Abstract
Death-associated protein kinase (DAPK) has been found associated with HSP90, and inhibition of HSP90 with 17-alkyl-amino-17-demethoxygeldanamycin reduced expression of DAPK. These results were extended to determine whether the degradation of DAPK in the absence of HSP90 activity is dependent on the ubiquitin-proteasome pathway. Our results show that treatment of cells with geldanamycin (GA) leads to degradation of DAPK, and this degradation is attenuated by the proteasome inhibitor, lactacystin. GA-induced DAPK degradation is also dependent on phosphorylation of DAPK at Ser308, and the cellular levels of phospho(Ser308)-DAPK dramatically increase in response to GA treatment. Expression of two distinct ubiquitin E3 ligases, carboxyl terminus of HSC70-interacting protein (CHIP) or DIP1/Mib1, enhanced DAPK degradation, and conversely, short interfering RNA depletion of either CHIP or DIP1/Mib1 attenuated DAPK degradation. In vitro ubiquitination assays confirmed that DAPK is targeted for ubiquitination by both CHIP and DIP. Consistent with these results, DAPK is found in two distinct immune complexes, one containing HSP90 and CHIP and a second complex containing only DIP1/Mib. Collectively, these results indicate that strict modulation of DAPK activities is critical for regulation of apoptosis and cellular homeostasis.
Death-associated protein kinase (DAPK)2 is a Ca2+/calmodulin (CaM)-regulated Ser/Thr kinase. DAPK has been shown to have multiple functions in cell survival, tumor suppression, cell motility, and cytoskeletal organization (1–6). DAPK activities can be regulated by cellular calcium levels (7), autophosphorylation (7), ubiquitination (8, 9), and promoter methylation (10–13). Mechanisms regulating cellular stabilization and turnover of DAPK, although not completely defined, appear to be critical for modulating DAPK activities. Our previous studies have shown that DIP1/Mib1 (DAPK-interacting protein 1/mind-bomb 1) can ubiquitinate DAPK both in vitro and in vivo to result in degradation of DAPK by the proteasome. Recently, Citri et al. (14) reported that 17-AAG, an inhibitor of heat shock protein 90 (HSP90), induced DAPK degradation and in addition showed an interaction between the αC-β loop, within the kinase domain of DAPK and HSP90 (14).
HSP90 is an important chaperone, controlling the stability of thousands of client proteins. Most HSP90 client proteins are key signaling proteins and protein kinases that regulate cell survival, proliferation metastasis, and angiogenesis (15). Dynamic assembly of client protein-HSP90 complexes by the chaperone machinery stabilizes client proteins, and inhibition of HSP90 by ansamycin antibiotics like 17-allyalamino-17-demethoxygeldanamycin (17-AAG), geldanamycin (GA), or radicicol enhances the degradation of the clients via the ubiquitin-proteasome pathway (15, 16). GA directly binds to the amino-terminal ATPase pocket of HSP90 at a higher affinity than ATP, locking this domain in its ADP-bound conformation. This conformational change inhibits HSP90 function and induces client protein degradation (15). Carboxyl terminus of HSC70-interacting protein (CHIP) is a U-box-containing E3 ubiquitin ligase that binds through its tetratricopeptide repeat (TPR) domain to independent TPR acceptor sites on HSP90 and HSP70 (17, 18). CHIP has been shown to facilitate the ubiquitination of HSP90 client proteins, such as cytochrome P450 2E1 (19), estrogen receptor-α (20), p53 (21), neuronal nitric-oxide synthase (22), nucleophosmin-anaplastic lymphoma kinase (23), and ErbB2 (24).
In the current study, we report that treatment of cells with HSP90 inhibitor GA leads to degradation of endogenous and recombinant DAPK. The GA-induced DAPK degradation was inhibited by proteasome inhibitor, lactacystin. In addition, transient transfection of CHIP and DIP into HEK293 cells induced degradation of recombinant DAPK, and co-immuno-precipitation analysis of cellular extracts showed an association between DAPK and HSP90, CHIP, and DIP, suggesting that these proteins form complexes in vivo. Collectively, these results demonstrate that cellular levels of DAPK are regulated by a complex of proteins, including HSP90 and two distinct ubiquitin ligases, CHIP and DIP.
EXPERIMENTAL PROCEDURES
Materials
Geldanamycin (GA), MG132, Histagged ubiquitin, anti-DAPK (DAP-55), anti-phospho-DAPK (phospho-Ser308), anti-vinculin, protease inhibitor mixture, and phosphatase inhibitor mixtures were purchased from Sigma. clasto-Lactacystin β-lactone, human ubiquitin-activating enzyme E1, and ubiquitin-conjugating enzyme 5a (UbcH5a) were purchased from Calbiochem. Anti-Omni probe (D-8) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). TrueBlot™ anti-mouse and anti-rabbit IgG beads were from eBioscience (San Diego, CA). Rabbit anti-CHIP antibody was purchased from Affinity BioReagents (Golden, CO). Mouse anti-HSP90 antibody was purchased from Stressgen Bioreagents (Ann Arbor, MI). Rabbit anti-His tag antibody and mouse anti-phosphomyosin light chain 2 (Ser19) were purchased from Cell Signaling Technology (Hercules, MA). Overnight Express™ instant TB medium was from Novagen (Madison, WI). Fugene 6 transfection reagent was purchased from Roche Applied Science. DharmaFect-1 siRNA transfection reagent was from Dharmacon (Lafayette, CO).
Plasmid Construction
The construction of pcDNA3.1/His-DAPK and p3×FLAG-DIP has been described previously (1, 8). The pcDNA-His6-CHIP, pcDNA-His6-CHIP(K30A), and pcDNA-His6-CHIP(H260Q) vectors have been previously described (20). The expression vector for human DAPK, pcDNA3-DAPK, was kindly provided by Dr. Adi Kimchi (Weizmann Institute of Science, Israel) Vectors for expression of hemagglutinin-tagged ubiquitin were provided by Dr. Dirk Bohmann (University of Rochester). The human CHIP cDNA was subcloned from pcDNA-His6-CHIP into the HindIII and XhoI sites of pET28a(+).
Cell Culture and Transient Transfection
HeLa cells and human embryonic kidney HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Transient transfection of HEK293 cells was carried out using equal amounts of total plasmid DNA (adjusted with the corresponding empty vectors) by using Fugene 6 transfection reagent according to the manufacturer’s guidelines. Short interfering RNAs (siRNAs) for CHIP were synthesized by Dharmacon using the published sequence (target sequence 5′-GGAGCAGGGCAAUCGUCUG-3′) (25). DIP siRNA was also from Dharmacon (target sequence 5′-UCAUUGGCAUUCGAUGGAA-3′). Nontargeting siRNAs were purchased from Dharmacon and used as negative control. DharmFect 1 was used for the siRNA transfection.
Western Blotting and Immunoprecipitation
Western blotting and immunoprecipitation were performed as described previously (1). Equivalent amounts of total cellular protein or immunoprecipitates were fractionated by electrophoresis through an SDS-polyacrylamide gel and transferred to nitrocellulose membrane. Immunoreactive proteins on Western blots were visualized using the Supersignal West Pico or West Femto detection systems (Pierce) according to the manufacturer’s directions. Cell extracts were prepared in a lysis buffer containing 0.1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 10 mM sodium phosphate, pH 7.2, 2 mM EDTA, 50 mM sodium fluoride, protease inhibitor mixture, and phosphatase inhibitor mixtures. For immunoprecipitation, the transiently transfected HEK293 cells expressing recombinant DAPK were washed with phosphate-buffered saline, and lysates were prepared in lysis buffer (50 mM Tris-HCl, pH 7.4, 0.1% Nonidet P-40, 1% Triton X-100, 10% glycerol, 1 mM EGTA, 1 mM EDTA, 0.15 M NaCl, 10 mM sodium fluoride, 2 mM sodium vanadate) containing protease inhibitor mixture. Cell lysates were clarified by centrifugation, and the supernatant was pre-cleared by incubation with Trueblot IgG (eBioscience) beads. For each immunoprecipitation, 1-ml aliquots of lysates were incubated with 4–8 μg of DAP-3 antibody, anti-phospho-DAPK antibody, anti-His antibody, or anti-DIP antibody at 4 °C for 2 h. The immune complexes were then isolated by the addition of 50 μl of Trueblot IgG beads and incubation for 1 h. The immune pellets were washed four times with lysis buffer containing a protease inhibitor mixture, and the immune complexes were resolved by electrophoresis and analyzed by Western blotting.
Expression and Purification of CHIP
His-CHIP was expressed in bacteria and purified by nickel-nitrilotriacetic acid-agarose following the manufacturer’s protocol (Qiagen). Protein concentrations of recombinant proteins were determined by the Bradford method and confirmed by SDS-polyacrylamide gel fractionation of samples followed by Coomassie Blue staining.
In Vitro Kinase Assays
Recombinant DAPK was immuno-precipitated from HEK293 cells and subjected to an in vitro kinase assay using purified chicken gizzard smooth muscle RLCs as described previously (1) with minor modifications. The amount of phosphorylated RLC was quantified from Western blotting of phospho-RLC using an antibody specific for RLC phosphorylated on Ser19.
In Vitro Ubiquitination Assays
Ubiquitination assays were performed as described previously (8). Briefly, recombinant DAPK was incubated in a purified system containing an E1 ubiquitin-activating enzyme (0.25 μM), an E2 UbcH5α (2 μM), Histagged ubiquitin (0.6 mM), ATP (4 μM), 5 mM MgCl2, 1 mM dithiothreitol, expressed as final concentrations, for 2 h at 30 °C in a total volume of 15 μl of 50 mM Tris-HCl, pH 7.5. Where indicated, CHIP purified from bacteria was added to the reactions. After incubation, 2× SDS-PAGE sample buffer was added, and reactions were analyzed by Western blotting to detect the ubiquitinated proteins.
RESULTS
Geldanamycin Induces Proteasome-dependent DAPK Degradation
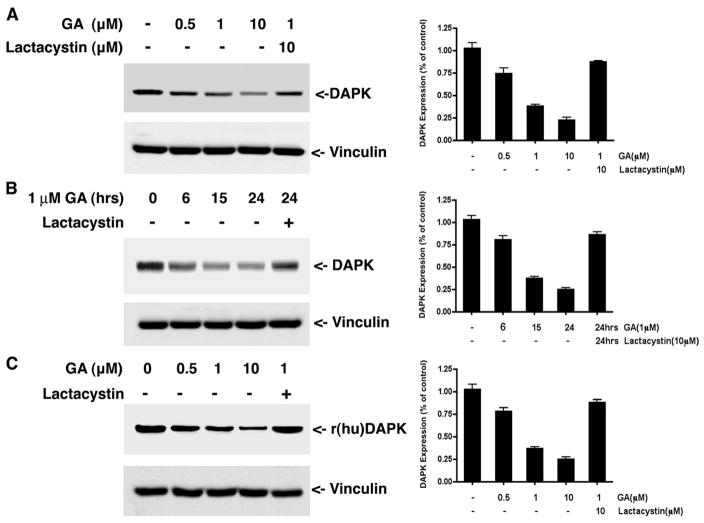
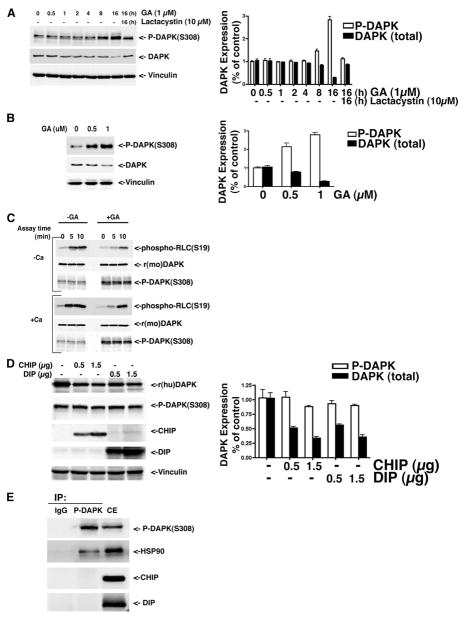
Our previous studies showed that the cellular levels of DAPK are post-translationally regulated by ubiquitin-mediated proteasomal degradation (8). That finding, together with the recent determination that DAPK is associated with HSP90 (14), led us to investigate whether the ubiquitin proteasome system is responsible for the degradation of DAPK when HSP90 activity is inhibited. To address this, HeLa cells were treated with varying concentrations of the HSP90 inhibitor, GA (1–10 μM for 24 h), and the levels of endogenous human DAPK were determined by Western blotting. GA induced degradation of endogenous DAPK in a dose-dependent manner (Fig. 1), and at 10 μM GA, the relative level of DAPK decreased to 28 ± 5% of the DAPK present in the untreated control. We then examined the time course of DAPK degradation in response to 1 μM GA. Maximal degradation of DAPK was observed between 15 and 24 h post-HSP90 inhibition by GA treatment (Fig. 1B). Treatment of HeLa cells with lactacystin (10 μM) antagonized GA-induced DAPK degradation and restored expression levels of DAPK to 86 ± 8% of the untreated control (Fig. 1B). This result is consistent with the proposal that in the absence of HSP90 activity, DAPK degradation is mediated by the ubiquitin proteasome pathway. To determine whether GA inhibition of HSP90 also induces degradation of exogenous DAPK, HEK293 cells were transfected with expression vectors for recombinant DAPK (rDAPK) and treated with GA in the presence or absence of lactacystin. As shown in Fig. 1C, GA treatment resulted in a 4-fold decrease in the expression levels of rDAPK, and lactacystin antagonized GA-induced rDAPK degradation. Together, these data confirm that DAPK is stabilized by HSP90 and suggest that in the absence of HSP90 activity, DAPK is degraded by the ubiquitin-proteasome pathway similar to other client proteins.
FIGURE 1. Geldanamycin treatment induces DAPK degradation through the ubiquitin proteasome pathway.
A, HeLa cells were treated with different concentrations of GA for 16 h in the presence or absence of lactacystin before lysis and analysis by Western blotting to detect the endogenous levels of human DAPK. Equal amounts of total cell protein were analyzed in each lane. B, HeLa cells were treated with 1 μM GA or 10 μM lactacystin for the indicated times and analyzed by Western blotting to detect the endogenous levels of human DAPK. C, HEK293 cells were transfected with 1 μg of pCDNA3-human DAPK. At 24 h, cells were then treated with Me2SO (vehicle) or the indicated concentrations of GA or 10 μM lactacystin for a further 16 h before analysis by Western blotting to detect recombinant human DAPK. Each blot is representative of three independent analyses, and vinculin is used as a loading control. r(hu)DAPK, recombinant human DAPK.
DAPK Forms Distinct Heterocomplexes with HSP90 and CHIP or DIP1/Mib
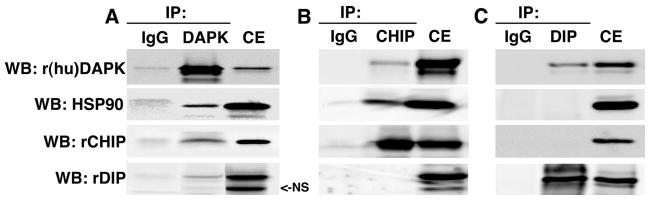
A recent study has shown that HSP90 recruits the U-box E3 ubiquitin ligase, CHIP, to target client proteins for ubiquitin-mediated proteasomal degradation (20), and our previous studies have shown that DAPK is associated with the RING E3 ligase DIP1/Mib1 (8). These findings suggested that the DAPK·HSP90 complex may be associated either with DIP1/Mib1 or CHIP or both E3 ligases. To define the components of the DAPK·HSP90 complex, HEK293 cells were co-transfected with pcDNA3-DAPK, pcDNA-His6-CHIP, and p3×FLAG-DIP1/Mib1. Endogenous human DAPK was immunoprecipitated using anti-DAPK antibody, and the immune complexes were analyzed by Western blotting using antibodies to detect DAPK, CHIP, DIP1/Mib1, and HSP90 (Fig. 2A). Consistent with the previous reports, DAPK co-immunoprecipitated with HSP90 (14) and DIP1/Mib1 (8). In addition to these two known partners for DAPK, CHIP was also detected in the immune complex, demonstrating that DAPK can interact with HSP90, CHIP, and DIP1/Mib1. In order to define the dynamic nature of the interactions between DAPK, CHIP, DIP1/Mib1, and HSP90, either CHIP or DIP were immunoprecipitated using anti-His and anti-DIP1/Mib1 antibodies (Fig. 2, B and C, respectively). The results in Fig. 2B show that when CHIP is immunoprecipitated, both DAPK and HSP90 are detectable in the immune complexes, but interestingly DIP1/Mib1 is not. Consistent with this, when DIP1/Mib1 is immunoprecipitated, only DAPK is detectable in the immune complexes, but HSP90 and CHIP are not (Fig. 2C). Together, these results suggest that DAPK forms two distinct complexes in cells; one complex includes both HSP90 and CHIP, and the second complex contains DIP1/Mib1 only.
FIGURE 2. DAPK forms two distinct heterocomplexes with HSP90 and CHIP or with DIP1/Mib1.
HEK293 cells were transfected with pCDNA3-human DAPK, pCDNA-His6-CHIP, or p3×FLAG-DIP. A, DAPK was immunoprecipitated (IP) with anti-DAPK antibody. B, CHIP was immunoprecipitated with anti-His tag antibody. C, DIP was immunoprecipitated with anti-DIP antibody. Western blots (WB) were probed with anti-DAPK, anti-HSP90, anti-CHIP, and anti-DIP1/Mib1 antibodies to detect recombinant human DAPK (r(hu)DAPK), recombinant CHIP (rCHIP), recombinant DIP1/Mib1 (rDIP), or endogenous HSP90. The blot is representative of three independent analyses. CE, cell extract; NS, nonspecific band.
Effect of CHIP Expression on DAPK Stabilization in HEK293 Cells
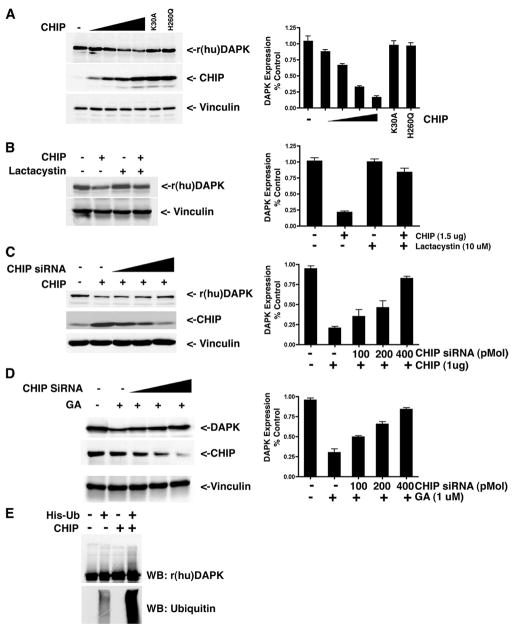
To investigate whether the U-box E3 ubiquitin ligase, CHIP, targets DAPK for proteasomal degradation, CHIP and human DAPK (rDAPK) were co-expressed in HEK293 cells. After a 48-h transfection period, the relative expression levels of CHIP and rDAPK were determined by immunoblotting of whole cell lysates. As shown in Fig. 3A, increasing the expression level of CHIP resulted in a decreased expression of rDAPK. Quantitation of the Western blots showed a 6-fold decrease in DAPK expression relative to the expression level in cells in the absence of exogenous CHIP. Expression of either of two mutant CHIP proteins, K30A or H260Q, was found to have little effect on DAPK expression levels. CHIP-K30A contains a point mutation in the TPR domain and does not bind HSP90 or CHIP-H260Q, which contains a point mutation in its U box domain and fails to bind its cognate E2, thus disabling its E3 ligase activity. Lactacystin treatment (10 μM during the last 16 h of the transfection) antagonized CHIP-induced DAPK degradation and stabilized DAPK expression (Fig. 3B), indicating that the exogenously expressed CHIP enhanced degradation of DAPK by ubiquitination and targeting to the proteasome. To investigate whether depletion of CHIP could restore DAPK expression, we used a previously described CHIP-specific siRNA (25). Transfection of increasing amounts of CHIP siRNA between 0 and 400 pM dramatically decreased the level of CHIP expression and attenuated CHIP-induced degradation of DAPK (Fig. 3C).
FIGURE 3. CHIP promotes DAPK degradation in HEK293 cells by ubiquitination.
A, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 μg of pMT-HA-Ub, and different amounts (0, 0.1, 0.5, 1.0, and 1.5 μg) of pCDNA-His6-CHIP or 1 μg of mutant CHIP constructs (K30A and H260Q) as indicated. The relative expression levels of recombinant human DAPK (r(hu)DAPK), CHIP, and vinculin (loading control) were determined using anti-DAPK, anti-CHIP, or anti-vinculin antibodies. B, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 g of pMT-HA-Ub, and 1.5 μg of pCDNA-His6-CHIP or 1.5 μg of pCDNA (vector control). At 24 h post-transfection, the cells were then treated with Me2SO or lactacystin for a further 16 h. The relative expression levels of recombinant human DAPK and vinculin (loading control) were determined by Western blotting. C, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 μg of pMT-HA-Ub, 1 μg of pCDNA-His6-CHIP, and different amounts of CHIP siRNA (0, 100, 200, and 400 pmol) or 400 pmol of nontargeting siRNA as control. The relative expression levels of recombinant human DAPK, CHIP, or vinculin (loading control) were determined by Western blotting using anti-DAPK, anti-CHIP, or anti-vinculin antibodies. D, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK and varying amounts of CHIP siRNA (0, 100, 200, and 400 pmol) or 400 pmol of nontargeting siRNA as control. At 24 h post-transfection, the cells were then treated with vehicle (Me2SO) or GA for a further 16 h and analyzed by Western blotting to detect recombinant human DAPK (r(hu)DAPK) protein levels. E, in vitro ubiquitination of DAPK by CHIP. Recombinant DAPK immunoprecipitated from HEK293 cells was incubated in a reaction mixture containing E1, E2, ATP, and purified bacterially expressed CHIP. The samples were analyzed by Western blotting (WB) using anti-DAPK or anti-ubiquitin antibodies. Each blot is representative of three independent analyses.
To determine whether CHIP plays a role in GA-induced DAPK degradation, CHIP siRNA was used to deplete endogenous levels of CHIP in HEK293 cells. Transfection of increasing levels of CHIP siRNA attenuated GA-induced DAPK degradation in a dose-dependent manner (Fig. 3D). To establish whether CHIP targets DAPK for ubiquitination, in vitro ubiquitination assays were performed as previously described (8). For these assays, HEK293 cells were transfected with X-press-tagged DAPK, and then DAPK was immunoprecipitated using an X-press tag antibody. These immune complexes were added to an in vitro ubiquitination assay consisting of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme UbcH5a (E2), Histagged ubiquitin, ATP, and bacterially expressed, purified CHIP. After incubation, ubiquitinated proteins were examined using Western blot analysis. As shown in Fig. 3E, inclusion of CHIP in the reaction increased ubiquitination of DAPK in vitro, a result consistent with the observed degradation of DAPK induced by expression of CHIP in vivo. Collectively, these results demonstrate that CHIP can target DAPK for ubiquitination and proteasome-mediated degradation.
DIP Overexpression Decreases and DIP Knockdown Increases DAPK Protein Level
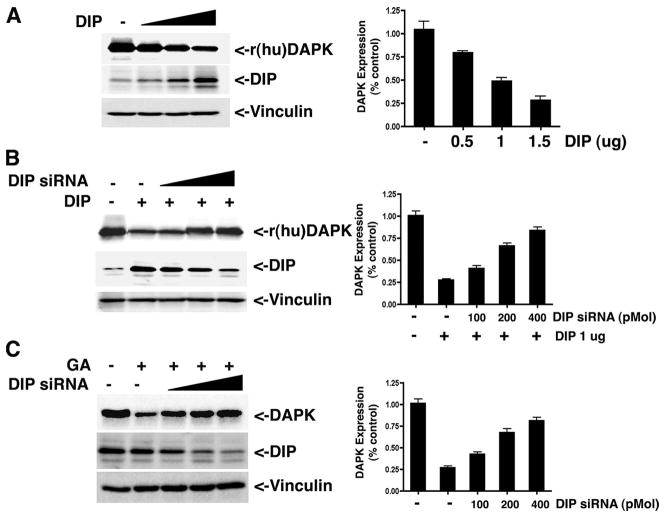
Previous studies using overexpression approaches have suggested that another E3 ubiquitin ligase, called DIP1/Mib1, targets DAPK for ubiquitination and proteasomal degradation (8). To confirm this, HEK293 cells were transfected with increasing amounts (0–1.5 μg) of plasmid DNA encoding DIP1/Mib1 and analyzed by Western blotting. As seen in Fig. 4A, DIP/Mib1 overexpression increased degradation of endogenous human DAPK in a dose-dependent manner to less than 30% of its original expression level, confirming that DAPK is targeted for ubiquitin proteasome-mediated degradation by DIP1/Mib1 (8).
FIGURE 4. DIP1/Mib1 promotes DAPK degradation in HEK293 cells by ubiquitination.
A, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 μg of pMT-HA-Ub, and different amounts (0, 0.5, 1.0, of 1.5 μg) of p3×FLAG-DIP1/Mib1. The relative expression levels of recombinant human DAPK (r(hu)DAPK) and DIP1/Mib1 were detected by Western blotting using anti-DAPK or anti-DIP1/Mib1 antibodies (8). B, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 μg of pMT-HA-Ub, 1 μg of p3×FLAG-DIP1/MIB1, and varying amounts of DIP1/MIB1 siRNA (0, 100, 200, and 400 pmol) or 400 pmol of nontargeting siRNA as control. The relative levels of recombinant human DAPK and DIP1/Mib1 were determined by Western blotting. C, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK and varying amounts of DIP1/MIB1 siRNA (0, 100, 200, and 400 pmol) or 400 pmol of nontargeting siRNA as control. At 24 h post-transfection, the cells were then treated with Me2SO or GA for a further 16 h. Cell lysates were analyzed by Western blotting to detect the expression levels of recombinant human DAPK or vinculin (loading control). Each blot is representative of three independent analyses.
To further establish that DIP1/Mib1 is a DAPK-specific ubiquitin E3 ligase, a DIP1/Mib1-specific siRNA was used to deplete exogenous DIP1/Mib1 in HEK293 cells. Depletion of DIP1/Mib1 attenuates ubiquitin-mediated degradation of DAPK induced by DIP (Fig. 4B). Because GA induces DAPK degradation through the ubiquitin proteasome pathway, and DIP1/Mib1 is an E3 ligase that can ubiquitinate DAPK, we investigated the role of DIP1/Mib1 in GA-induced DAPK degradation. As shown in Fig. 4C, DIP1/Mib1 depletion by its specific siRNA attenuated GA-induced DAPK degradation in a dose-dependent manner, restoring expression to 79% of its original level.
Inhibition of HSP90 Activity Induces DAPK Degradation and Decreases DAPK Activity
The catalytic activity of DAPK is inhibited by autophosphorylation of serine 308, which resides within the calmodulin-binding domain of this calcium-regulated protein kinase. Our previous studies have determined that, following activation, DAPK is ubiquitinated and degraded (9). Other studies have linked alterations of the phosphorylation status of proteins to ubiquitination (26–28). To extend these results, we investigated whether or not there is a potential link between the phosphorylation status of DAPK and its association with HSP90. To test this, HeLa cells were treated with 1 μM GA for 0.5–16 h in the presence or absence of 10 μM lactacystin. Whole cell lysates were prepared and then analyzed by Western blotting using antibodies to detect endogenous phospho(Ser308)-DAPK or total DAPK. Interestingly, these results show that the level of detectable phospho(Ser308)-DAPK was increased 3-fold within 16 h of GA treatment, whereas total DAPK levels diminished 4-fold (Fig. 5A). Interestingly, phospho(Ser308)-DAPK migrates as a doublet on these gels, suggesting the possibility that there may be additional modifications to the phospho(Ser308)-DAPK or multiple phosphorylation sites. Furthermore, there was a dose-dependent increase in the level of detectable phospho(Ser308)-DAPK observed in HeLa cells treated with GA (Fig. 5B), suggesting that the phosphorylated, inactive form of DAPK accumulates in the absence of HSP90 activity.
FIGURE 5. Inhibition of HSP90 activity by geldanamycin enhances DAPK phosphorylation and decreases DAPK catalytic activity.
A, HeLa cells were treated with 1 μM GA for the indicated times in the presence of absence of 10 μM lactacystin. Cell lysates were collected and analyzed by Western blotting to detect endogenous phosphorylated human DAPK (phospho(Ser308)-DAPK) or total endogenous DAPK using anti-phospho-DAPK and anti-DAPK antibodies. Vinculin was used as loading control. B, HeLa cells were treated with Me2SO or varying amounts of GA as indicated. At 16 h, cell lysates were collected and analyzed by Western blotting to detect endogenous phosphorylated DAPK (phospho(Ser308)-DAPK or total endogenous DAPK using anti-phospho(Ser308)-DAPK and anti-DAPK antibodies. C, HEK293 cells were transfected with pCDNA3.1/HisMouseDAPK (r(mo)DAPK). At 24 h post-transfection, the cells were then treated with Me2SO or GA for a further 16 h, and the cells were lysed and mouse DAPK was immunoprecipitated using anti-Omni probe antibody. The immune complexes were then utilized in an in vitro kinase assay in the absence (−) or presence (+) of calcium (Ca) as described under “Experimental Procedures.” Myosin II RLC was used as a substrate, and DAPK activity was determined by detection of phosphorylated RLC using anti-phospho-RLC (Ser19) and anti-Omni or anti-phospho(Ser308)-DAPK antibodies. Each blot is representative of three independent analyses. D, HEK293 cells were transfected with 0.5 μg of pCDNA3-human DAPK, 0.5 μg of pMT-HA-Ub, and different amounts (0, 0.5, and 1.5 μg) of pCDNA-His6-CHIP or p3×FLAG-DIP as indicated. The relative expression levels of total recombinant human DAPK (r(hu)DAPK), phospho-DAPK, CHIP, DIP1/Mib1, and vinculin (loading control) were determined using anti-DAPK, anti-phospho-DAPK, anti-His, anti-DIP, or anti-vinculin antibodies. E, HEK293 cells were transfected with pCDNA3-human DAPK, pCDNA-His6-CHIP, p3×FLAG-DIP. Phospho-DAPK was immunoprecipitated (IP) with anti-phospho-DAPK antibody. Western blots were probed with anti-phospho-DAPK, anti-phospho-HSP90, anti-His, and anti-DIP1/Mib1 antibodies, respectively. Each blot is representative of three independent analyses. CE, cell extract.
A previous report has demonstrated that autophosphorylation at Ser308 inhibited DAPK catalytic activity and that dephosphorylation of Ser308 was required for activation of DAPK (7). To determine whether GA inhibition of HSP90 activity would alter the catalytic activity of DAPK, an in vitro kinase assay using purified chicken gizzard myosin II regulatory light chains (RLCs) as the substrate was performed (1). HEK293 cells transfected with X-press-tagged DAPK were treated with 1 μM GA for 16 h prior to cell lysis and immunoprecipitation using an anti-X-press antibody. The immunoprecipitated DAPK was washed in assay buffer and added to the in vitro kinase reaction. Western blot analysis was carried out to determine the level of DAPK RLC phosphorylation activity using an anti-phospho-RLC antibody for Ser19 phosphorylation. Because DAPK is activated by Ca2+/CaM binding, the kinase assay was performed both in the presence and absence of Ca2+/CaM. In the absence of Ca2+/CaM, DAPK activity was much lower than the activity in the presence of Ca2+/CaM, a result consistent with previous studies (7, 9). In addition, we found that DAPK immunoprecipitated from GA-treated cells had much lower activity than DAPK in control cells irrespective of inclusion of Ca2+/CaM in the assay. The results of these in vitro kinase assays suggested that treatment of cells with GA significantly increases the cellular level of phospho(Ser308)-DAPK and results in a corresponding decrease in DAPK catalytic activity.
To further elucidate the mechanism resulting in accumulation of the phosphorylated DAPK in the presence of GA, HEK293 cells were transfected with different amounts of CHIP or DIP1/Mib1 DAPK together with a constant amount of human DAPK, and total DAPK or phosphorylated DAPK levels were determined by Western blotting. As shown in Fig. 5D, total DAPK levels were significantly decreased in the presence of CHIP or DIP1/Mib1, whereas there was no significant change in the expression level of phosphorylated DAPK. This suggests that both CHIP and DIP1/Mib1 target only the dephosphorylated, active DAPK and not phosphorylated inactive DAPK. Consistent with this suggestion, co-immunoprecipitation analysis also showed that there is no direct interaction between phosphorylated inactive DAPK and CHIP or DIP1/Mib1, although HSP90 still can be co-immunoprecipitated with phosphorylated DAPK (Fig. 5E). Collectively, these data indicate that GA inhibition of HSP90 activities induces preferential degradation of the dephosphorylated, active DAPK to result in accumulation of auto-phosphorylated, inactive DAPK and attenuation of DAPK catalytic activity.
DISCUSSION
Previous studies have established that the catalytic activity of DAPK is regulated by Ca2+/CaM and by autophosphorylation of Ser308 within its calmodulin-binding domain. It has also been shown that the expression levels of DAPK are modulated by ubiquitin-mediated proteasomal degradation (8) and that DAPK interacts with HSP90 (14). Those studies suggest that an intricate network of protein modifications and interactions acts to regulate the activities of this fascinating Ser/Thr protein kinase. In the present study, by identifying novel components of the DAPK signaling complex and defining their roles in modulating the cellular levels of DAPK, we have extended our understanding of how DAPK activities are regulated.
It has been established that in the presence of HSP90 inhibitors like GA or 17-AAG, many client proteins are destabilized and subsequently degraded via the ubiquitin proteasome pathway (29). In addition, other studies have linked phosphorylation status of proteins to ubiquitination and degradation (26–28). Along these lines, we have recently reported that subsequent to its dephosphorylation and activation, DAPK is rapidly degraded through the ubiquitin proteasome pathway (9). The recent determination that DAPK and HSP90 are associated (14) prompted us to further investigate the events regulating the cellular levels of DAPK. We show for the first time that GA inhibition of HSP90 results in DAPK degradation by the ubiquitin proteasome pathway and that DAPK degradation is mediated by the activities of two distinct E3 ligases, CHIP and DIP1/Mib1. This conclusion is supported by studies demonstrating that the proteasome inhibitor lactacystin attenuates GA-induced DAPK degradation (Fig. 1). However, we also noted that lactacystin does not completely restore expression of DAPK to control levels, suggesting the possibility that additional proteolytic systems may operate to modulate the cellular levels of DAPK.
The ubiquitin proteasome degradation pathway is an important triage pathway that enforces quality control on protein activities in cells and consists of activating (E1), conjugating (E2), and ligating (E3) enzymes (30). The E3 ligases are a large group of diverse molecules that function to identify target substrates and mediate the covalent ligation of ubiquitin by direct interaction with the target (31). We previously identified a novel RING E3 ubiquitin ligase, called DIP1, which was later also identified as mindbomb (Mib1) (32). Those studies showed that DIP1/Mib1 directly interacts with and ubiquitinates DAPK to promote its proteasomal degradation (8). In the present study, we have extended those results and determined that siRNA-mediated depletion of DIP1/Mib1 expression rescues DAPK from proteasome-mediated degradation (Fig. 4). In addition, we show that siRNA-mediated depletion of DIP1/Mib1 attenuates GA-induced DAPK degradation, linking DIP1/Mib1-directed ubiquitination and degradation of DAPK to DAPK degradation induced in response to HSP90 inhibitors (Fig. 4).
Previous studies have demonstrated that CHIP, a U-box-containing E3 ligase, binds through its TPR domain to TPR acceptor sites on HSP90 to mediate ubiquitination of at least some HSP90 client proteins (18, 33). Based on those reports and our finding that DAPK is targeted for proteasome degradation by ubiquitination, we asked whether CHIP could also target DAPK for degradation. The present studies demonstrate that overexpression of CHIP induces the degradation of DAPK and that inhibition of the proteasome with the proteasome inhibitor, lactacystin, attenuates the DAPK degradation caused by CHIP overexpression (Fig. 3). Consistent with this, we also show, using an in vitro ubiquitination assay, that CHIP increases ubiquitination of DAPK (Fig. 3). Finally, by using siRNA to deplete levels of CHIP, we can rescue DAPK from CHIP-induced degradation. Together, these results suggest that CHIP can also target DAPK for ubiquitination and degradation by the proteasome.
Since CHIP has been previously reported to associate directly with HSP90 in a heterocomplex (18), we asked whether CHIP, DIP1/Mib1, and DAPK co-associate in a heterocomplex with HSP90. These studies revealed that two distinct DAPK hetero-complexes are present in cells. In one complex, HSP90, CHIP, and DAPK are associated (Fig. 2, A and B). In a second complex, however, only DAPK and DIP1/Mib1 are associated (Fig. 2C). The finding that two E3 ligases are associated with the DAPK is surprising; however, it may suggest that regulation of DAPK activities by proteasomal degradation is critical and that when DAPK is complexed with HSP90 it can be ubiquitinated by CHIP and when released from HSP90 it can be ubiquitinated by DIP1/Mib1. This heightened surveillance of DAPK activities by HSP90, CHIP, and DIP1/Mib1 may also reflect the central importance of DAPK in regulation of cellular death and survival pathways (1, 3). Consistent with this suggestion are the recent findings that Cdt-1, a DNA licensing and replication protein, and p27Kip1, a DNA replication inhibitor, are targeted for degradation by two E3 ubiquitin ligases, DDB1-Cul4 and SCF-Skp2 (34, 35). One question these results raise that is currently being investigated is whether CHIP and DIP1/Mib1 utilize the same or distinct ubiquitination sites within DAPK and the location of these ubiquitination sites.
In the current report, we also show that treatment of cells with GA results in accumulation of inactive phospho (Ser308)-DAPK, consistent with the low catalytic activity found using immunoprecipitated total DAPK in an in vitro kinase assay (Fig. 5). Together, these data suggest that inhibition of HSP90 activity not only induces DAPK degradation, it also attenuates DAPK catalytic activity by the selective degradation of active DAPK. Unexpectedly, we found that GA also attenuated degradation of DAPK by both E3 ligases (Figs. 3 and 4), although HSP90 does not appear to be associated with the DAPK·DIP1/Mib1 complex. This finding suggests that the effect of GA on DIP1/Mib1-mediated degradation of DAPK is not directly related to its association with HSP90. If this is true, then following dephosphorylation and activation, DAPK would be released from the HSP90·CHIP complex, phosphorylate substrates such as myosin II RLC, and then become ubiquitinated through its association with DIP1/Mib1. Prior to its release from HSP90, any misfolded DAPK that is unable to achieve a native conformation could be targeted for degradation by CHIP. Alternatively, it is also possible that a fraction of HSP90 is in fact associated with DIP1/Mib1·DAPK heterocomplexes, but its association is weak or below the level that is detectable by Western blotting.
The finding that DAPK is a target for at least two E3 ubiquitin ligases (DIP1/Mib1 and CHIP), one of which is found complexed with HSP90, suggests that these two heterocomplexes function to scrutinize the folding and activation of DAPK as well as to modulate DAPK activities in cells. The finding that GA inhibition of HSP90 enhances degradation of the active, dephosphorylated DAPK suggests a link between activation and degradation (Fig. 5) and is consistent with our previous results showing that tumor necrosis factor treatment results in DAPK dephosphorylation and activation, which is subsequently attenuated by the ubiquitin-mediated proteasomal degradation of DAPK (9).
In summary, we find that inhibition of HSP90 results in degradation of active dephosphorylated DAPK via the ubiquitin proteasome pathway. The discovery that GA treatment dramatically increases the expression levels of the inactive form of DAPK, phospho(Ser308)-DAPK, whereas total DAPK decreases them, suggests that HSP90 activity is essential for stabilization of activated DAPK. The finding that DAPK can be isolated from cells in heterocomplexes composed of HSP90 and CHIP or DIP1/Mib1 and that DAPK is targeted for proteasomal degradation through the activities of two distinct E3 ubiquitin ligases (CHIP, a U-box E3 ligase, and DIP1/Mib1, a RING E3 ligase) indicates that heightened surveillance and modulation of DAPK activities is critical to accurate regulation of apoptosis and cellular homeostasis.
Footnotes
The abbreviations used are: DAPK, death-associated protein kinase; phospho(Ser308)-DAPK, DAPK phosphorylated at Ser308; GA, geldanamycin; RLC, myosin II regulatory light chain, CaM, calmodulin; Ub, ubiquitin; CHIP, carboxyl terminus of HSC70-interacting protein; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; siRNA, short interfering RNA.
This work was supported by NHLBI, National Institutes of Health (NIH), Grants HL54118 and DK062810 (to P. J. G.), NCI, NIH, Grant CA 085289, American Cancer Society Research and Alaska Run for Woman Grant TBE-104125, and the Walther Cancer Institute (to K. P. N.).
References
- 1.Jin Y, Blue EK, Dixon S, Hou L, Wysolmerski RB, Gallagher PJ. J Biol Chem. 2001;276:39667–39678. doi: 10.1074/jbc.M101886200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bialik S, Kimchi A. Annu Rev Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Gallagher PJ. J Biol Chem. 2003;278:51587–51593. doi: 10.1074/jbc.M309165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider-Stock R, Roessner A, Ullrich O. Int J Biochem Cell Biol. 2005;37:1763–1767. doi: 10.1016/j.biocel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Kuo JC, Wang WJ, Yao CC, Wu PR, Chen RH. J Cell Biol. 2006;172:619–631. doi: 10.1083/jcb.200505138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen RH, Wang WJ, Kuo JC. J Biomed Sci. 2006;13:193–199. doi: 10.1007/s11373-005-9063-5. [DOI] [PubMed] [Google Scholar]
- 7.Shohat G, Spivak-Kroizman T, Cohen O, Bialik S, Shani G, Berrisi H, Eisenstein M, Kimchi A. J Biol Chem. 2001;276:47460–47467. doi: 10.1074/jbc.M105133200. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, Blue EK, Gallagher PJ. J Biol Chem. 2006;281:39033–39040. doi: 10.1074/jbc.M605097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan AW, Chan MW, Lee TL, Ng EK, Leung WK, Lau JY, Tong JH, Chan FK, To KF. Oncol Rep. 2005;13:937–941. [PubMed] [Google Scholar]
- 11.Collins Y, Dicioccio R, Keitz B, Lele S, Odunsi K. Int J Gynecol Cancer. 2006;16(Suppl 1):195–199. doi: 10.1111/j.1525-1438.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 12.Chim CS, Fung TK, Wong KF, Lau JS, Liang R. J Hum sGenet. 2006;51:832–838. doi: 10.1007/s10038-006-0029-x. [DOI] [PubMed] [Google Scholar]
- 13.Voso MT, Gumiero D, D’Alo F, Guidi F, Mansueto G, Di Febo AL, Massini G, Martini M, Larocca LM, Hohaus S, Leone G. Haematologica. 2006;91:1108–1113. [PubMed] [Google Scholar]
- 14.Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 15.Georgakis GV, Younes A. Future Oncol. 2005;1:273–281. doi: 10.1517/14796694.1.2.273. [DOI] [PubMed] [Google Scholar]
- 16.Isaacs JS, Xu W, Neckers L. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 17.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 19.Morishima Y, Peng HM, Lin HL, Hollenberg PF, Sunahara RK, Osawa Y, Pratt WB. Biochemistry. 2005;44:16333–16340. doi: 10.1021/bi0515570. [DOI] [PubMed] [Google Scholar]
- 20.Fan M, Park A, Nephew KP. Mol Endocrinol. 2005;19:2901–2914. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- 21.Esser C, Scheffner M, Hohfeld J. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 22.Peng HM, Morishima Y, Jenkins GJ, Dunbar AY, Lau M, Patterson C, Pratt WB, Osawa Y. J Biol Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 23.Bonvini P, Dalla Rosa H, Vignes N, Rosolen A. Cancer Res. 2004;64:3256–3264. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- 24.Zhou P, Fernandes N, Dodge IL, Reddi AL, Rao N, Safran H, DiPetrillo TA, Wazer DE, Band V, Band H. J Biol Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 25.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doucet C, Gutierrez GJ, Lindon C, Lorca T, Lledo G, Pinset C, Coux O. BMC Biochem. 2005;6:27. doi: 10.1186/1471-2091-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees I, Lee S, Kim H, Tsai FT. Biochim Biophys Acta. 2006;1764:1073–1079. doi: 10.1016/j.bbapap.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Burrows F. J Mol Med. 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A, Ciechanover A, Varshavsky A. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 31.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 32.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 33.Cyr DM, Hohfeld J, Patterson C. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 34.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, Raychaudhuri P. Mol Cell Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu E, Li X, Yan F, Zhao Q, Wu X. J Biol Chem. 2004;279:17283–17288. doi: 10.1074/jbc.C300549200. [DOI] [PubMed] [Google Scholar]