halo of pericentriolar material (PCM). The events surrounding centrosome duplication have been best described in mitotic cells where duplication initiates with the disengagment (or separation) of the two centrioles of a pair. This event is thought to license centrioles for a single round of duplication (Tsou and Stearns, 2006a; Tsou and Stearns, 2006b). In the next step, which is linked to S phase, daughter centrioles are assembled next to each pre-existing centriole. Ultimately, the two pairs of centrioles separate, with each enveloped by a cloud of PCM from which microtubules are nucleated.
The molecular pathway underlying centriole assembly is conserved among animals and is dependent on a group of ‘core’ centriole replication factors that were first identified in the nematode Caenorhabditis elegans. The kinase ZYG-1 is a key upstream regulator of this pathway and is related to polo-like kinase 4 (Plk4) in humans and SAK in Drosophila (Bettencourt-Dias et al., 2005; Kleylein-Sohn et al., 2007; O'Connell et al., 2001). ZYG-1 is one of the first proteins to localize to sites of centriole assembly and, like Plk4, is required to recruit the coiled-coil proteins SAS-6 and SAS-4 to the assembling centriole (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006). SAS-6 is thought to be required for formation of a molecular scaffold that directs the overall assembly process (Dammermann et al., 2004; Leidel et al., 2005; Nakazawa et al., 2007; Pelletier et al., 2006; Rodrigues-Martins et al., 2007a), and SAS-4, whose recruitment is dependent on SAS-6, is required for assembly of the nine sets of microtubules that form the outer wall of the centriole (Delattre et al., 2006; Kirkham et al., 2003; Kleylein-Sohn et al., 2007; Leidel and Gonczy, 2003; Pelletier et al., 2006). In addition, worms possess two other core replication factors: the coiled-coil proteins SPD-2 and SAS-5 function to localize ZYG-1 and SAS-6, respectively (Delattre et al., 2006; Pelletier et al., 2006). However, the involvement of SPD-2 in centrosome duplication in other species remains controversial and SAS-5 homologs have not yet been identified (Dix and Raff, 2007; Giansanti et al., 2008; Gomez-Ferreria et al., 2007; Zhu et al., 2008).
Although a rudimentary understanding of the steps involved in centriole assembly has been obtained, questions remain about how the assembly process is regulated so that only one round of centriole duplication occurs per cell cycle. Overexpression of either SAK/Plk4 or SAS-6 drives centriole overduplication, indicating that precise duplication is achieved in some cells by carefully regulating the activities of duplication factors (Habedanck et al., 2005; Peel et al., 2007; Rodrigues-Martins et al., 2007b; Strnad et al., 2007). However, this does not appear to be true for all cells; Peel and colleagues (Peel et al., 2007) found that the ability of overexpressed SAK/Plk4, SAS-6 or SAS-4 to drive centriole overduplication differs greatly among cell types, suggesting that different cell types might employ different strategies to control centriole duplication.
In C. elegans, core duplication factors are also required for centriole duplication during the male meiotic divisions (Delattre et al., 2004; O'Connell et al., 2001). Like other species, C. elegans spermatocytes undergo two successive divisions to produce four haploid sperm (L'Hernault, 2006). Both meiosis I and II spindles possess a pair of centrioles at each pole (Albertson and Thomson, 1993), indicating that a single round of duplication takes place between meiosis I and II. Unlike mitotic centriole duplication, which is tightly linked to S phase, meiotic centriole duplication is not accompanied by DNA synthesis, suggesting that at some level, control of mitotic and meiotic centriole duplication is likely to differ. However, such differences have not yet been investigated. Following meiosis II, spermatids form in the vicinity of each spindle pole and incorporate a single haploid nucleus and centriole pair (Wolf et al., 1978). The two centrioles are donated to the egg at fertilization, where they direct assembly of the first two zygotic centrosomes.
Here, we show that C-terminal truncations of ZYG-1 block the replication of mitotic centrosomes but result in amplification of meiotic centrosomes. We also show that the extra meiotic centrioles induce the formation of supernumerary spermatids. Our data reveal differences in the regulatory mechanisms that control mitotic and meiotic centrosome duplication and establish a role for centrosomes in spermatid formation.
Results
The ZYG-1 C-terminus is necessary and sufficient for centrosome targeting
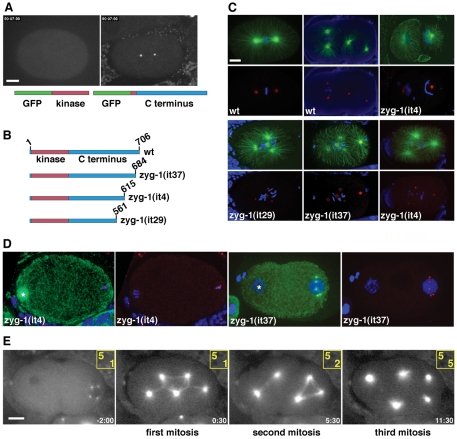
The C. elegans kinase ZYG-1 and its putative vertebrate homolog Plk4 are upstream regulators of centrosome duplication (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006). Elevated levels of Plk4 induce centrosome amplification, indicating that the activity of this kinase must be carefully regulated to ensure the once-and-only-once-per-cell-cycle nature of duplication (Cunha-Ferreira et al., 2009; Habedanck et al., 2005; Peel et al., 2007; Rodrigues-Martins et al., 2007b; Rogers et al., 2009). However, the full extent to which ZYG-1 and Plk4 are regulated is not clear. Substitution of single amino acid residues within the C-terminus of ZYG-1 can completely block centrosome duplication during embryogenesis (O'Connell et al., 2001), indicating that this region plays an important regulatory role. In fact, all known loss-of-function mutations of ZYG-1 affect the C-terminus and produce a temperature-sensitive (ts) block to centrosome duplication (O'Connell et al., 2001). We now refer to these ts loss-of-function mutations as class I mutations, the strongest of which also inhibit centriole duplication during the male meiotic divisions and block most post-embryonic cell divisions, indicating that ZYG-1 is required in many different cell types in the worm. To further characterize the C-terminus of ZYG-1, we fused amino acids 217-706 to GFP and found that this region is sufficient to target GFP to centrosomes (Fig. 1A and supplementary material Movie 1). By contrast, GFP fused to the kinase domain (amino acids 13-249) localizes to the cytoplasm (Fig. 1A and supplementary material Movie 1). Thus, the C-terminus of ZYG-1 plays an important role in duplication, and is both necessary and sufficient to target the protein to centrosomes.
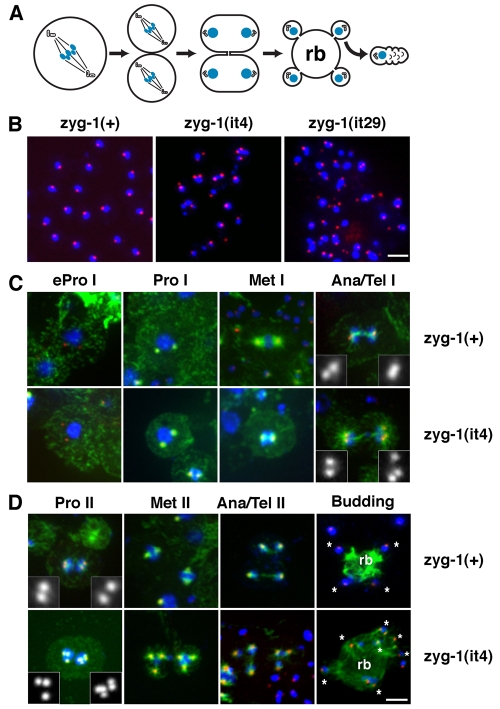
Fig. 1.
Elements within the C-terminus of ZYG-1 are important for localization to the centrosome and maintaining proper centrosome number. (A) GFP fused to the ZYG-1 C-terminus localizes to the centrosome (right), whereas ZYG-1 fused to the kinase domain is cytoplasmic (left). Scale bar: 10 μm. (B) Presumptive structure of the wild-type ZYG-1 protein and the three class II mutants. Due to mutation of a 3′ splice site, the zyg-1(it29) allele probaby produces a mixture of properly and improperly spliced messages. (C) Class II embryos stained for microtubules (green), DNA (blue) and centrosomes (red). Embryos are shown with (top) and without (bottom) microtubules. In the top set of panels, centrosomes are stained for SPD-2. The zyg-1(it4) embryo has failed to duplicate the sperm centrioles and as a result assembles monopolar spindles at the two-cell stage. In the bottom set of panels, the zyg-1(it29) embryo is stained for SPD-2, the zyg-1(it37) embryo is stained for SPD-5, and the zyg-1(it4) embryo is stained for SAS-4. Scale bar: 10 μm. (D) Young class II mutant embryos showing extra centrosomes exclusively associated with the male pronucleus. Centrosomes are stained for SPD-2. In the zyg-1(it4) embryo, the female meiotic spindle is visible (asterisk) but is not associated with centrosomes. In the zyg-1(it37) embryo, the female pronucleus (asterisk) lacks centrosomes. (E) Frames from a recording of zyg-1(it29) embryo expressing GFP-tubulin. Though the embryo inherits five centrosomes, they do not duplicate during the embryonic divisions. Numbers in yellow indicate the number of centrosomes (top) and cells (bottom) at each point in the recording. Time (minutes:seconds) is relative to first metaphase. Scale bar: 10 μm (D,E).
The embryonic lethality of class II mutants is due to both maternal and paternal defects
To further characterize the C-terminal region, we analyzed mutants that express truncated versions of ZYG-1 (Fig. 1B). Three alleles (it4, it29 and it37) were identified in a screen for maternal-effect lethal mutations (Kemphues et al., 1988). On sequencing, we found that it4 and it37 are nonsense mutations that delete the last 91 and 22 amino acids, respectively. The it29 mutation disrupts the 3′ splice site of the fifth intron, thereby impeding production of a protein containing the last 145 amino acids. As shown below, all three alleles behave in a similar manner yet are genetically distinct from previously characterized class I alleles. Thus, we collectively refer to it4, it29 and it37 as class II mutations.
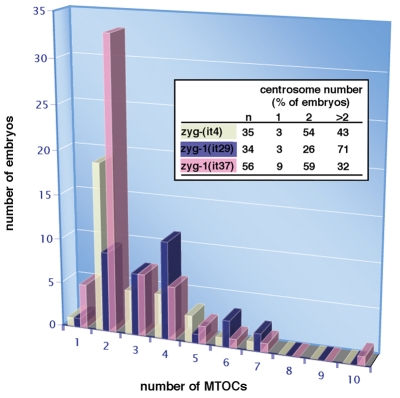
Although class I mutations are ts and can be maintained as homozygotes, class II mutations are not ts and must be maintained as balanced heterozygotes. Homozygotes are viable and typically develop into fertile hermaphrodites that produce only dead embryos (Kemphues et al., 1988). We found that the embryonic lethal phenotype of all three alleles is largely recessive, with only it4 showing a weak degree of dominance (Table 1). This embryonic lethal phenotype is not rescued by mating mutant hermaphrodites to wild-type males (Kemphues et al., 1988), indicating that lethality is the result of a defect in maternal control of the embryonic cell divisions. However, we found that mating either zyg-1(it4) or zyg-1(it29) homozygous mutant males to wild-type females also produces significant embryonic lethality (Table 1). Thus, class II mutants possess both maternal-effect and paternal-effect embryonic lethal phenotypes.
Table 1.
Maternal- and paternal-effect lethality of class 2 mutants
Class II mutants possess two distinct centrosome defects
To determine what effects these truncations have on centrosome behavior, we immunostained embryos for centrosomes, microtubules and DNA. Imaging revealed that these mutations affect centrosome number in a complex manner. Some of the mutant embryos display monopolar spindles at the two-cell stage (Fig. 1C), a phenotype diagnostic for a maternal block in centrosome duplication (O'Connell et al., 2001). In striking contrast, other embryos possess supernumerary centrosomes and multipolar spindles (Fig. 1C,D). In general, these extra centrosomes appear similar in size and morphology to wild-type centrosomes, and they contain PCM and centriole components such as SPD-2, SPD-5 and SAS-4 (Fig. 1C,D).
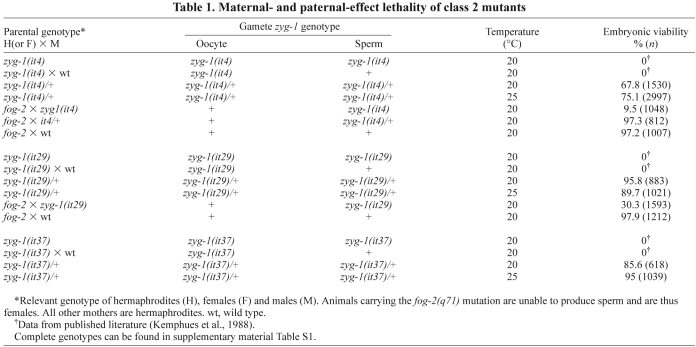
Although they differ in potency, all three alleles have similar effects on embryonic centrosome number, which typically varies between one and seven but occasionally can be more (Fig. 2). The it29 allele produces the strongest increase in centrosome number as 71% of embryos (n=34) possess more than two centrosomes. In comparison, 43% of it4 embryos (n=35) and 32% it37 embryos (n=56) have more than two centrosomes. All three mutants produce both embryos containing odd or even numbers of supernumerary centrosomes; among all class II embryos, the probability of having an odd or even number of supernumerary centrosomes is equal (51% odd versus 49% even, n=57). We found the presence of odd numbers of supernumerary centrosomes to be particularly revealing because they rule out some potential causative defects such as cytokinesis failure, which would double the number of centrioles by failing to partition the centriole pairs to separate daughter cells, and polyspermy, which would increase centriole number by factors of two, a result of each fertilizing sperm donating a centriole pair to the zygote.
Fig. 2.
The constancy of zygotic centrosome number is disrupted in class II zyg-1 mutants. For each of the class II alleles, the distribution of centrosome number (MTOCs, microtubule-organizing centers) in live and fixed one-cell embryos was determined. For each allele, the percentage of embryos with one, two, or more than two centrosomes is given in the inset. Although not shown, wild-type one-cell embryos always have two centrosomes.
The extra centrosomes have a paternal origin
Cytological examination of newly fertilized zygotes revealed that the extra centrosomes are invariably found next to the paternal pronucleus (Fig. 1D). By contrast, we never observed centrosomes associated with the nascent female pronucleus, the female meiotic spindle (Fig. 1D), or in unfertilized oocytes (not shown). To further address the origin and behavior of the extra centrosomes, we recorded the early development of embryos expressing GFP-tubulin. In five of eight zyg-1(it4) and five of nine zyg-1(it29) one cell embryos, we could detect more than two centrosomes (Fig. 1E and supplementary material Movies 2 and 3). In embryos imaged prior to the period of pronuclear migration, all centrosomes were invariably found associated with the male pronucleus. Similarly, we detected extra centrosomes in three of 13 zyg-1(it37) embryos expressing GFP—SPD-2 (supplementary material Movie 3). Interestingly, centrosomes did not duplicate in most class II mutant embryos and as a consequence their number did not increase over the first few cell cycles (Fig. 1E). Thus, class II mutant embryos possess two diametrically opposed phenotypes; they frequently begin life with too many centrosomes but are unable to duplicate these organelles.
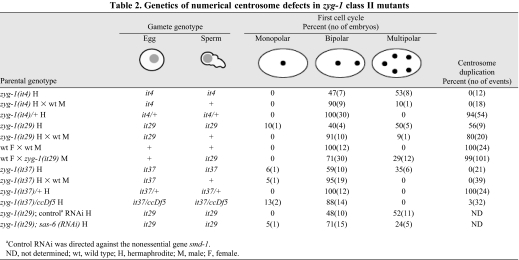
Next, the class II alleles were subject to genetic analysis. Centrosome duplication was followed in the progeny of self-fertilizing homozygous mutant hermaphrodites using four-dimensional differential interference contrast imaging (4D-DIC). Multipolar spindles were observed during the first cell cycle in 53% of zyg-1(it4), 50% of zyg-1(it29) and 35% of zyg-1(it37) embryos grown at 20°C (Table 2). We also noticed that about a third (6 out of 19) of the embryos with extra centrosomes also had one or two extra sperm pronuclei, suggestive of male meiotic defects. Again, centrosome duplication invariably failed in both zyg-1(it4) and zyg-1(it37) embryos (Table 2). By contrast, we observed several successful centrosome duplication events in zyg-1(it29) embryos. Thus, although it29 is the strongest of the three alleles with respect to the extra centrosome phenotype, it is also the weakest of the three alleles with respect to the centrosome duplication phenotype. To determine whether the extra centrosomes were of paternal origin, we repeated this analysis after mating class II mutant hermaphrodites to wild-type males. In C. elegans, male sperm out-compete hermaphrodite sperm. Thus, after such matings, most progeny will be outcross progeny (i.e. the product of a mutant egg and a wild-type sperm). Strikingly, mating to wild-type males nearly eliminated the multipolar spindle phenotype (Table 2); among the progeny of class II mutant hermaphrodites, extra centrosomes were only found in 10% of zyg-1(it4), 9% of zyg-1(it29) and 0% of zyg-1(it37) embryos. By contrast, mating did not appreciably affect the frequency of centrosome duplication failure in class II mutants. The ability of wild-type sperm to rescue the extra centrosome phenotype but not the duplication phenotype reveals that the inheritance of extra centrosomes is a paternal phenotype and that the duplication failure is a maternal phenotype.
Table 2.
Genetics of numerical centrosome defects in zyg-1 class II mutants
We next examined centrosome behavior in the progeny of zyg-1(it37) heterozygotes. 4D-DIC imaging revealed that centrosome number and duplication are normal in all progeny (n=12) of zyg-1(it37)/+ hermaphrodites, indicating that both phenotypes are recessive. We also examined the progeny of zyg-1(it37)/ccDf5 animals. The chromosomal deficiency ccDf5 removes the zyg-1 locus and thus serves as a complete loss-of-function allele. As with zyg-1(it37) homozygotes, centrosome duplication fails in the progeny of zyg-1(it37)/ccDf5 hermaphrodites (31 out of 32 cases; Table 2), confirming that the block in centrosome duplication is due to a loss of zyg-1 function. Interestingly, although the progeny of zyg-1(it37)/ccDf5 animals display a strong centrosome duplication defect, they lack multipolar spindles (0 out of 16), indicating that the multipolar spindle phenotype arises from a gain-of-function effect.
In summary, our genetic data indicate that the duplication failure is a maternal loss-of-function phenotype and that the extra centrosome defect is a paternal gain-of-function phenotype. Mutant alleles with such tissue-specific effects are rare and indicate that the gene or its product are regulated differently in different cells.
The extra centrosomes arise during male meiosis
Because our analysis indicates that the extra centrosomes of class II mutants are of paternal origin, we examined male testes for defects in mitosis and/or meiosis. Wild-type and mutant male gonads were fixed and stained for tubulin, SPD-2 as a centrosome marker and DNA. The male gonad has a distal-to-proximal polarity with a mitotic population of syncytial stem cell nuclei at the distal end (supplementary material Fig. S1A). As these nuclei move proximally, they transition into meiotic prophase, cellularize, and differentiate into primary spermatocytes. At the proximal end, spermatocytes undergo two successive meiotic divisions to produce four spermatids that subsequently differentiate into spermatozoa.
Through most of their length, wild-type and class II mutant germ lines appear similar (supplementary material Fig. S1B,C and data not shown). We did not observe any obvious defects in the mitotic and early prophase I regions of class II mutant germ lines. However, striking defects in spindle structure were observed in class II mutant germ line nuclei undergoing the meiotic divisions. In the wild type, spermatocytes undergo two rounds of meiotic division, each with a bipolar spindle (supplementary material Fig. S1B, Fig. S3A). In class II mutants, striking multipolar spindles are evident (supplementary material Fig. S1C). The number of spindle poles varies greatly with some having just two poles and others having more than ten. On the basis of spindle size and position in the gonad, the multipolar figures appeared to be meiosis II spindles (supplementary material Fig. S1C). This gross defect in spindle structure does not block progression of the spermatogenesis program, and highly abnormal sperm are produced. Unlike wild-type sperm, which are uniform with respect to size, DNA content and centrosome number, class II mutant sperm are variable in size and DNA content and often appear to have more than one centrosome (Fig. 3B). Despite these defects, mutant sperm apparently remain competent to fertilize oocytes and, as a result, are able to donate these extra centrioles to the zygote.
Fig. 3.
The meiotic centrosome cycle is abnormal in zyg-1 class II mutants. (A) Schematic of C. elegans spermatogenesis showing the arrangement of chromosomes and centrioles during meiosis I (MI), meiosis II (MII), spermatid formation, and in sperm. Spermatids form in the vicinity of spindle poles through budding, which produces a residual body (rb) that is later degraded. (B) Wild-type sperm contain a nucleus of uniform size and a single focus of SPD-2 (red), which corresponds to a centriole pair. Class II mutant sperm are heterogeneous with respect to DNA content and the number and intensity of SPD-2 foci. Scale bar: 5 μm. (C,D) zyg-1(+); him-8(e1489) and zyg-1(it4); him-8(e1489) spermatocytes at various stages of meiosis I and meiosis II. Spermatocytes are stained for microtubules (green), DNA (blue) and SPD-2 (red). Scale bar: 5 μm. (C) Meiosis I is mostly normal in zyg-1(it4) spermatocytes. Extra centrosomes first become apparent in anaphase or telophase I when more than two SPD-2 foci are detected at the poles of the spindle. (D) In zyg-1(it4) spermatocytes, meiosis II often occurs in the presence of too many centrosomes and DNA segregates on multipolar spindles. The extra centrosomes give rise to abnormal budding patterns in which more than four spermatids (asterisks) are formed from a single residual body. Notice that more than one SPD-2 focus is apparent in some of the budding mutant spermatids.
To more carefully characterize the meiotic defect, we examined centrosome behavior in staged spermatocytes. We first characterized the events of the meiotic centrosome cycle in wild-type spermatocytes. At the time primary spermatocytes enter the meiotic divisions, each spermatocyte nucleus is associated with two centrosomes (Fig. 3C). These migrate away from each other over the surface of the nucleus and begin to organize microtubules into two astral arrays. The size of the meiotic centrosomes peaks in metaphase I. At this time, each pole possesses a single focus of SPD-2. Late in meiosis I (anaphase or telophase), the first visible manifestation of centrosome duplication arises: the single focus of SPD-2 staining at each pole resolves into two foci (Fig. 3C). This probably corresponds to the initial event of centriole duplication — disengagement of mother and daughter centrioles. During meiosis II (Fig. 3D), each pair of duplicated centrosomes organizes a smaller spindle and, in contrast to meiosis I, only a single focus of SPD-2 is found at the pole of spindles in late anaphase and telophase II. This focus probably corresponds to the centriole pair that will be inherited by spermatids (Wolf et al., 1978). In C. elegans, spermatids form through a budding process in which each haploid nucleus and associated centriole pair becomes incorporated into the forming spermatid that pinches off from the cell (Fig. 3A,D) (L'Hernault, 2006). As spermatids form, microtubules are segregated out of the nascent bud, ending up in an anucleate cell remnant, called a residual body; this structure later decays. The asymmetric segregation of microtubules is so complete that, with the exception of the microtubules that form the centriole walls, C. elegans spermatids and sperm lack microtubules (Fig. 3D).
We next examined class II mutants, and found that they appear normal throughout most of meiosis I (Fig. 3C). Typically, mutant primary spermatocytes assemble a morphologically normal bipolar spindle in meiosis I. The first obvious defect in centrosome number arises late (anaphase I or telophase I) in the division cycle when wild-type spermatocytes are approaching interkinesis, the period between meiosis I and II. In some class II mutant primary spermatocytes, more than two SPD-2 foci are detected at the poles during anaphase and telophase I, suggesting centrosome overduplication during the preceding cell cycle (Fig. 3C). Due to the presence of extra centrosomes, we frequently detected multipolar meiosis II spindles in the mutants, and abnormal budding configurations in which more than four spermatids were associated with a single residual body (Fig. 3D). In many cases, the mutant spermatids that formed following such abnormal meioses contained more than one SPD-2 focus.
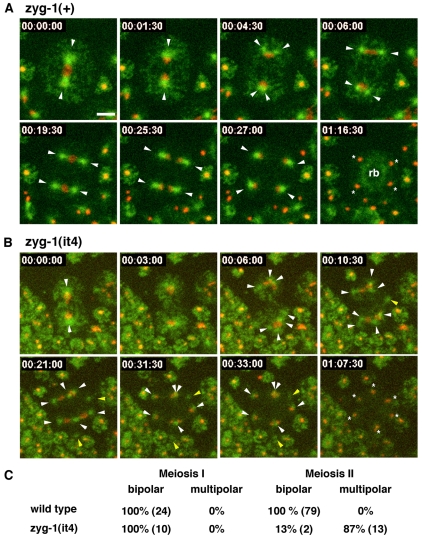
To further define this phenotype, we undertook live imaging of the meiotic divisions in spermatocytes expressing GFP-tubulin and mCherry-histone. Although the small size of spermatocytes (5 μm) presented a challenge for imaging, we managed to develop a reliable method for recording the behavior of spindles and chromatin during the two successive male meiotic divisions. As expected, all wild-type spermatocytes assembled bipolar spindles during meiosis I (n=24) and meiosis II (n=79) (Fig. 4C). Our analysis revealed that the microtubule cytoskeleton undergoes a rapid rearrangement during the transition between the two meiotic divisions (Fig. 4A and supplementary material Fig. S2, Movies 4 and 5). During late anaphase I or early telophase I, each of the spindle poles abruptly splits into two distinct MTOCs that immediately begin to organize the poles of the second meiotic spindle. The second meiotic division yields four haploid nuclei, each of which is associated with a single centrosome. As spermatids bud, each nucleus and one centriole pair is incorporated into a spermatid while the microtubules are transported to the residual body (Fig. 4 and supplementary material Movie 5).
Fig. 4.
Selected frames from recordings of zyg-1(+) and zyg-1(it4) spermatocytes expressing GFP-tubulin and mCherry-histone (Movie 5). Arrowheads indicated centrosomes and asterisks indicate spermatids. (A) A zyg-1(+) primary spermatocyte undergoing two meiotic divisions with bipolar spindles. At 00:04:30, each pole of the meiosis I spindle splits to form two centrosomes. At 00:27:00, meiosis is complete and each haploid nucleus is associated with a single aster. At 01:16:30, the four spermatids have finished budding. The asters have disassembled and the tubulin has relocated to the residual body (rb). (B) A zyg-1(it4) spermatocyte assembles multipolar spindles during meiosis II. In the first frame, two centrosomes are apparent at the end of meiosis I. Beginning at 00:06:00, these centrosomes begin to split into three centrosomes (top) and five centrosomes (bottom). The yellow arrowheads indicated centrosomes not associated with DNA. The last frame shows that six spermatids form. Time is in hours:minutes:seconds. Scale bar: 5 μm and applies to all panels. (C) The percentages of bipolar and multipolar spindles observed during meiosis I and II in live zyg-1(+) and zyg-1(it4) spermatocytes are shown. The actual numbers observed are shown in parentheses.
Throughout most of meiosis I, zyg-1(it4) spermatocytes behave similarly to the wild type in that they assemble a bipolar spindle and segregate chromatin to the poles (Fig. 4B,C and supplementary material Fig. S3, Movies 5 and 6). Strikingly however, 87% (n=15) of the time, multiple centrosomes arise from the poles of the meiosis I spindle during late anaphase or early telophase. The time of appearance of the extra centrosomes in the mutant is similar to the time at which two centrosomes arise at the poles of wild-type spindles. The number of centrosomes that form at each pole does not appear to follow a particular pattern, and both odd and even numbers of centrosomes were observed. Most of the extra centrosomes participate in spindle assembly during meiosis II, thus giving rise to the multipolar figures. However, some centrosomes migrate to the outer regions of the cell and do not function in spindle assembly. A similar observation has been made in Drosophila, in which centriole amplification (driven by overexpression of the ZYG-1-related kinase SAK) results in the formation of some abnormal centrosomes that do not participate in spindle assembly (Basto et al., 2008).
The presence of extra centrosomes distorts the budding pattern such that more than the normal number of spermatids are formed. As seen in Fig. 4B and supplementary material Movie 5, the extra centrosomes lead to the formation of at least six spermatids. Close examination of supplementary material Movie 5 reveals that some of these forming spermatids inherit more than one centrosome and have different amounts of DNA. Similarly, DIC imaging of dividing class II mutant spermatocytes reveals defects in the spermatid budding pattern. Although wild-type secondary spermatocytes produce two equal-sized spermatids (supplementary material Movie 7), mutant secondary spermatocytes frequently produce more than two spermatids (supplementary material Movie 8). These are heterogeneous in size, with some abnormally large and others abnormally small. As shown above, they do not contain the wild-type chromosome complement.
Centriole overduplication most probably accounts for the centriole amplification phenotype of zyg-1 class II mutants
The meiotic phenotype of zyg-1 class II mutants is quite distinct from that of class I mutants; as shown previously, loss of ZYG-1 function during the meiotic divisions results in the production of sperm carrying a single centriole, presumably the consequence of a meiotic block in centriole duplication (O'Connell et al., 2001). Consistent with our genetic data, the class II mutants are likely to represent gain-of-function mutants and might drive the formation of extra centrosomes either through centriole overduplication or centriole fragmentation. We reasoned that if centrioles were over-replicating, partial depletion of SAS-6, a key component in the centriole replication pathway (Delattre et al., 2006; Pelletier et al., 2006; Zhu et al., 2008), might suppress the multipolar spindle defect. To determine this, we attempted to deplete SAS-6 during spermatogenesis by feeding hermaphrodites sas-6 dsRNA during larval development. To assess the affect of RNAi on spermatogenesis, we used 4D-DIC to determine the frequency of multipolar spindles in the progeny of treated hermaphrodites. sas-6(RNAi) did not have a detectable effect on centrosome duplication in wild-type spermatocytes (data not shown). We found that such embryos always possess two centrosomes rather than one, which would be expected if centrosome duplication had been blocked during spermatogenesis (O'Connell et al., 2001). Thus, spermatocytes are relatively resistant to sas-6 RNAi, a finding that is not surprising given that RNAi does not efficiently inhibit spermatocyte-expressed genes (Reinke et al., 2004). Nevertheless, we found that sas-6 RNAi had a significant effect on the frequency of multipolar spindles in class II mutants (Table 2). Multipolar spindles were observed in 52% (n=21) of control-treated zyg-1(it29) embryos. However, when exposed to sas-6 dsRNA, only 24% (n=21) of zyg-1(it29) animals possessed multipolar figures. Thus, the multipolar spindle phenotype of class II zyg-1 mutants appear to be at least partially dependent on SAS-6 activity, suggesting that the extra centrosomes arise from centriole overduplication.
To shed additional light on the mechanism by which extra centrioles are generated, we sought to determine whether the centrioles produced in class II zyg-1 mutant male germ cells possess an inherent ability to duplicate. We reasoned that structurally intact centrioles (as would be produced by centriole overduplication) would duplicate properly, although structurally compromised centrioles (as would be produced by fragmentation) would not. To carry out this experiment we needed to introduce class II mutant sperm centrioles into wild-type eggs, which are competent for centriole duplication. This avoided the maternal block to centrosome duplication present in class II mutant eggs. For this purpose, we constructed a strain (OC297) that expresses GFP-tubulin and carries the fog-2(q71) mutation, which eliminates hermaphrodite sperm (Schedl and Kimble, 1988). The fog-2(q71) mutation therefore effectively converts hermaphrodites to females. We then mated either wild-type or zyg-1(it29) males to OC297 females and analyzed centrosome behavior among the offspring by imaging GFP-tubulin fluorescence. When wild-type control males were used in such matings, all resulting embryos contained two centrosomes during the first cell cycle and all invariably duplicated to produce four centrosomes during the second cell cycle (Table 2). By contrast, when zyg-1(it29) males were used, one-cell embryos with extra centrosomes were observed among the offspring, confirming the paternal nature of this phenotype (Table 2). Strikingly, in the presence of maternally provided wild-type ZYG-1, nearly all (100 out of 101) of the zyg-1(it29) sperm-derived centrosomes duplicated (Table 2 and supplementary material Movie 9). Thus, nearly all centrioles produced during male meiosis in zyg-1(it29) animals do not possess structural defects that impair their ability to duplicate.
To further address this issue, we examined centrioles in zyg-1(it4) spermatids by electron microscopy. As sperm centrioles are normally embedded in an electron-dense RNA-containing material surrounding the DNA, it is difficult to reliably detect these structures in both wild-type and mutant cells. However, we managed to observed 12 centrioles in nine zyg-1(it4) spermatocytes (supplementary material Fig. S4). In all cases, the mutant centrioles appeared similar in structure to their wild-type counterparts. Although subtle structural defects could have been missed, we did not detect any gross abnormalities or evidence of fragmentation in the mutant centrioles, suggesting that most centrioles in zyg-1(it4) sperm lack major structural defects. Together, our data suggest that the extra centrioles of class II mutants are structurally and functionally intact.
ZYG-1 and PPH-4.1 control different aspects of meiotic centrosome behavior
C. elegans embryos with extra paternally derived centrosomes have been described by Sumiyoshi and colleagues (Sumiyoshi et al., 2002), who found that multipolar spindles form in the offspring of males depleted of PPH-4.1, a protein phosphatase 2A family member. Given that ZYG-1 is a kinase, one intriguing possibility is that centriole assembly during male meiosis might be controlled by a balance of ZYG-1 and PPH-4.1 activities, and that disruption of this balance by depletion of PPH-4.1, or expression of a hyperactive ZYG-1 protein, might lead to centriole over-replication. To address this, we closely examined the phenotype of a strain carrying the pph-4.1(tm1598) deletion allele, reasoning that if loss of PPH-4.1 affects centriole behavior in the same manner as class II mutants, then the phenotypes should look similar. As with zyg-1 class II mutants, we found that pph4.1(tm1598) sperm often possess more than one focus of SPD-2 staining, indicating the presence of extra centrioles (supplementary material Fig. S5). Further, as previously reported, we found that hermaphrodites lacking PPH-4.1 produce embryos with multipolar spindles. However, these embryos strictly possess either two or four centrosomes (40% tetrapolar, 60% bipolar; n=15). Unlike zyg-1 class II mutants, we never observed embryos with an odd number of centrosomes in the pph-4.1 deletion strain. Thus, the zyg-1 and pph-4.1 mutations apparently affect the inheritance of paternal centrosomes in distinctly different ways, suggesting that ZYG-1 and PPH-4.1 govern different aspects of centrosome behavior during spermatogenesis.
Sequences at the C-terminus of ZYG-1 are required for localization to mitotic but not meiotic centrosomes
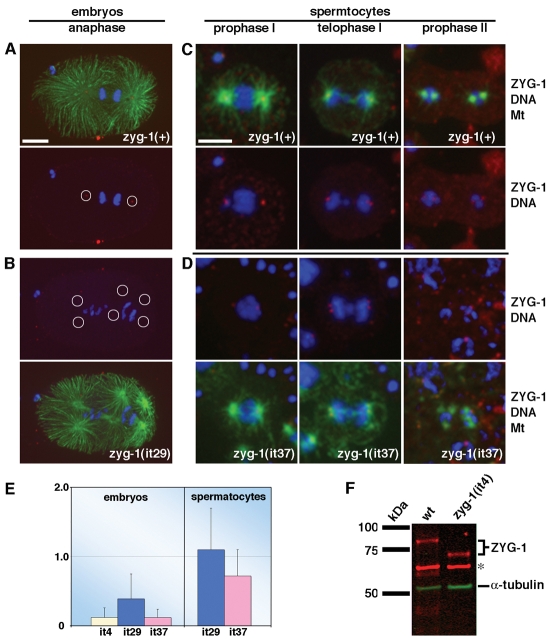
Finally, to understand how deletion of the ZYG-1 C-terminus can have different effects on centrosome duplication during mitosis and meiosis, we examined the subcellular distribution of the truncated forms of ZYG-1 in embryos and spermatocytes. In wild-type embryos, ZYG-1 localizes to the centrosome in a cell-cycle-dependent manner, attaining a peak level at anaphase (Fig. 5A; (Delattre et al., 2006; O'Connell et al., 2001). We found that all three truncated forms of ZYG-1 still localize to centrosomes in the embryo, but at greatly diminished levels compared to the wild-type (Fig. 5B). By quantitative immunofluorescence microscopy (Fig. 5E), we found that, on average, centrosomes in zyg-1(it4) embryos (n=43 centrosomes; 13 embryos) and zyg-1(it37) embryos (n=21 centrosomes; 10 embryos) possess only 12% as much ZYG-1 as their wild-type counterparts (n=46 centrosomes; 23 embryos). The zyg-1(it29) mutation had a weaker effect, reducing the level of ZYG-1 at centrosomes to 39% of controls (n=31 centrosomes; 8 embryos). That the zyg-1(it29) mutation only partially inhibits ZYG-1 localization probably explains why centrosome duplication is only partially blocked in zyg-1(it29) embryos (Table 2). To determine whether the lower level of centrosome-associated ZYG-1 in these mutants reflects a decrease in the overall levels of ZYG-1 or a decreased ability of the truncated proteins to localize to centrosomes, we measured total levels of ZYG-1 in wild-type and zyg-1(it4) embryos. Quantitative immunoblotting revealed that wild-type and mutant embryos possess similar levels of ZYG-1 (Fig. 5F). Thus, recruitment of ZYG-1 to mitotic centrosomes is impeded by truncations of the C-terminus.
Fig. 5.
Truncation of ZYG-1 specifically disrupts recruitment to mitotic centrosomes. (A-D) Embryos (A,B) and spermatocytes (C,D) stained for ZYG-1 (red), microtubules (green) and DNA (blue). A zyg-1(+) embryo (A) and a zyg-1(it29) embryo (B) are shown. Relative to full-length ZYG-1, significantly less truncated ZYG-1 is detected at mitotic centrosomes. Circles indicate the positions of centrosomes. Embryos in A and B are shown at same magnification. Scale bar: 10 μm. zyg-1(+) (C) and zyg-1(it37) spermatocytes (D) are shown. Similar levels of ZYG-1 are detected at meiotic centrosomes. All spermatocytes are shown at same magnification. Scale bar: 5 μm. (E) Quantitative immunofluorescence microscopy reveals that ZYG-1 levels at mitotic, but not meiotic, centrosomes are diminished in class II mutants. (F) Quantitative immunoblotting demonstrates that wild-type (wt) and zyg-1(it4) embryos possess similar amounts of ZYG-1 protein. α-tubulin and a nonspecific band (asterisk) serve as loading controls.
In contrast to mitosis, truncation of ZYG-1 did not have a significant affect on the centrosome-associated levels of ZYG-1 during meiosis. Meiotic centrosomes in wild-type (37 centrosomes; 19 spermatocytes), zyg-1(it29) (36 centrosomes; 17 spermatocytes) and zyg-1(it37) (32 centrosomes; 16 spermatocytes) animals possess similar levels of ZYG-1 (Fig. 5C-E). This difference in the behavior of truncated ZYG-1 probably accounts for the different effects of zyg-1 class II mutations on mitotic and meiotic centrosome duplication.
Discussion
Over the past decade, research on the centrosome has culminated in the discovery of a conserved centriole assembly pathway (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006). Many species, including algae, flies, worms, fish and humans, appear to share the same set of core centriole duplication factors (Bettencourt-Dias et al., 2005; Dammermann et al., 2004; Kemp et al., 2004; Leidel et al., 2005; Leidel and Gonczy, 2003; Nakazawa et al., 2007; O'Connell et al., 2001; Pelletier et al., 2004; Rodrigues-Martins et al., 2007a; Yabe et al., 2007; Zhu et al., 2008). Further, within a species, these factors function in a variety of cell types: in Drosophila for instance, SAK, SAS-6 and SAS-4 have been shown to be required for centrosome duplication in embryonic cells, brain cells and spermatocytes (Basto et al., 2006; Bettencourt-Dias et al., 2005; Peel et al., 2007; Rodrigues-Martins et al., 2007a). The broad requirement for these factors suggests that all centrioles assemble via the same core molecular mechanism.
A universal centriole assembly mechanism does not mean that this process is regulated in precisely the same manner in all cells. This much was rendered obvious from early studies on the role of protein synthesis in centrosome duplication, which showed that in the absence of translation, centrosomes could repeatedly double in embryonic cells (Gard et al., 1990; Sluder et al., 1990) but not in somatic cells (Phillips and Rattner, 1976). Thus, translational controls limit centrosome duplication in some but not all cells. More recent studies have also suggested that regulatory differences exist among cell types. In particular, overexpression of SAK or SAS-6 in flies drives centriole amplification in brain cells but not in spermatocytes (Peel et al., 2007), and inhibition of SPD-2 in flies has no effect on centriole number in somatic cells but results in centriole amplification in spermatocytes (Dix and Raff, 2007). Although an explanation for any of these results has not been established, they illustrate that cell-type specific differences in the regulation of centrosome duplication do exist.
One reason why duplication control mechanisms might vary according to cell type is differences in the cell cycle. As centrosome duplication is intimately linked to the cell cycle, variations in cell-cycle format could necessitate variations in centrosome duplication control. Thus, one would expect control mechanisms to differ between mitotic and meiotic cells. In mitotic cells, centriole assembly is linked to S phase and the timing of events in the centriole cycle mirrors that of DNA. For instance, separation of engaged centrioles and sister chromatids is controlled by the protease separase (Nasmyth et al., 2000; Tsou and Stearns, 2006b) and reduplication of centrioles and DNA is inhibited by Cullin-containing E3 ubiquitin ligases (Cunha-Ferreira et al., 2009; DePamphilis et al., 2006; Rogers et al., 2009). Although centriole and chromosome dynamics are linked in mitosis, this linkage is broken in meiosis; during the interkinesis separating meiosis I and II, centrioles, but not DNA, duplicate.
In class II zyg-1 mutants, extra centrioles first become apparent as spermatocytes approach interkinesis, yet it is not clear when these extra centrioles are generated. It seems unlikely that they are formed during the germ-line stem-cell divisions because we did not observe multipolar spindles in the mitotic region of the germ line. A multipolar stem-cell division would lead to gross aneuploidy and abnormal centrosome number in downstream regions of the germ line. However, nuclear content and centrosome number appear normal during all stages of prophase I. Thus, centrioles probably overduplicate following the mitotic divisions but remain closely associated, and typically only become apparent when they separate to assemble the meiosis II spindle. Although centrosome overduplication could occur at any point in meiosis I, at least some of these events occur early in the meiotic program because we have occasionally noticed multipolar meiosis I spindles in zyg-1(it4) spermatocytes. These meiosis I defects might represent instances where centrioles separated early.
Two recent studies have shown that the E3 ubiquitin ligase SCFSlimb regulates Plk4/SAK levels during mitosis and that mutation of a Slimb-binding element in the C-terminus of Plk4/SAK abrogates this regulation, leading to centriole overduplication (Cunha-Ferreira et al., 2009; Rogers et al., 2009). ZYG-1 also possesses a consensus Slimb-binding sequence (334-DSGRGT-339) downstream of the kinase domain, but this element is not deleted in class II alleles. Thus, regulation via SCFSlimb is clearly distinct from the mitotic-meiotic regulatory mechanism described in this study, in which the relevant control elements reside at the very C-terminus. In fact, the product of the zyg-1(it37) allele lacks only the last 22 amino acids. Although the sequence of this region does not provide any insight into the nature of the regulatory mechanism, this region somehow differentiates between the two modes of nuclear division, playing a positive regulatory role during mitosis and a negative one during meiosis.
We propose the following model to explain how these truncated forms of ZYG-1 behave so differently in mitosis and meiosis. We attribute to the C-terminus of ZYG-1, at least two distinct regulatory activities: an inhibitory activity that prevents centriole amplification, and an activity involved in targeting the protein to the centrosome, with the latter activity playing a more important role in mitosis than in meiosis. Thus, truncation of the C-terminus would render ZYG-1 overactive in both mitosis and meiosis, but at the same time would cripple the ability of the protein to localize to mitotic centrosomes. This would explain both the mitotic block to centrosome duplication (a hyperactive protein that cannot localize to sites of centriole assembly) and the meiotic overduplication defect (a hyperactive protein that can localize). Thus, the differing effects of class II mutants on mitosis and meiosis might simply reflect different requirements for localization to sites of centriole assembly during the two modes of division. Further analysis of class 2 alleles should allow us to test this model.
Our study also provides insight into the role of centrosomes in spermatid formation. In worms, spermatids form at the end of meiosis II via a highly specialized type of asymmetric division. Spermatids bud from an anucleate residual body and inherit a single nucleus, a centriole pair, mitochondria and Golgi-derived fibrous-body—membranous-organelles (FB-MO). Although these components are almost exclusively segregated to the spermatids, other components such as tubulin, actin and ribosomes are segregated to the residual body (L'Hernault, 2006). We have found that, in the presence of extra centrosomes, the asymmetry of budding is more or less normal in that tubulin, centrioles and nuclei segregate properly. However, extra centrosomes disrupt the wild-type budding pattern so that more than four spermatids can form per residual body. Furthermore, regardless of the number of nascent buds in a budding figure, all are associated with at least one centriole (Fig. 3D). These results indicate that centrosomes specify the budding pattern, possibly by acting as an organizing center around which spermatid components assemble. One such component might be the FB-MOs, which are involved in spermatid formation (Zhu and L'Hernault, 2003).
In summary, we have shown that centriole duplication is subject to different controls in meiosis and meiosis and that these different regulatory mechanisms are affected in part through the core centriole duplication factor ZYG-1. Our results also show that centrosomes play an important role in spermatid formation in C. elegans. Further analysis of these class II zyg-1 alleles should provide insight into the mechanisms governing mitotic and meiotic centrosome duplication and the generation of spermatids.
Materials and Methods
Genetics
All worm strains were derived from the wild-type Bristol N2 strain and are listed in supplementary material Table S1. Unless otherwise indicated, worms were maintained at 20°C on MYOB plates seeded with E. coli OP50 (Church et al., 1995). For RNAi, worms were fed bacteria expressing dsRNA (clones from Geneservice) corresponding to sas-6 or, as a negative control, the nonessential gene smd-1 beginning at the L1 stage (Timmons and Fire, 1998).
Molecular biology
Gateway technology was used to create the plasmids encoding GFP-tagged versions of the ZYG-1 kinase domain (amino acids 13-249) and C-terminus (amino acids 217-706). Transgenic lines were produced by microparticle bombardment as described (Praitis et al., 2001).
Spermatocyte culture
Spermatocytes were mounted for live imaging in sperm medium (SM: 50 mM HEPES pH 7.0, 50 mM NaCl, 25 mM KCl, 5 mM CaCl2, 1 mM MgSO4) (Ward et al., 1981) supplemented with 50 mM glucose and 1 mg/ml BSA. Briefly, a small amount of vacuum grease was applied to each corner of an 18×18 mm cover glass. This was placed greased-side up on the base of a dissecting microscope and a drop (5-6 μl) of SM was applied to the center of the coverglass. One to five young adult males were picked into the drop of SM and dissected with a hypodermic needle. A microscope slide containing a thin pad of 2% agarose in SM plus 50 mM glucose was gently placed on the cover glass so that the greased corners contacted the agarose pad. The slide was turned over, gentle pressure applied to the corners, and the edges sealed with molten petroleum jelly. Wild-type spermatocytes [designated zyg-1(+)] were obtained from either fog-2(q71), him-5 (e1490) or him-8(e1489) males.
Cytology
Methods for 4D-DIC microscopy, indirect immunofluorescent staining and spinning disk confocal microscopy have been described previously (Kemp et al., 2007). For 4D-DIC, embryos or spermatocytes were mounted on a Zeiss Axiovert 200M microscope equipped with a Hamamatsu ORCA-ER camera. IPLab 4.0 was used to acquire series of image stacks at intervals of 30-60 seconds.
For immunostaining, the following antibodies were used at 1:1000 dilutions: DM1A, an anti-α-tubulin monoclonal antibody (Sigma-Aldrich); 969LA, an affinity-purified rabbit anti-SPD-2 polyclonal antibody (Kemp et al., 2004); α-ZYG-1201-402, an affinity-purified rabbit polyclonal antibody (Kemp et al., 2007); α-SPD-5 (Hamill et al., 2002); Alexa Fluor 488 goat anti-rabbit; and 568 goat anti-mouse secondary antibodies (Invitrogen).
For spinning-disk confocal microscopy, we used a Nikon Eclipse TE2000U microscope equipped with a 60× 1.4 N.A. Plan Apo objective. This system is outfitted with a Spectral Applied Research LMM5 laser merge module to control the output of four diode lasers (excitation at 405, 491, 561 and 655 nm), a Yokogawa CSU10 spinning disk unit and a Hamamatsu C9100-13 EM-CCD camera. Confocal images were acquired using Openlab 4.0 and processed with Adobe Photoshop CS. Gamma adjustments were occasionally made to reduce the intensity of the tubulin signal at the centrosome relative to single microtubules. Fluorescence intensity measurements were made using ImageJ 1.40g.
Electron microscopy of him-8(e1489) and zyg-1(it4); him-8(e1489) males was performed as previously described (Song et al., 2008).
Immunoblotting
Total embryonic levels of ZYG-1 were determined as previously described (Song et al., 2008). Antibody DM1A was used at 1:500 and α-ZYG-1201-402 was used at 1:100. To improve the signal-to-noise ratio, α-ZYG-1201-402 was pre-absorbed with an acetone powder prepared from the nematode Caenorhabditis Remanei (http://cshprotocols.cshlp.org/cgi/content/full/2009/5/pdb.rec11801?rss=1).
Supplementary Material
Acknowledgments
We thank Jenny Hinshaw and Jurgen Heymann for help with electron microscopy, Nina Peel, Nicholas Miliaras, and Andy Golden for comments on the manuscript, and Catherine Kemp for technical assistance. Strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR), and by Shohei Mitani and the National Bioresource Project-Japan. This work was supported by the Intramural Research Program of the National Institutes Health (NIH) and by the National Institute of Diabetes and Digestive and Kidney Diseases. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/5/795/DC1
References
- Albertson D. G., Thomson J. N. (1993). Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1, 15-26 [DOI] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B., Kimble J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. (2006). Flies without centrioles. Cell 125, 1375-1386 [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J. W. (2008). Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199-2207 [DOI] [PubMed] [Google Scholar]
- Church D. L., Guan K. L., Lambie E. J. (1995). Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121, 2525-2535 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D. M., Bettencourt-Dias M. (2009). The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19, 43-49 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815-829 [DOI] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., Schnabel H., Feichtinger R., Schnabel R., Gonczy P. (2004). Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656-664 [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gonczy P. (2006). Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844-1849 [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Blow J. J., Ghosh S., Saha T., Noguchi K., Vassilev A. (2006). Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 18, 231-239 [DOI] [PubMed] [Google Scholar]
- Dix C. I., Raff J. W. (2007). Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759-1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. (2005). Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411-434 [DOI] [PubMed] [Google Scholar]
- Gard D. L., Hafezi S., Zhang T., Doxsey S. J. (1990). Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. J. Cell Biol. 110, 2033-2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Bucciarelli E., Bonaccorsi S., Gatti M. (2008). Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303-309 [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria M. A., Rath U., Buster D. W., Chanda S. K., Caldwell J. S., Rines D. R., Sharp D. J. (2007). Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960-1966 [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. (2005). The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140-1146 [DOI] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C., Bowerman B. (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3, 673-684 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S. (1979). Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91, 67-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C. A., Kopish K. R., Zipperlen P., Ahringer J., O'Connell K. F. (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511-523 [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Song M. H., Addepalli M. K., Hunter G., O'Connell K. (2007). suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics 176, 95-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Kusch M., Wolf N. (1988). Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics 120, 977-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Muller-Reichert T., Oegema K., Grill S., Hyman A. A. (2003). SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575-587 [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. (2007). Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190-202 [DOI] [PubMed] [Google Scholar]
- Leidel S., Gonczy P. (2003). SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431-439 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. (2005). SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115-125 [DOI] [PubMed] [Google Scholar]
- L'Hernault S. W. (2006). Spermatogenesis. WormBook, 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J. (2006). Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175, 881-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. (2007). SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17, 2169-2174 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Peters J. M., Uhlmann F. (2000). Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288, 1379-1385 [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. (2001). The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547-558 [DOI] [PubMed] [Google Scholar]
- Peel N., Stevens N. R., Basto R., Raff J. W. (2007). Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17, 834-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., Ozlu N., Hannak E., Cowan C., Habermann B., Ruer M., Muller-Reichert T., Hyman A. A. (2004). The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863-873 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. (2006). Centriole assembly in Caenorhabditis elegans. Nature 444, 619-623 [DOI] [PubMed] [Google Scholar]
- Phillips S. G., Rattner J. B. (1976). Dependence of centriole formation on protein synthesis. J. Cell Biol. 70, 9-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K. (2004). Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311-323 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D. M. (2007a). DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17, 1465-1472 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M., Bettencourt-Dias M. (2007b). Revisiting the role of the mother centriole in centriole biogenesis. Science 316, 1046-1050 [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Roberts D. M., Peifer M., Rogers S. L. (2009). The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184, 225-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Kimble J. (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119, 43-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G., Miller F. J., Cole R., Rieder C. L. (1990). Protein synthesis and the cell cycle: centrosome reproduction in sea urchin eggs is not under translational control. J. Cell Biol. 110, 2025-2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Muller-Reichert T., O'Connell K. F. (2008). The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell 15, 901-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gonczy P. (2007). Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell 13, 203-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi E., Sugimoto A., Yamamoto M. (2002). Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J. Cell Sci. 115, 1403-1410 [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. (1998). Specific interference by ingested dsRNA. Nature 395, 854 [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. (2006a). Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18, 74-78 [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. (2006b). Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947-951 [DOI] [PubMed] [Google Scholar]
- Ward S., Argon Y., Nelson G. A. (1981). Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91, 26-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N., Hirsh D., McIntosh J. R. (1978). Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J. Ultrastruct Res. 63, 155-169 [DOI] [PubMed] [Google Scholar]
- Yabe T., Ge X., Pelegri F. (2007). The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev. Biol. 312, 44-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Lawo S., Bird A., Pinchev D., Ralph A., Richter C., Muller-Reichert T., Kittler R., Hyman A. A., Pelletier L. (2008). The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18, 136-141 [DOI] [PubMed] [Google Scholar]
- Zhu G. D., L'Hernault S. W. (2003). The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics 165, 145-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.