Abstract
Background
The regulation of gene expression via a 3′ untranslated region (UTR) plays essential roles in the discrimination of the germ cell lineage from somatic cells during embryogenesis. This is fundamental to the continuation of a species. Mouse NANOS3 is an essential protein required for the germ cell maintenance and is specifically expressed in these cells. However, the regulatory mechanisms that restrict the expression of this gene in the germ cells is largely unknown at present.
Methodology/Principal Findings
In our current study, we show that differences in the stability of Nanos3 mRNA between germ cells and somatic cells is brought about in a 3′UTR-dependent manner in mouse embryos. Although Nanos3 is transcribed in both cell lineages, it is efficiently translated only in the germ lineage. We also find that the translational suppression of NANOS3 in somatic cells is caused by a 3′UTR-mediated mRNA destabilizing mechanism. Surprisingly, even when under the control of the CAG promoter which induces strong ubiquitous transcription in both germ cells and somatic cells, the addition of the Nanos3-3′UTR sequence to the coding region of exogenous gene was effective in restricting protein expression in germ cells.
Conclusions/Significance
Our current study thus suggests that Nanos3-3′UTR has an essential role in translational control in the mouse embryo.
Introduction
The manner in which genes are regulated to produce the correct combination of proteins for every cell type remains a fundamental question in biology. In many cases, gene expression is primarily regulated via transcription under the control of enhancer and promoter sequences. However, it is now becoming clear that post-transcriptional regulation mediated via a 3′ untranslated region (UTR) plays key roles in the control of mRNA stability and/or translation. A critical step in the establishment of elaborate germ cell lineages during early embryogenesis in nematodes, fly, fish and frog is the temporal and spatial regulation of several proteins via mechanisms that are dependent on the 3′UTR of maternal mRNAs including nanos [1], [2], [3], [4].
The nanos genes are evolutionarily conserved among many organisms and play important roles during germ cell development [5], [6], [7], [8], [9], [10], [11]. During germ cell specification in Drosophila, maternal nanos mRNA becomes localized in the germ plasm in the posterior part of the egg [12], [13]. This localization is inefficient, however, and translational repression is therefore essential for the restricted production of Nanos protein in the posterior region. This repression is mediated by a 90-nucleotide translational control element (TCE) in the 3′UTR of nanos mRNA [14], [15], [16], [17], [18] to which Smaug (Smg) or Glorund (Glo) bind [19], [20], [21]. On the other hand, the localization in the germ plasm and subsequent translational activation of nanos mRNA is regulated by the Oskar (Osk) protein via 3′UTR-dependent mechanisms [19], [22], [23]. In Danio rerio, maternal nanos1 mRNA is also present in a whole oocyte, but only a portion is localized to the germ plasm and translated specifically in the PGC. The translation of the bulk of nanos1 mRNA in somatic cells is then rapidly degraded during embryogenesis. The regulation of nanos1 both in the PGC and somatic cells depends on three elements within the nanos1-3′UTR: (1) a site required for its localization to the germ plasm [24]; (2) two miR430 sites responsible for mRNA degradation in somatic cells; and (3) the binding site for the Dead end 1(Dnd1) protein that is expressed only in the PGC and protects mRNA from miR430-dependent degradation [2], [25].
In Mus musculus, primordial germ cells (PGCs) are induced from a population of pluripotent epiblast cells [26], [27]. Following their induction, these PGC precursors translocate to the base of the allantois by E7.25 and once formed, migrate to the endoderm (E7.5), travel through the hindgut (from E8.0), dorsal mesentery and dorsal body wall, and reach the genital ridge at around E10.5 to E11.5. Following the sex differentiation of the somatic gonads, PGCs themselves differentiate into male or female germ cells at around E12.0 [28]. Three Nanos homologs (Nanos1-3) have been identified in mice, and Nanos2 and Nanos3 have been implicated in germ cell development [10]. NANOS2 is specifically expressed in the mouse male germ cells after their colonization of the gonads and is essential for their development. In our previous study, we reported that the Nanos2-3′UTR promotes the efficient translation of this protein in the male germ cell after E13.5 via an unknown mechanism [29]. Nanos3 is expressed in the PGCs after their formation until shortly after their settlement in the gonads (E14.5 in male, E13.5 in female), and is re-expressed after birth in the testes [10]. Nanos3 knockout mice are thus sterile because of the loss of migrating PGCs during embryogenesis. These data suggest that NANOS3 plays an important role in the maintenance and survival of PGCs [10], [30]. However, the regulatory mechanism of NANOS3 expression and the function of the Nanos3-3′UTR had not been fully investigated as yet.
In our present report, we show that Nanos3 mRNA is transcribed in both germ cells and somatic cells, although NANOS3 protein is expressed specifically in germ cells. By applying a transgenic mouse strategy, we show that the translation of NANOS3 in somatic cells is suppressed via an mRNA destabilizing mechanism mediated by the Nanos3-3′UTR.
Results
The Nanos3-3′UTR Is Required for Suppression of Nanos3 Expression in Somatic Cells
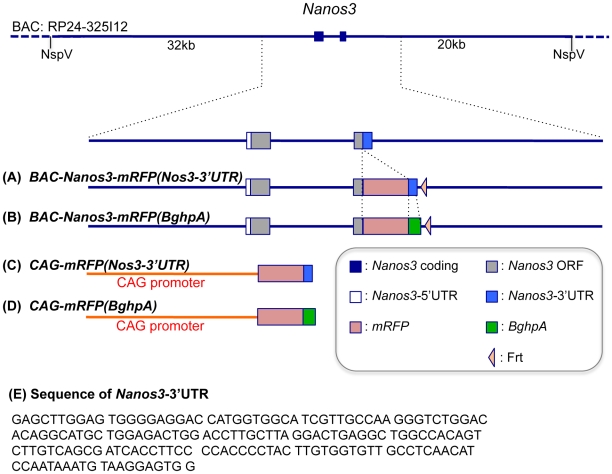
To elucidate whether the Nanos3-3′UTR is involved in NANOS3 expression in mouse, we generated two BAC transgenic mouse lines; BAC-Nanos3-mRFP(Nos3-3′UTR) containing the endogenous Nanos3-3′UTR (Fig. 1A and E), and BAC-Nanos3-mRFP(BghpA) harboring an exogenous 3′UTR, Bovine growth hormone poly(A) signal (BghpA; Fig. 1B). We first confirmed that either BAC-Nanos3-mRFP(Nos3-3′UTR) or BAC-Nanos3-mRFP(BghpA) could rescue the Nanos3−/− embryonic phenotype (Fig. S1), indicating that this BAC construct contains regulatory elements that are sufficient to maintain endogenous NANOS3 expression and the NANOS3-mRFP protein was functional. The NANOS3-mRFP expression in germ cells in both transgenic embryos showed a similar pattern to the endogenous protein (Fig. 2 and S2) exhibiting cytoplasmic localization as seen for NANOS2 [31]. The small difference between Nanos3 and Nanos3-mRFP was that the former was clearly localized to the cytoplasm whereas the latter was less clear and slightly localized in nuclei also. It was consistent with a previous report [32].
Figure 1. Schematic representation of the transgenes used in this study.
The top line represents BAC RP24-325I12 which contains the Nanos3 gene, the second line is a larger scale schema of a portion of this construct. (A–D) Different modifications of the transgene. Blue lines denote sequences derived from the BAC RP24-325I12 construct and the red lines those of the CAG promoter. The meanings of each box is indicated. (E) Sequence of Nanos3-3′UTR we used.
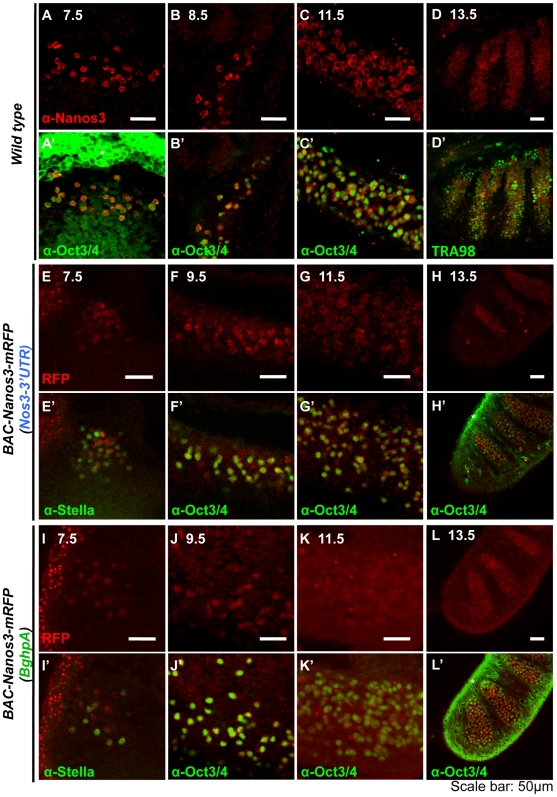
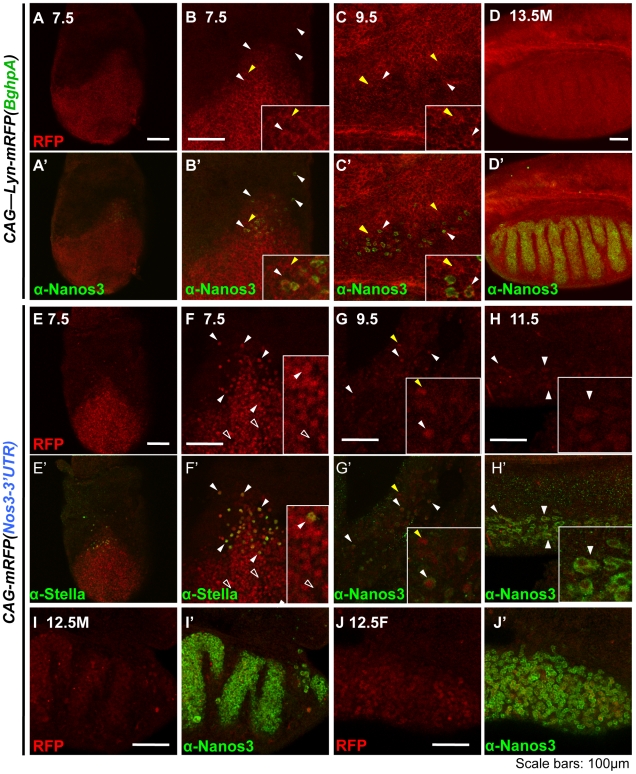
Figure 2. Nanos3-mRFP is expressed dominantly in germ cells in BAC transgenic mouse lines.
Confocal images of embryos of the wild-type (A–D), BAC-Nanos3-mRFP(Nos3-3′UTR) (E-H) or BAC-Nanos3-mRFP(BghpA) (I–L). Panels (A–D) show images of immunostaining for anti-NANOS3. Panels (E–L) show images of mRFP fluorescence (not immunostaining) and (A′–L′) are merged images that include immunostaining for the germ cell marker anti-OCT3/4, anti-Stella/PGC7 or TRA98 (green signal). The developmental stage associated with each figure is indicated above each panel: E7.5 (A, E, and I), E8.5 (B), E9.5 (F and J), E11.5 (C, G and K), E13.5 male gonad (D, H and L). Asterisks represent non-specific signals by the secondary antibody. Scale bars, 50 µm.
Interestingly, however, in the embryo harboring BAC-Nanos3-mRFP(BghpA), the intensity of NANOS3-mRFP was gradually increased in the somatic tissues at later embryonic stage (Fig. 2K-L). In the E14.5 male, a striped pattern was observed for BAC-Nanos3-mRFP(Nos3-3′UTR) reflecting germ cell localization in the testis cords in gonads, whereas the pattern was unclear in BAC-Nanos3-mRFP(BghpA), indicating strong expression in the surrounding somatic tissues (compare Figs. 2H and 2L, 3A–D). In addition, the whole body of the BAC-Nanos3-mRFP(BghpA) embryos expressed NANOS3-mRFP (Fig. 3D). These results suggest that Nanos3 is transcribed in many embryonic tissues and that the Nanos3-3′UTR is required to suppress translation in somatic tissues.
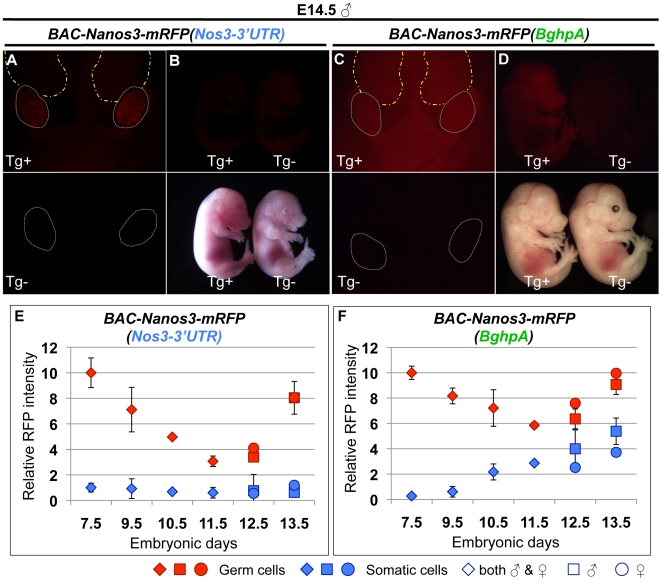
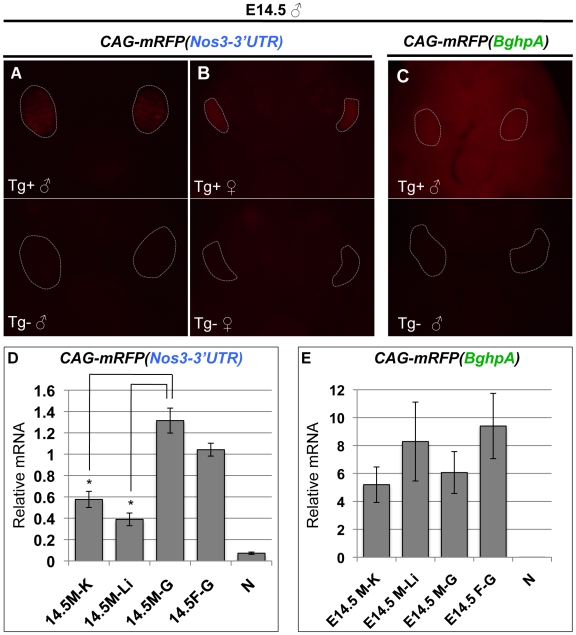
Figure 3. Replacement of Nos3-3′UTR with BghpA results in the upregulation of NANOS3-mRFP protein in somatic tissues.
(A–D) Fluorescence images of male embryos derived from BAC-Nanos3-mRFP(Nos3-3′UTR) (A–B) and BAC-Nanos3-mRFP(BghpA) (C–D) transgenic embryos at E14.5. (A and C) The upper images are of the abdomens of embryos harboring the transgene (Tg+), whereas the lower images are of the same tissues from embryos with no transgene (Tg−). The broken gray lines indicate the gonads and broken yellow lines indicate the kidneys. (B and C) Whole body of Tg+ and Tg- embryos are shown. The image in the upper panel shows the mRFP fluorescence pattern, whilst the lower panels are the corresponding bright field images. (E and F) Developmental changes in the relative mRFP intensities in germ cells (red) and somatic cells (blue) derived from BAC-Nanos3-mRFP(Nos3-3′UTR) (E) and BAC-Nanos3-mRFP(BghpA) (F) transgenic embryos. Error bars represent the s.e.m.
To evaluate the suppressive effects of 3′UTR in somatic cells quantitatively, we compared the abundance of Nanos3-mRFP protein based on the intensities of mRFP signals in both the germ cells and surrounding somatic cells in each transgenic embryo from E7.5 to E13.5 using imageJ software (Fig. 3E–F). The changes in the relative mRFP intensities in the germ cells were similar between the two transgenic lines. These were gradually decreased after E7.5, reached their lowest level at E11.5 and then rapidly increased from E12.5. Contrary to the data found in the germ cells, the mRFP intensities in the surrounding somatic cells showed a clear difference between the two lines. In the embryo harboring Nanos3-3′UTR, the mRFP intensity was maintained at very low levels throughout embryogenesis. However, in the embryo containing the BghpA elements, this expression gradually increased from E10.5 and at E13.5 reached 60% of the intensity seen in the germ cells, although it was maintained at low levels at E7.5 and E9.5. These data suggest that the translation of NANOS3 is upregulated after E9.5 in somatic tissues and that the Nanos3-3′UTR is required to suppress this activity.
The Accumulation of Somatic Nanos3 mRNA Is Suppressed by the Nanos3-3′UTR
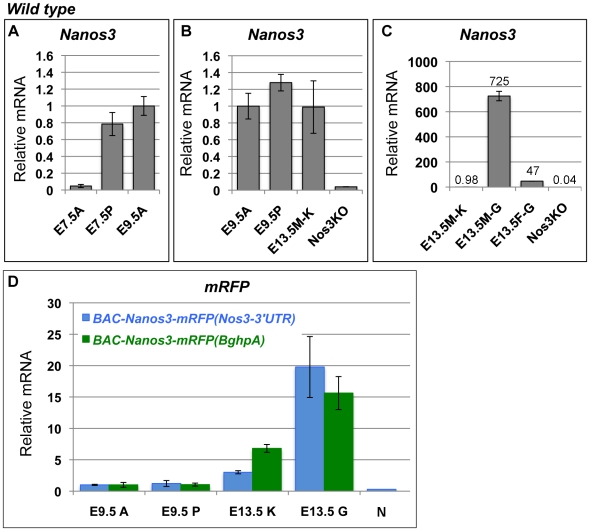
We next examined the endogenous Nanos3 mRNA levels in somatic tissues between E9.5 and E13.5 by quantitative RT-PCR (qRT-PCR) and found transcripts even in somatic tissues (Fig. 4A–C), consistent with the above data. The anterior half of the embryo at E9.5 (9.5A) and the kidney at E13.5 (13.5K) do not contain any germ cells, but Nanos3 mRNA was detected (Fig. 4B–C). The level of Nanos3 mRNA in somatic tissue from E9.5 to E13.5 was maintained at very low levels compared with the gonads which containing many germ cells (Fig. 4C). Interestingly, Nanos3 expression was not detected in the anterior half of the embryo at E7.5 (E7.5A), suggesting the transcription of this gene is restricted to the PGCs when they are formed and may be slightly increased in the somatic cells at the later stage. It is consistent with previous reports, which include single–cell PCR analyses demonstrating Nanos3 expression exclusively in the PGCs [33] and our lineage study using Nanos3-cre [30].
Figure 4. The accumulation of somatic Nanos3 mRNA is suppressed by the Nanos3-3′UTR.
The levels of Nanos3 (A–C) or mRFP (D) mRNA were compared by quantitative RT-PCR using RNA samples derived from wild-type embryos (A–C) and BAC-Nanos3-mRFP(Nos3-3′UTR) (blue in D) and BAC-Nanos3-mRFP(BghpA) (green in D) transgenic embryos at E7.5, E9.5 and E13.5. Data were normalized by G3PDH in each sample. The relative mRNA levels in the E9.5A sample (an anterior part of E9.5 embryo) were assigned the reference value of 1.0. A, anterior part of the embryo; P, posterior part of the embryo; M–K or K, the male kidney; M–G or G, the male gonad; F–G, the female gonad; N, the posterior part of a Nanos3 knockout embryo at E9.5. Error bars represent the s.d.
In embryos harboring BAC-Nanos3-mRFP(Nos3-3′UTR), the levels of mRFP at both stages was low, similar to Nanos3 mRNA in the wild-type embryo (Fig. 4B and D). In contrast, in embryos harboring BAC-Nanos3-mRFP(BghpA), the relative mRFP levels became two-fold higher at E13.5 than those of BAC-Nanos3-mRFP(Nos3-3′UTR) (Fig. 4D). Taken together, these data suggest that Nanos3 is transcribed in both germ cells and somatic tissues by at least E9.5 and that Nanos3-3′UTR is required to suppress Nanos3 accumulation in somatic tissues.
The Nanos3-3′UTR Is Sufficient for the Establishment of Germ-Cell Specific Expression Pattern in the Mouse Embryo
To further investigate whether the Nanos3-3′UTR affects the transcription or stability of mRNA, we generated two additional transgenic mice. We utilized the CAG promoter, a known strong promoter-enhancer that drives the ubiquitous transcription of mRFP with either Nanos3-3′UTR or BghpA (Fig. 1C–D). Surprisingly, Nanos3-3′UTR proved to be effective in restricting the mRFP expression in the germ cells at E14.5 (Fig. 5A–B), whilst mRFP was always expressed ubiquitously with no pattern observed in the CAG-mRFP(BghpA) embryo (Fig. 5C). The same expression pattern was observed in CAG-lyn-mRFP(BghpA) transgenic embryo that had been previously established in our laboratory [34]. The lyn-mRFP is an mRFP that contains the lyn kinase at its N-terminus, which serves as a membrane localization signal but does not affect neither transcription and translation [34]. Therefore, we considered CAG-lyn-mRFP(BghpA) is compatible with CAG-mRFP(BghpA). By qRT-PCR analyses, we further revealed that the relative amounts of mRFP mRNA in somatic tissues (a kidney and a hind limb) were significantly lower than those in the gonads in CAG-mRFP(Nos3-3′UTR) embryos whereas not significantly altered in CAG-lyn-mRFP(BghpA) (Fig. 5D and E). The results suggest that Nanos3-3′UTR is sufficient to suppress protein expression in somatic cells by destabilizing mRNA and establishing a germ-cell specific expression pattern.
Figure 5. Nanos3-3′UTR is sufficient to establish the germ cell-specific expression pattern in the mouse embryo.
(A–C) Fluorescence images of CAG-mRFP(Nos3-3′UTR) (A, male; B, female) and CAG-mRFP(BghpA) (C, male) transgenic embryos at E14.5. The upper images are of the abdomens of embryos harboring transgenes (Tg+), whereas the lower panels show corresponding images from embryos lacking a transgene (Tg−). Broken gray lines indicate gonads. (D–E) Quantitative RT-PCR analysis of mRFP in CAG-mRFP(Nos3-3′UTR) (D) or CAG-Lyn-mRFP(BghpA) (E) embryos at E14.5. The data were normalized using G3PDH. M–K, male kidney; M-Li, male limb; M–G, male gonad; F–G, female gonad; N, wild-type embryo. Error bars represent the s.d. Student t-test was used to calculate P values. *, P<0.05.
To determine the point at which Nanos3-3′UTR begins to function and contribute to the establishment of the germ cell-specific pattern for NANOS3, we compared the mRFP expression patterns between CAG-mRFP(Nos3-3′UTR) and CAG-lyn-mRFP(BghpA) transgenic embryos. In CAG-lyn-mRFP(BghpA), mRFP was expressed in all embryonic tissues at all stages (Fig. 6A–D). In CAG-mRFP(Nos3-3′UTR) however, mRFP was expressed in all embryonic cells prior to germ cell formation at E7.0, similar to the profile found in the CAG-mRFP(BghpA) embryo (data not shown). After PGC formation, mRFP was still found to be expressed in most embryonic cells at E7.5, but in somatic cells this expression is gradually reduced (E9.5) and the germ-cell specific pattern is almost established by E11.5 (Fig. 6E–H). At E12.5, mRFP expression in the germ cells became notably stronger than in the somatic cells of both male and female embryos (Fig. 6I–J). This germ cell specific mRFP pattern was maintained until at least E16.5 at which stage endogenous Nanos3 expression is almost lost. These observations suggest that the Nanos3-3′UTR might function in all embryonic cells from E7.5 to E16.5. It is possible also that the Nanos3-3′UTR is involved in translational activation in germ cells. These different functions of this regulatory element in germ cells and in somatic cells might therefore contribute to the establishment of germ cell-specific NANOS3 protein expression.
Figure 6. The Nanos3-3′UTR may function in both germ cells and somatic tissues after generation of the PGC.
Confocal images of CAG-Lyn-mRFP(BghpA) (A–D) and CAG-mRFP(Nos3-3′UTR) (E–J) transgenic embryos. (A–J) mRFP fluorescence; (A′–J′) merged images of mRFP fluorescence (red) and immunostaining signals (green) for NANOS3 (A′–D′ G′–J′), STELLA/PGC7 (E′–F′). Insets are high magnification of each panel. The embryonic stage for each sample is indicated. White arrowheads, germ cells; white open arrowheads, somatic cells that do not express mRFP; yellow arrowheads, somatic cells that express mRFP. Scale bars, 100 µm.
It is noteworthy that the addition of the 3′UTRs of other germ cell specific genes such as Nanos2 and Stella/PGC7 was not sufficient to establish germ cell-specific expression patterns i.e. the transgenic embryos CAG-mRFP(Nanos2-3′UTR) and CAG-mRFP(Stella-3′UTR) showed strong and ubiquitous mRFP expression (Fig. S3). This suggests that the function of Nanos3-3′UTR is relatively unique in the mouse germ cell, unlike fly and nematodes in which 3′UTRs of many genes each have significant responsibility for the temporal and spatial control of a specific protein in their germlines [4], [35].
Discussion
The regulatory mechanisms underlying gene expression remains one of the most fundamental and significant themes in biology. In our current study, we analyzed the mechanisms underlying NANOS3 expression in vivo using BAC modification and transgenic technologies. Although Nanos3 is transcribed both in germ cells and in many somatic tissues, efficient translation of NANOS3 protein occurs only in germ cells.
It has been shown previously that the expression of maternal mRNAs depends on the corresponding 3′UTR in many animal species. The 3′UTRs of nanos homologs play essential roles in the respective mRNA localization, translation and degradation in C. elegans, Drosophila, and Zebrafish [1], [9], [36]. We have found in our present experiments that the Nanos3-3′UTR of Mus musculus also has a regulatory function during embryogenesis, even though NANOS3 is transcribed zygotically and germ cell formation in mice is quite different from other animals. There have been several reported examples of 3′UTR regulation of zygotic mRNA [37], [38], [39]. However, in all these cases, the transcripts were driven by tissue-specific promoters. Hence, Nanos3-3′UTR is the first example of a regulator of zygotic mRNA that can establish a tissue-specific gene expression pattern even if the mRNA is transcribed by a ubiquitous promoter.
The mechanisms of 3′UTR-dependent nanos mRNA regulation have been addressed previously in fishes and flies, in which miR430 and the Dnd1 protein, or the Smg, Glo and Osk proteins are involved in mRNA regulation via the nanos-3′UTR. In mice, one ortholog of Dnd1 and two orthologs of Smg have now been identified [40], [41]. We examined the possible effects of these proteins on the translation of an mRNA harboring the Nanos3-3′UTR by a luciferase assay in the NIH3T3 cell line. The stability of luciferase mRNA is also affected by Nanos3-3′UTR. However, the addition of both proteins did not result in any effects on reporter activities (data not shown). It is possible that they need co-factors which are not expressed in this cell line. It is also possible that the abundant expression of endogenous Smg in NIH3T3 cells caused no effect. In addition, the sequence of Nanos3-3′UTR has almost no similarity to the 3′UTRs of nanos orthologs and has no significant match with any miRNA target sites. Although several stem-loop structures have been predicted using the ‘mfold’ program (Zuker, 2003), these are not similar to the Drosophila TCE (data not shown). Hence, the mechanism of Nanos3-3′UTR dependent regulation is still unclear and is an essential project for a future study.
The somatic expression of BAC-Nanos3-mRFP(BghpA) did not affect mouse development unlike in the case of fly and the biological significance of Nanos3-3′UTR-dependent regulation also remains unclear. Since the regulation of gene transcription appears not always to be strict, Nanos3-3′UTR may prevent the accumulation of waste materials in the cell by promoting mRNA degradation.
Materials and Methods
Mice
The methods used to generate Nanos3-L-3′UTR (Nanos3+/−) mice and their subsequent characterization has been previously described [10]. All mice were an MCH background (closed colony derived from an ICR strain, CREA, Japan).
Generation of Nanos3-BAC Transgenic Mice
A Nanos3-BAC clone, RP24-325I12 (Invitrogen) was used for modification via the λ red recombination method as described previously [42], [43]. The vectors were constructed as follows: the Nanos3-3′UTR or BghpA sequence was inserted into the pBSIIKS vector harboring the mRFP gene (kindly provided by Dr. Roger Tsien [44]) and a cassette containing the kanamycin resistance gene flanked by two FRT sequences. The primers used for the BAC modifications are as follows (primer sequences are listed in Methods S1):
BAC-Nanos3/mRFP-F and BAC-Nanos3(fusion-3′UTR)-R
for BAC-Nanos3-mRFP(Nos3-3′UTR), and BAC-Nanos3-mRFP(BghpA);
BAC-Nanos3(ATG)/mRFP-F and BAC-Nanos3(fusion-3′UTR)-R
for BAC-ΔNos3-mRFP(Nos3-3′UTR) and BAC-ΔNos3-mRFP(BghpA). The primers used to confirm recombination were N3-IN-F1 and N3-LA-KR1. All PCR reactions were performed using Prime STAR DNA polymerase (Takara).
Cloning of 3′UTR Sequences
The Nanos3-3′UTR was cloned by PCR using a DNA template prepared from the tail of a C57BL6/J mouse. The primers used were N3-stop-SalI-F1 and N3-3′U-HindIII-R1.
Generation of Transgenic Mice
All BAC constructs were digested with Csp45I and PmacI (Takara) and then gel purified. Transgenic mice were then generated by the microinjection of DNA into fertilized eggs. The injected eggs were then transferred into the oviducts of pseudopregnant foster females. The genotypes of the mice or embryos were identified by PCR using isolated genomic DNA from the tail or yolk-sac. The primers used were as follows:
RFP-F2 and N3-3U-R1 for BAC-Nanos3-mRFP(Nos3-3′UTR) and BAC-ΔNos3-mRFP(Nos3-3′ UTR);
mRFP-F2 and bghpA-R2 for BAC-Nanos3-mRFP(BghpA) and BAC-ΔNos3-mRFP(BghpA). The primer sequences were described in Method S1.
Immunofluorescence
The mouse embryos and gonads were fixed in 4% PFA for 2 hours at 4°C and washed three times for 5 min each with PBS. After blocking with PBS containing 3% skim milk or 10% FBS for 1 hour at RT, samples were rinsed and incubated overnight with primary antibodies in PBS containing 0.1% TritonX-100 (PBS-Tr) at 4°C or RT. The following day, samples were washed 6 times for 15 min each in PBS-Tr and were incubated for 2 hours at RT with secondary antibodies in PBS-Tr. After the samples had been washed 6 times for 15 min each with PBS-Tr, they were mounted on MAS-coated slide glasses or a glass-bottom dish (Matsunami) and enclosed with PBS by manicure. The samples were then analyzed by confocal laser microscopy (Zeiss).
Primary antibodies were used at the following dilutions: 1∶500 for rabbit anti-NANOS3 [45], 1∶50000 for anti-PGC7 (Sato et al. 2002), 1∶500 for mouse anti-Oct3/4 (C-10) (Santa Cruz sc-5279) and 1∶8000 for rat TRA98 [46]. Secondary antibodies were all used at a 1∶200 dilution (Alexa-488 conjugated donkey anti-rabbit IgG, Alexa-488 conjugated donkey anti-mouse IgG, Alexa-488 conjugated donkey anti-rat IgG, Alexa-594 conjugated donkey anti-mouse IgG and Alexa-594 conjugated donkey anti-rat IgG).
Evaluation of mRFP Intensity
Samples were fixed in 4%PFA and immunostained with anti-Oct4 and Alexa-488 conjugated donkey anti-mouse IgG to determine germ cells. Then the images were taken using confocal laser microscopy (Zeiss). mRFP intensity in the immunostained embryo did not show significant difference from the unfixed embryo (data not shown). The mRFP intensity of each cell was measured using imageJ software (NIH, Bethesda, MD) and the mean gray values were regarded as the intensity of cell. Three embryos for each stage were examined. All data were normalized using the intrinsic background intensity of wild type embryos at each embryonic stage. The intensity of the E7.5 PGCs was assigned a value of 10 and the data were plotted accordingly.
Quantitative RT-PCR
Total RNAs were prepared with RNeasy (Qiagen) and used for reverse transcription by Super script III (Invitrogen). Quantitative RT-PCR was performed on the Mini Opticon Real-Time PCR System (Bio-RAD) using SYBR Premix Ex Taq (Takara). Samples were prepared as a pool of cDNA derived from 2–3 pieces of embryos, 4–8 gonads or 4–6 kidneys and each sample was analyzed in triplicate. mRNA levels were calculated with an absolute quantification method and normalized by the amount of G3PDH for each sample. The primers used were as follows: mNos3-F2 and N3-cod-R1 for Nanos3 mRNA, RFP-F2 and RFP-R2 for mRFP mRNA (including those fused with Nanos3), G3PDH-F and G3PDH-R for G3pdh mRNA. The primer sequences were described in Method S1.
Supporting Information
Both BAC-Nanos3-mRFP(Nos3-3′UTR) and BAC-Nanos3-mRFP(BghpA) transgenes rescue defects of Nanos3−/−. HE-stained sections of adult testes (A–D) and ovary (E–H) derived from Nanos3+/− (A), Nanos3+/+ (E), Nanos3−/− (B and F), Nanos3+/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (C), Nanos3+/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (G) and Nanos3−/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (D and H) are shown. Scale bar indicates 250 µm. Immunofluorescence images of E14.5 male (I–L) and female (M–P) gonads derived from Nanos3+/− (I and M), Nanos3−/− (J and N), Nanos3+/− harboring BAC-Nanos3-mRFP(BghpA) (K and O) and Nanos3−/− harboring BAC-Nanos3-mRFP(BghpA) (L and P) are shown. Masenta represents germ cells (TRA98) and blue represents DNA (DAPI). Scale bar indicates 100 µm. Although Nanos3−/− had no germ cell, Nanos3−/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) or harboring BAC-Nanos3-mRFP(BghpA) had many germ cells.
(10.04 MB TIF)
Nanos3-mRFP protein showed cytoplasmic localizaion in germ cells as well as Nanos3 protein. Confocal images of embryos of the wild-type at E7.5 (A and B) and E9.5 (C, C′, D and D′). Panels (A–D) show immunostaining with anti-mRFP antibody and the merged images with immunostaining for the germ cell marker anti-OCT3/4 antibody are shown in (C′–D′). Scale bar indicates 100 µm.
(2.28 MB TIF)
3′UTR of other germ cell specific genes was not sufficient for establishing the germ cell-specific expression pattern. The fluorescence images of male embryos derived from CAG-mRFP(Nos2-3′UTR) (A), CAG-mRFP(Stella-3′UTR) (B) and CAG-mRFP(TubulinB1-3′UTR) at E13.5 male (A–B) or female (C). Top images represent the abdomens of embryos harboring transgene (Tg+), whereas bottom images represent those harboring no transgene (Tg−). Broken gray lines indicate gonads.
(2.28 MB TIF)
Supplementary methods.
(0.03 MB DOC)
Acknowledgments
We thank Dr. M. Yamaji (Kyoto University) for information for BAC construction strategy. We also thank R. Yorozu, and Y. Kamimura for general technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the National BioResource Project (NBRP) (http://www.nbrp.jp/); the Genome Network Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan; and Research Fellowships of the Japan Society for the Promotion of Science (JSPS) for Young Scientists to A.S. (http://www.jsps.go.jp/english/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 4.Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, et al. Temporal and spatial control of germ-plasm RNAs. Curr Biol. 2009;19:72–77. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- 6.Pilon M, Weisblat DA. A nanos homolog in leech. Development. 1997;124:1771–1780. doi: 10.1242/dev.124.9.1771. [DOI] [PubMed] [Google Scholar]
- 7.Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 8.Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T. Expression and evolutionary conservation of nanos-related genes in Hydra. Dev Genes Evol. 2000;210:591–602. doi: 10.1007/s004270000105. [DOI] [PubMed] [Google Scholar]
- 9.Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, et al. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 11.Kurokawa H, Aoki Y, Nakamura S, Ebe Y, Kobayashi D, et al. Time-lapse analysis reveals different modes of primordial germ cell migration in the medaka Oryzias latipes. Dev Growth Differ. 2006;48:209–221. doi: 10.1111/j.1440-169X.2006.00858.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- 13.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 14.Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- 15.Gavis ER, Lunsford L, Bergsten SE, Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- 16.Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 17.Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 18.Crucs S, Chatterjee S, Gavis ER. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell. 2000;5:457–467. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- 19.Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, et al. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 21.Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- 23.Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 24.Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 26.Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 27.Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 28.McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 29.Tsuda M, Kiso M, Saga Y. Implication of nanos2-3′UTR in the expression and function of nanos2. Mech Dev. 2006;123:440–449. doi: 10.1016/j.mod.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, Tsuda M, Kiso M, Saga Y. Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol. 2008;318:133–142. doi: 10.1016/j.ydbio.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev. 2008;22:430–435. doi: 10.1101/gad.1612708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaji M, Tanaka T, Shigeta M, Chuma S, Saga Y, et al. Functional reconstruction of Nanos3 expression in the germ cell lineage by a novel transgenic reporter reveals distinct subcellular localizations of Nanos3. Reproduction. 2009 doi: 10.1530/REP-09-0373. [DOI] [PubMed] [Google Scholar]
- 33.Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- 34.Okamura Y, Saga Y. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech Dev. 2008;125:663–673. doi: 10.1016/j.mod.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Loya A, Pnueli L, Yosefzon Y, Wexler Y, Ziv-Ukelson M, et al. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008;14:1352–1365. doi: 10.1261/rna.867208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 37.Morales CR, Kwon YK, Hecht NB. Cytoplasmic localization during storage and translation of the mRNAs of transition protein 1 and protamine 1, two translationally regulated transcripts of the mammalian testis. J Cell Sci. 1991;100 (Pt 1):119–131. doi: 10.1242/jcs.100.1.119. [DOI] [PubMed] [Google Scholar]
- 38.Giorgini F, Davies HG, Braun RE. MSY2 and MSY4 bind a conserved sequence in the 3′ untranslated region of protamine 1 mRNA in vitro and in vivo. Mol Cell Biol. 2001;21:7010–7019. doi: 10.1128/MCB.21.20.7010-7019.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giorgini F, Davies HG, Braun RE. Translational repression by MSY4 inhibits spermatid differentiation in mice. Development. 2002;129:3669–3679. doi: 10.1242/dev.129.15.3669. [DOI] [PubMed] [Google Scholar]
- 40.Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem. 2005;280:43131–43140. doi: 10.1074/jbc.M508374200. [DOI] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oginuma M, Hirata T, Saga Y. Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology. Mech Dev. 2008;125:432–440. doi: 10.1016/j.mod.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki A, Tsuda M, Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- 46.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Both BAC-Nanos3-mRFP(Nos3-3′UTR) and BAC-Nanos3-mRFP(BghpA) transgenes rescue defects of Nanos3−/−. HE-stained sections of adult testes (A–D) and ovary (E–H) derived from Nanos3+/− (A), Nanos3+/+ (E), Nanos3−/− (B and F), Nanos3+/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (C), Nanos3+/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (G) and Nanos3−/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) (D and H) are shown. Scale bar indicates 250 µm. Immunofluorescence images of E14.5 male (I–L) and female (M–P) gonads derived from Nanos3+/− (I and M), Nanos3−/− (J and N), Nanos3+/− harboring BAC-Nanos3-mRFP(BghpA) (K and O) and Nanos3−/− harboring BAC-Nanos3-mRFP(BghpA) (L and P) are shown. Masenta represents germ cells (TRA98) and blue represents DNA (DAPI). Scale bar indicates 100 µm. Although Nanos3−/− had no germ cell, Nanos3−/− harboring BAC-Nanos3-mRFP(Nos3-3′UTR) or harboring BAC-Nanos3-mRFP(BghpA) had many germ cells.
(10.04 MB TIF)
Nanos3-mRFP protein showed cytoplasmic localizaion in germ cells as well as Nanos3 protein. Confocal images of embryos of the wild-type at E7.5 (A and B) and E9.5 (C, C′, D and D′). Panels (A–D) show immunostaining with anti-mRFP antibody and the merged images with immunostaining for the germ cell marker anti-OCT3/4 antibody are shown in (C′–D′). Scale bar indicates 100 µm.
(2.28 MB TIF)
3′UTR of other germ cell specific genes was not sufficient for establishing the germ cell-specific expression pattern. The fluorescence images of male embryos derived from CAG-mRFP(Nos2-3′UTR) (A), CAG-mRFP(Stella-3′UTR) (B) and CAG-mRFP(TubulinB1-3′UTR) at E13.5 male (A–B) or female (C). Top images represent the abdomens of embryos harboring transgene (Tg+), whereas bottom images represent those harboring no transgene (Tg−). Broken gray lines indicate gonads.
(2.28 MB TIF)
Supplementary methods.
(0.03 MB DOC)