Abstract
IRF4 and 8 are members of the interferon regulatory factor family of transcription factors and have been shown to be essential for the development and function of T cells, macrophages and dendritic cells. A series of recent studies have further demonstrated critical functions for IRF4 and 8 at several stages of B cell development including pre-B cell development, receptor editing, germinal center reaction and plasma cell generation. Collectively, these new studies provide molecular insights into the function of IRF4 and 8 and underscore a requirement for IRF4 and 8 throughout B cell development. This review focuses on the recent advances on roles of IRF4 and 8 in B cell development.
Introduction
The interferon regulatory factor family of transcription factor (IRF) consists of nine mammalian members that are important regulators of both immunity and other physiological processes [1]. Two members of this family, IRF4 (also known as Pip, LSIRF, LCSAT and MUM1) and IRF8 (also known as ICSBP) are unique in that they are highly homologous to each other rather than to other IRF family members and they are expressed exclusively in the immune system. IRF4 and 8 interact with other transcription factors to either stimulate or repress gene expression in the immune system. The best characterized interacting partners of IRF4 and 8 are the Ets transcription factor family members PU.1 and Spi-B. It has been shown that IRF4 and 8 dimerize with PU.1 or Spi-B and bind to the Ets-IRF composite element (EICE; GGAAnnGAAA) located in immunoglobulin (Ig) light chain κ 3’ (Ek3’) and λ gene enhancer regions [2-4]. More recently, another IRF-Ets composite sequence {IECS; GAAANN(N)GGAA} has also been identified on the target genes of IRF8[5]. The presence of EICE or IECS sites on many putative targets of IRF4 and 8 suggests that interactions of IRF4 and 8 with PU.1 and Spi-B are likely to be common regulatory mechanisms in the immune system. In addition, IRF4 and 8 have also been found to interact with the other key transcription factors E2A, NFAT and as well as other members of the IRF family [6-8]. The roles of IRF4 and 8 in immune system development and function have been well-documented. For example, IRF4 is critical for Th2 and Th17 cell development whereas IRF4 and IRF8 together are essential for macrophage and dendritic cell (DC) development and function [9-12]. Recently, a series of studies have established an additional role for IRF4 and 8 as essential regulators of B cell development, in which they have been shown to be critical for pre-B cell development, receptor editing, germinal center (GC) reaction and plasma cell differentiation (Figure 1)[13-21].
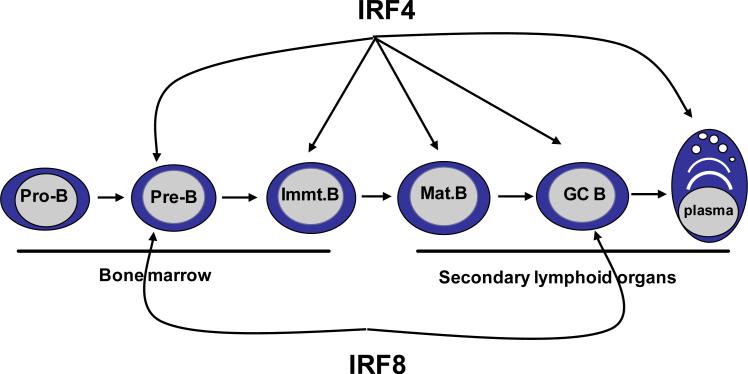
Figure 1. IRF4 and 8 are required throughout B cell development.
IRF4 and 8 function redundantly during pre-B cell development. However, IRF4, but not IRF8, is uniquely required for receptor editing at the immature B cell stage. IRF4 is likely also required for the generation of mature B cells. IRF8 promotes the early stages of the germinal center (GC) reaction whereas IRF4 is critical for later stages of GC reaction and the development of plasma cell. IRF4 and 8 show distinct expression pattern in B cell development. Expression of IRF4 is low in pro-B cells but its expression is significantly elevated in pre-B cells. The expression of IRF4 is moderate/low in naïve B cells but is induced significantly by BCR and CD40 signaling in GC B cells. The expression of IRF4 remains high in plasma cells. The expression of IRF8 remains stable throughout B cell development but its expression can be further induced by BCR signaling.
IRF4 and 8 function redundantly to control pre-B cell development
The identification of IRF4 and 8 as the transcription factors that bind to an EICE motif located in the immunoglobulin Ek3’ and λ gene enhancer regions provided the early evidence that IRF4 and 8 might play an overlapping role in pre-B cell development[3,4]. Indeed, B cell development is almost completely blocked at the large pre-B cell stage in IRF4 and 8 compound mutant mice (IRF4,8−/−)[18]. Moreover, IRF4 and IRF8 were found to be sufficient individually to rescue the development of IRF4,8−/− pre-B cells, confirming that they function redundantly in pre-B cell development[15]. Interestingly, IRF4,8−/− pre-B cells are hyperproliferative, suggesting that IRF4 and 8 negatively regulate pre-B cell proliferation. Indeed, recent findings from both ourselves and others have demonstrated that IRF4 and 8 are critical not only for light chain rearrangement but also for limiting pre-B cell expansion (Figure 2)[15,17,20,22].
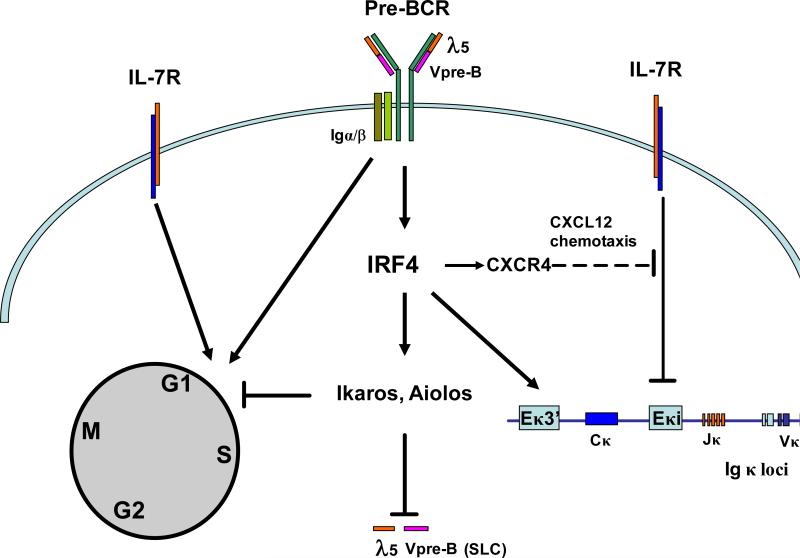
Figure 2. IRF4 and 8 limit pre-B cell expansion and promote light chain rearrangement.
IRF4, whose expression is turned on by pre-BCR signaling, induces expression of Ikaros and Aiolos to suppress surrogate light chain (SLC) expression. Besides downregulating the pre-BCR, Ikaros and Aiolos also limit pre-B cell expansion by directly inhibiting the G1-S transition. In addition, IRF4 and 8 also promote light chain rearrangement and transcription, either through direct activation of Ig light chain enhancers or indirectly through attenuation of IL-7 signaling, the latter of which is achieved via a CXCL12 mediated chemotaxis which requires an IRF4-dependent induction of CXCR4 in pre-B cells. Expression of IRF8 is not induced by the pre-BCR; however, it is still possible that the activity of IRF8 can be modulated by pre-BCR signaling.
IRF4 and 8 orchestrate the transition from cycling pre-B to small, resting pre-B by inducing the expression of Ikaros and Aiolos
Pre-B cells consist of two subsets: large pre-B and small pre-B (see Box 1). Recent studies have demonstrated that IRF4 and 8 orchestrate the transition from large pre-B to small pre-B by inducing the expression of the Ikaros family transcription factors Ikaros and Aiolos[17]. Ikaros and Aiolos have been shown to downregulate the pre-B cell receptor (BCR) by suppressing the expression of the surrogate light chain (SLC) ([23,24]. Expression of Ikaros and Aiolos is known to be elevated in pre-B cells and at least, expression of Aiolos has been shown to be induced by pre-BCR signaling[23,25]. Interestingly, the expression of Ikaros and Aiolos is induced by IRF4 and 8 in pre-B cells and expression of IRF4 itself is also induced by pre-BCR signaling[23]. Thus, pre-BCR signaling induces the expression of IRF4 which in turn stimulates the expression of Ikaros and Aiolos (Figure 2). Downregulation of the pre-BCR was originally thought to be critical in limiting pre-B cell expansion, however, a recent study has indicated that this is not essential for terminating pre-B cell expansion and that pre-BCR signaling likely only plays a role in the expansion of IL-7 receptor (IL-7R) expressing large pre-B cells[26]. That said, IRF4,8−/− pre-B cells undergo cell cycle arrest in the presence of IRF4, indicating that IRF4 does play a role in limiting pre-B cell expansion[17]. The growth inhibitory effect of IRF4 is also found to be dependent on Ikaros and Aiolos[17]. These findings suggest that besides targeting pre-BCR, Ikaros and Aiolos must use other means to inhibit pre-B cell expansion. Indeed, it has been shown that Ikaros directly inhibits the cell cycle's G1-S transition in lymphocytes[27]. The molecular mechanisms through which Ikaros inhibits cell cycle progression remain to be determined. It has been suggested that the effects of Ikaros on the cell cycle are achieved either directly through inhibiting DNA replication or indirectly by inducing the expression of cell cycle inhibitors[25,28]. Thus, IRF4 and 8, by inducing Ikaros and Aiolos, target two critical events to limit pre-B cell expansion: downregulation of pre-BCR and inhibition of the G1-S transition (Figure 2).
IRF4 and 8 promote light chain locus activation and rearrangement
Besides limiting pre-B cell expansion, IRF4 and 8 also promote immunoglobulin light chain rearrangement and transcription during pre-B cell development[15,20]. The importance of IRF4 and 8 in Igκ (kappa) loci activation is demonstrated by the findings that reconstituting the expression of IRF4 and 8 in IRF4,8−/− pre-B cells induces the expression of the κ germline transcript and promotes light chain rearrangement and transcription[15]. Igκ loci undergo dynamic changes in B cell development (see Box 2). A recent study [20] has suggested that IRF4 might play a role in positioning Igκ allele away from pericentrometic heterochromatin. Results of this study has shown that both Igκ alleles are associated with pericentromeric heterochromatin in a high proportion of IRF4,8−/− pre-B cells, however, in the presence of IRF4, the percentage of IRF4,8−/− pre-B cells that are biallelic associated with pericentromeric heterochromatin was dramatically reduced. IRF4 and Ikaros appear to play opposite roles in the positioning of the Igκ loci to pericentromeric heterochromatin. Paradoxically, IRF4 and 8 induces the expression of Ikaros and Aiolos in pre-B cells. The reason why IRF4 and 8 play this dichotomic role in Igκ rearrangement is not clear however one study has demonstrated that only about 5 % of Igκ alleles are active and undergo rearrangement in pre-B cells[29]. They proposed that Igκ rearrangement is controlled by a variegated expression of one or more rate-limiting transcription factors. IRF4 could be one of those developmentally regulated rate-limiting factors except that the amount of IRF4 might not be limiting in pre-B cells, as its expression is significantly induced by pre-BCR signaling. Should IRF4 be the rate-limiting factor that controls light chain rearrangement, a much higher proportion of pre-B cells would be expected to undergo light chain activation and rearrangement. Therefore, one possible explanation for this apparent paradox would be that pre-B cells use elevated levels of Ikaros and Aiolos to dampen activity of IRF4 on the activation of light chain loci so that only a small percentage of pre-B cells can undergo light chain rearrangement in the presence of IRF4 at any given time.
IRF4 attenuates IL-7 signaling
IL-7 signaling is essential for pre-B cell survival and proliferation, however, attenuation of IL-7 signaling is also needed for efficient light chain rearrangement. Activation of the Igκ locus is controlled by two redundant enhancer complexes, namely Ek3’ and the kappa intronic enhancer (Eki)[30]. The transcription factor E2A has been found to bind to both enhancers and thus is critical for Igκ rearrangement[31]. Recent evidence has indicated that IRF4 also regulates the activity of both enhancers to promote light chain rearrangement[20]. IRF4 directly binds to and activates the Ek3’ enhancer. In addition, IRF4 attenuates IL-7 signaling which induces the expression of recombinase activating genes (RAGs) and the activation of Eki, presumably through enhancing the binding of E2A to the RAG promoter and Eki. IRF4 was found to induce the expression of the chemokine receptor CXCR4 in pre-B cells[20]. CXCL12, the ligand of CXCR4, is mainly expressed by bone marrow stromal cells that are localized away from IL-7 producing stroma[32]. Therefore, it has been suggested that attenuation of IL-7 signaling in pre-B cells is actually achieved indirectly via a CXCL12 mediated chemotaxis in which pre-B cells expressing CXCR4 would migrate along the CXCL12 gradient away from IL-7 producing stroma[20]. It should be noted that CXCL12-mediated chemotaxis does not appear to be necessary for terminating pre-B cell expansion, as induction of Ikaros and Aiolos by IRF4 and 8 is alone sufficient to shut down pre-B cell proliferation. Thus, in pre-B cells, IRF4 and 8 induce the expression of Ikaros, Aiolos, and CXCR4 to attenuate the effects of IL-7 signaling: Ikaros and Aiolos directly antagonize the effect of IL-7 signaling on cell cycle progression, whereas CXCR4 triggered chemotaxis nullifies the inhibitory effect of IL-7R signaling on light chain rearrangement by moving the B cells away from sources of IL-7 (Figure 2). The molecular mechanism through which IL-7 signaling interferes with the binding of E2A to Eki remains to be determined. In addition, it is also possible that IRF4 might regulate, either directly or indirectly, the binding of E2A to Eki in pre-B cells independent of IL-7 signaling. This notion is supported by a recent study which showed that knockdown of IRF4 expression in a pre-B cell line also reduces the binding of E2A to Eki [22].
IRF4 and the sequential rearrangement of Ig heavy and light chain loci
Ig heavy (H) and light (L) chain loci are sequentially rearranged in B cell development. Recent studies have established IRF4 as key components of a regulatory circuit that orchestrates the sequential Ig H and L chain rearrangement. At the pro-B stage, IL-7 signaling promotes Ig H rearrangement but inhibits light chain rearrangement[20,33]. In addition, a low level of IRF4 at the pro-B stage fails to activate Ek3’, which also disfavors light chain rearrangement. At the pre-B stage, expression of IRF4 is rapidly induced by pre-BCR signaling, which in turn, binds and activates Ek3’. Elevated IRF4 further promotes light chain rearrangement by inducing cell cycle exit and by attenuation of IL-7 signaling[17,20]. Thus, IL-7 signaling and IRF4 together orchestrate the sequential rearrangement of Ig H and L chain in B cell development.
Role of IRF4 and 8 in receptor editing
Although IRF4 and 8 function redundantly to control pre-B cell development, IRF4, but not IRF8, is uniquely required for receptor editing[21]. Receptor editing is the primary means through which B cells revise antigen receptors and maintain self-tolerance. Receptor editing at the immature B cell stage can be induced by a self-reactive BCR[34]. Interestingly, expression of IRF4 but not IRF8 is rapidly induced by self-antigen in immature B cells suggesting that IRF4 plays a unique role in receptor editing[21]. Indeed, the secondary immunoglobulin gene rearrangements triggered by membrane-bound antigen during receptor editing were defective in IRF4 but not IRF8 deficient mice[21]. Importantly, induction of RAGs by self-reactive BCR was minimally affected in the absence of IRF4. BCR signaling at the immature B cell stage not only induces the expression of RAGs but could also promotes and maintains the activation of light chain loci for efficient receptor editing[35]. Accordingly, the expression of light chain germline transcripts is low in immature B cells deficient for IRF4, indicating that the impairment of secondary immunoglobulin gene rearrangements in IRF4 deficient mice is caused by defect in the activation of light chain loci[21]. Moreover, in the absence of IRF4, the defect in secondary rearrangement appears to be more severe at the λ than at the κ light chain loci[21]. This finding is consistent with a recent report showing that rearrangement of Igλ loci is more dependent on IRF4 than the Igκ loci[20].Thus, IRF4 functions as a nuclear effector of a BCR-initiated signaling pathway that promotes secondary rearrangement of immunoglobulin genes during the immature B cell stage. Therefore by initiating receptor editing, IRF4 plays an important role in the maintenance of B cell self-tolerance.
Role of IRF4 and 8 in the germinal center reaction and plasma cell differentiation
Germinal centers (GC) are transient structures existing in the secondary lymphoid organs that are critical for T-cell dependent immune responses (see Box 3). Recently, a series of studies have demonstrated that IRF4 and 8 regulate the expression of the transcription factors Bcl-6, AID and Blimp-1 and therefore play a critical role in the GC reaction and plasma cell generation [13,14,16,19]. In contrast to their role in pre-B cell development where IRF4 and 8 function in an overlapping fashion, IRF4 and 8 appear to play critical and non-redundant roles in the GC reaction and plasma cell development (Figure 3).
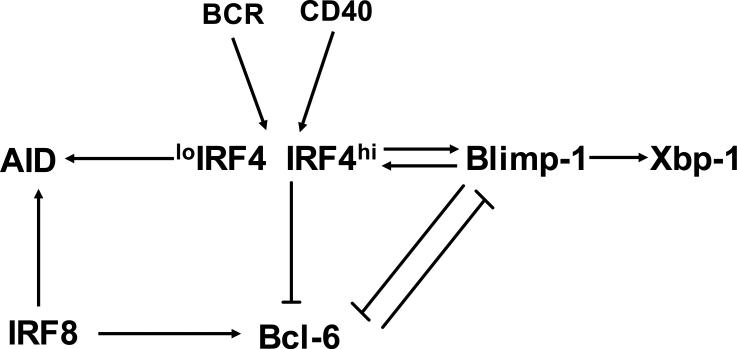
Fig. 3. IRF4 and 8 regulate the germinal center reaction and plasma cell development.
IRF8 modulates the germinal center (GC) reaction by inducing the expression of Bcl-6 and activation-induced cytidine deaminase (AID) in centroblasts. IRF4, whose expression can only be detected in subsets of centrocytes, is critical for CSR and the downregulation of Bcl6 at the end stage of the GC reaction. Expression of IRF4 can be induced in GC B cells by signals emanating from the B cell receptor (BCR), CD40, and cytokines. It is proposed that a low level of IRF4 induces AID to promote CSR whereas a high level of IRF4 turns on Blimp-1 to favor plasma cell development. Blimp-1 is also found to induce IRF4. This autoregulatory loop between IRF4 and Blimp-1 is suggested to be necessary for overcoming Bcl-6 mediated repression of the Blimp-1 gene.
IRF8 directly induces Bcl6 and AID to promote GC formation
Expression of IRF8 is high in centroblasts but low in plasma cells, suggesting that IRF8 might play a role in the GC reaction [16,36]. Supporting this notion is the finding that IRF8 directly regulates the expression of Bcl-6 and AID in GC B cells. In its absence, the expression of Bcl-6 and AID in GC B cells is reduced [16]. However, the GC, although structurally disorganized, can still be generated in the absence of IRF8, indicating that IRF8 modulates but is not essential for the GC reaction.
IRF4 is critical for class switch recombination
Early evidence linking IRF4 with the GC reaction and plasma cell development came from studies analyzing IRF4 germline mutant mice; the IRF4 germline mutant mice fail to form GC and are defective in the generation of plasma cells[37]. In a subsequent study, the IRF4 gene was conditionally deleted in GC B cells by the Cre recombinase whose expression is driven by the promoter of IgH Cλ1 switch transcript[19]. This elegant model demonstrated that IRF4 is critical for class switch recombination (CSR) in GC B cells and further showed that the impaired CSR is a result of failed induction of AID. Independently, another study [13] demonstrated that CSR is impaired in the IRF4 germline mutant B cells, a defect which could be rescued by ectopic expression of AID. Surprisingly, GC can be readily detected in mice where IRF4 is deleted specifically in GC B cells, suggesting that IRF4 is dispensable for GC formation [19]. Consistent with this view, immunohistochemistry shows that IRF4 protein expression cannot be detected in the majority of GC B cells[36,38]. Thus, the failure to form GC in IRF4 germline mutant mice is most likely due to defects in the generation of the naïve B cell pool which supply the GC.
IRF4 downregulates Bcl-6 in GC B cells
Expression of Bcl-6 is downregulated in post GC B cells concomitantly with the induction of plasma cell development. A recent study has identified IRF4 as transcriptional repressor that directly downregulates Bcl-6 expression in GC B cells[14]. Furthermore, CD40 signaling was found to directly induce the expression of IRF4 in GC B cells which in turn, binds to the promoter region of the Bcl6 gene to repress its expression. Thus, induction of IRF4 at the end stage of the GC downregulates Bcl-6 and likely contributes to the termination of the GC reaction. Constitutive expression of Bcl-6 gene caused by chromosomal translocation or mutations is often found in diffuse large B cell lymphoma (DLBCL). Interestingly, in subsets of DLBCL, the chromosomal translocation or mutations disrupt the binding motif for IRF4 on the Bcl-6 regulatory region[14].
IRF4 is essential for plasma cell development
Recent studies have established IRF4 as a critical regulator of plasma cell development[13,19]. Analyzing lipopolysaccahride-induced plasma cell differentiation, one study [13] revealed that IRF4 directly induces the expression of Blimp-1 to promote plasma cell development. Interestingly, expression of IRF4 can also be induced by Blimp-1 in plasma cells[39]. This autoregulatory loop between IRF4 and Blimp-1 is suggested to be necessary for overcoming Bcl-6 mediated repression of the Blimp-1 gene, an important event for initiation of plasma cell development[13]. This study further revealed that a higher level of IRF4 is required for plasma cell development and led to the proposal that low levels of IRF4 induce the expression of AID and CSR whereas high levels of IRF4 induce Blimp-1 to facilitate plasma cell development (Figure 3). Plasma cell differentiation is regulated by BCR affinity and in that regard, graded levels of IRF4 established by BCR of different affinity would coordinate BCR signaling strength with the GC reaction and plasma cell generation[13,40]. It should be noted that expression of Blimp-1 was not found to be affected in a separate study using GC specific IRF4 mutant B cells[19]. The findings of this study instead suggest that IRF4, working in parallel with Blimp-1, induces the expression of Xbp-1 to promote plasma cell development. The reason for this discrepancy is not clear, however, the detection of binding by IRF4 to the Blimp-1 gene by a chromatin immunoprecipitation assay strongly suggests that IRF4 regulates Blimp-1 expression[13]. Finally, expression of IRF4 in B cells can be negatively regulated by MITF (microphthalamia-associated transcription factor) through an unknown mechanism[41]. MITF mutant B cells express high levels of IRF4 and undergo spontaneous plasma cell differentiation, indicating that MITF is a negative regulator of plasma cell development.
Concluding remarks
The past couple years have witnessed tremendous progress in our understanding of IRF4 and 8 in B cell development. It has become clear that IRF4 and 8 are essential transcriptional regulators throughout B cell development. Numerous putative targets of IRF4 and 8 in B cells have been identified and the lists of genes are sure to increase in the coming years. Additional efforts, however, are required to determine which of those targets are directly regulated by IRF4 and 8. Furthermore, it would be important to distinguish the genes that can be regulated by both IRF4 and 8 from those that are regulated independently by IRF4 and IRF8. IRF4 and 8 functions are known to be regulated by factors with which they interact. The proteins that have been found to interact with IRF4 or IRF8 in B cells so far include PU.1, Spi-B, E47, NFAT, Stat6, Bcl-6 and Blimp-1[4,6,42-44]. Efforts to identify the stage specific interacting partners of IRF4 and 8, and to understand how they modify the function of IRF4 and 8 at different stages of B cell development would provide valuable insights towards our comprehension of the roles of IRF4 and 8 in B cell development.
Box 1. Large pre-B and small pre-B cells.
After a productive immunoglobulin heavy chain rearrangement at the pro-B stage, heavy chain protein μ pairs with the surrogate light chain (SLC) λ5 and Vpre-B. Together with the signaling molecules Igα and Igβ, they form the pre-B cell receptor (pre-BCR) on the B cell surface[45]. Pre-B lymphocytes consist of two distinct cell populations: large pre-B cells are newly generated pre-B cells that express pre-B cell receptor (pre-BCR) on the surface and are highly proliferative; small pre-B cells are derived from large pre-B cells that have downregulated pre-BCR and withdrawn from cell cycle. Light chain rearrangement, transcription, and receptor editing take place primarily in these quiescent pre-B cells.
Box 2. Dynamic changes of Igκ loci during B cell development.
At the pro-B stage, Igκ loci move away from the nuclear periphery and become centrally localized, an event that is believed to facilitate light chain rearrangement[46]. However, at the pre-B stage, one Igκ allele becomes associated with pericentromeric heterchromatin, which represses V(D)J rearrangement on this allele and instead favors rearrangement on the allele located away from pericentromeric chromatin[47]. The Ikaros family of transcription factors have been shown to inhibit Igκ rearrangement in pre-B cells via targeting Igκ loci to pericentromeric heterochromatin through a the cis-acting element Sis, an event that has been linked to monoallelic silencing of Igκ loci[48].
Box 3. Germinal center reaction and its essential regulators.
Upon engaging cognate antigen, naïve B cells enter primary follicles of secondary lymphoid organs such as spleens, lymph nodes, Peyer's patches and tonsils, undergo very rapid cell proliferation, and form a microscopically distinguishable structure called a germinal center (GC) [49]. The GC consists of dark and light zones; the proliferating cells also known as centroblasts mainly reside in the dark zone whereas the nonproliferating cells also called centrocytes are concentrated in the light zone. Somatic hypermutation (SHM) of the immunoglobulin genes mainly takes place in the dark zone centroblasts, whereas the later stages of the B cell response e.g. class switch recombination (CSR), affinity maturation and initiation of plasma cell development occur in the light zone centrocytes. Prior to the initiation of plasma cell development, GC centrocytes compete for binding to antigen associated with follicular dendritic cells (FDC) and for T cell help to undergo affinity maturation and CSR[50]. Post GC plasma cell development is initiated in a small subset of GC centrocytes.
Bcl-6, a member of POZ/BTB zinc finger protein family of transcriptional repressors, is a master regulator of the GC reaction[50]. Within the B cell lineage, expression of Bcl6 protein can only be detected in the GC B cells. Mice lacking Bcl-6 cannot form GC and fail to mount the T cell dependent immune response. In GC B cells, Bcl-6 is required to suppress apoptosis and promote proliferation of centroblasts.
AID (activation-induced cytidine deaminase) catalyzes the targeted deamination of deoxycytidine residues on DNA. The expression of AID is induced in GC B cells and is essential for SHM and CSR of immunoglobulin gene in centroblasts and centrocytes respectively.
Blimp-1 (B lymphocyte induced maturation protein 1; also known as PRDI-BF1 or PRDM1) is a zink figer containing transcriptional repressor. Expression of Blimp-1 is induced in subsets of GC centrocytes and in plasma cells. Blimp-1 promotes plasma cell development by suppressing genes associated with the GC program and induces the expression of those essential for plasma cell development [51].
XBP-1 (X-box binding protein 1) is a bZip transcriptional activator. Expression of XBP-1 is induced by ER stress response (unfolded protein response). Xbp-1 is expressed at a high level in plasma cells and is essential for maintaining the secretory phenotype of the plasma cells.
ACKNOWLEDGEMENTS
The author wishes to thank Dr. Neena Haider for critical reading of the manuscript. R. Lu is supported by National Institutes of Health grant AI67891.
REFERENCES
- 1.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 2.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 3.Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. Embo J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 5.Tamura T, Thotakura P, Tanaka TS, Ko MS, Ozato K. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 2005;106:1938–1947. doi: 10.1182/blood-2005-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagulapalli S, Atchison ML. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol Cell Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A, Meraro D. ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J Interferon Cytokine Res. 2002;22:153–160. doi: 10.1089/107999002753452764. [DOI] [PubMed] [Google Scholar]
- 9.Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 10.Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci U S A. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 13.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 16.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 20.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Pathak S, Ma S, Trinh L, Lu R. A role for interferon regulatory factor 4 in receptor editing. Mol Cell Biol. 2008;28:2815–2824. doi: 10.1128/MCB.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol Cell Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. Embo J. 2001;20:2812–2822. doi: 10.1093/emboj/20.11.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. Embo J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Loo PF, Dingjan GM, Maas A, Hendriks RW. Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity. 2007;27:468–480. doi: 10.1016/j.immuni.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Gomez-del Arco P, Maki K, Georgopoulos K. Phosphorylation controls Ikaros's ability to negatively regulate the G(1)-S transition. Mol Cell Biol. 2004;24:2797–2807. doi: 10.1128/MCB.24.7.2797-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol. 2005;25:1645–1654. doi: 10.1128/MCB.25.5.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang HE, Hsu LY, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin kappa locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 30.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 31.Inlay MA, Tian H, Lin T, Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa Jkappa rearrangement. J Exp Med. 2004;200:1205–1211. doi: 10.1084/jem.20041135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 34.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 35.Pelanda R, Torres RM. Receptor editing for better or for worse. Curr Opin Immunol. 2006 doi: 10.1016/j.coi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Cattoretti G, Shaknovich R, Smith PM, Jack HM, Murty VV, Alobeid B. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 37.Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 38.Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- 39.Sciammas R, Davis MM. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J Immunol. 2004;172:5427–5440. doi: 10.4049/jimmunol.172.9.5427. [DOI] [PubMed] [Google Scholar]
- 40.Benson MJ, Erickson LD, Gleeson MW, Noelle RJ. Affinity of antigen encounter and other early B-cell signals determine B-cell fate. Curr Opin Immunol. 2007;19:275–280. doi: 10.1016/j.coi.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin L, Gerth AJ, Peng SL. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J Exp Med. 2004;200:115–122. doi: 10.1084/jem.20040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagulapalli S, Goheer A, Pitt L, McIntosh LP, Atchison ML. Mechanism of e47-Pip interaction on DNA resulting in transcriptional synergy and activation of immunoglobulin germ line sterile transcripts. Mol Cell Biol. 2002;22:7337–7350. doi: 10.1128/MCB.22.20.7337-7350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta S, Jiang M, Anthony A, Pernis AB. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J Exp Med. 1999;190:1837–1848. doi: 10.1084/jem.190.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S, Anthony A, Pernis AB. Stage-specific modulation of IFN-regulatory factor 4 function by Kruppel-type zinc finger proteins. J Immunol. 2001;166:6104–6111. doi: 10.4049/jimmunol.166.10.6104. [DOI] [PubMed] [Google Scholar]
- 45.Martensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 47.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, Garrard WT. A recombination silencer that specifies heterochromatin positioning and ikaros association in the immunoglobulin kappa locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 51.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]