Abstract
Voltage-gated potassium (Kv) channels play important roles in regulating the excitability of myocytes and neurons. Kv4.2 is the primary α-subunit of the channel that produces the A-type K+ current in CA1 pyramidal neurons of the hippocampus, which is critically involved in the regulation of dendritic excitability and plasticity. K+ channel-interacting proteins, KChIPs (KChIP1-4), associate with the N-terminal of Kv4.2 and modulate the channel's biophysical properties, turnover rate and surface expression. In the present study, we investigated the role of Kv4.2 C-terminal PKA phosphorylation site S552 in the KChIP4a-mediated effects on Kv4.2 channel trafficking. We found that while interaction between Kv4.2 and KChIP4a does not require PKA phosphorylation of Kv4.2S552, phosphorylation of this site is necessary for both enhanced stabilization and membrane expression of Kv4.2 channel complexes produced by KChIP4a. Enhanced surface expression and protein stability conferred by co-expression of Kv4.2 with other KChIP isoforms did not require PKA phosphorylation of Kv4.2 S552. Finally, we identify A-kinase anchoring proteins (AKAPs) as Kv4.2 binding partners, allowing for discrete local PKA signaling. These data demonstrate that PKA phosphorylation of Kv4.2 plays an important role in the trafficking of Kv4.2 through its specific interaction with KChIP4a.
Introduction
Voltage-gated potassium (Kv) channels play a critical role in regulating the excitability of neurons by preventing membrane depolarization and providing repolarization. Rapidly inactivating, A-type K+ channels of the Kv4 subfamily are highly expressed in the dendrites of hippocampal CA1 pyramidal neurons where they regulate signal propagation and synaptic plasticity (Kim and Hoffman, 2008). Kv4.2 has six transmembrane domains (S1-S6) and N- and C-terminal cytoplasmic domains. The Kv4.2 N-terminus contains a T1 domain that mediates subfamily specification (Papazian, 1999), and also binds to auxiliary subunits (Gulbis et al., 2000; Sewing et al., 1996). Kv4.2 C-terminal phosphorylation sites modulate the channel's trafficking and gating (Anderson et al., 2000) and we have recently shown that PKA phosphorylation is necessary for activity-dependent Kv4.2 internalization (Hammond et al., 2008).
The two main classes of Kv4 auxiliary subunits are the single transmembrane dipeptidylpeptidase-like (DPPx) proteins and the K+ channel interacting proteins (KChIPs) (Jerng et al., 2004a). KChIPs are encoded by at least four genes, KChIP1-4. All four are highly expressed in the brain, whereas only KChIP2 is abundant in the heart. KChIPs belong to the neuronal calcium sensor and EF-hand protein families (Berridge et al., 2000; Burgoyne and Weiss, 2001) and have been shown to influence Kv4 channel assembly, phosphorylation status and stability (An et al., 2000; Kunjilwar et al., 2004; Shibata et al., 2003). The association between KChIPs and Kv4 subunits does not require calcium binding, but the effects on channel gating are calcium dependent or at least are highly sensitive to point mutations within the EF-hand domains (An et al., 2000).
The KChIP4a isoform, which has a unique KIS (K-channel inactivation suppressor) domain (Holmqvist et al., 2002), has been reported to reduce fast inactivation of Kv4 currents in various cell types, and, unlike other KChIPs, has previously been found to not significantly enhance Kv4 channel surface expression. A recent report suggests that multiple KChIP isoforms express this KIS sequence which may be a transmembrane domain important for both trafficking and gating (Jerng and Pfaffinger, 2008).
PKA modulation of A-type K+ channels requires formation of a supramolecular complex with KChIPs (Hoffman and Johnston, 1998; Schrader et al., 2002) and we have recently found that PKA phosphorylation of Kv4.2 channels at site S552 is required for their activity-dependent internalization (Hammond et al., 2008). Therefore, in this study we investigated the roles of KChIP4a and Kv4.2S552 PKA phosphorylation in the trafficking of Kv4.2. Our results indicate that KChIP4a can be integral to both the trafficking and stabilization of Kv4.2 channels and, furthermore, that PKA phosphorylation of Kv4.2S552 is uniquely necessary for the trafficking effects regulated by KChIP4a. Finally, we show that A-kinase anchoring proteins (AKAPs) associate with Kv4.2, enhancing surface expression of the Kv4.2/KChIP4a complex.
Results
Enhanced surface expression of Kv4.2 by KChIP4a requires S552 phosphorylation
Activation of PKA leads to a prompt downregulation of dendritic A-type K+ currents in CA1 pyramidal neurons of the hippocampus, resulting in enhanced action potential back-propagation (Hoffman and Johnston, 1998). Although phosphorylation of the Kv4.2 α-subunit at site S552 is necessary for electrical remodeling, PKA modulation of Kv4.2's kinetic properties additionally requires formation of a supramolecular complex with KChIP auxiliary subunits (Schrader et al., 2002). More recently, we have shown that S552 PKA phosphorylation of Kv4.2 is required for rapid, activity-dependent channel internalization (Hammond et al., 2008). Together with the observation that KChIP subunits generally affect Kv4 channel trafficking (Jerng et al., 2004a), these data suggest that PKA phosphorylation of the Kv4.2 α-subunit at site S552 is coordinated with KChIP binding for the regulation of channel properties and surface expression. We questioned whether phosphorylation at this site was also necessary for modifications produced by co-expression with KChIP4a.
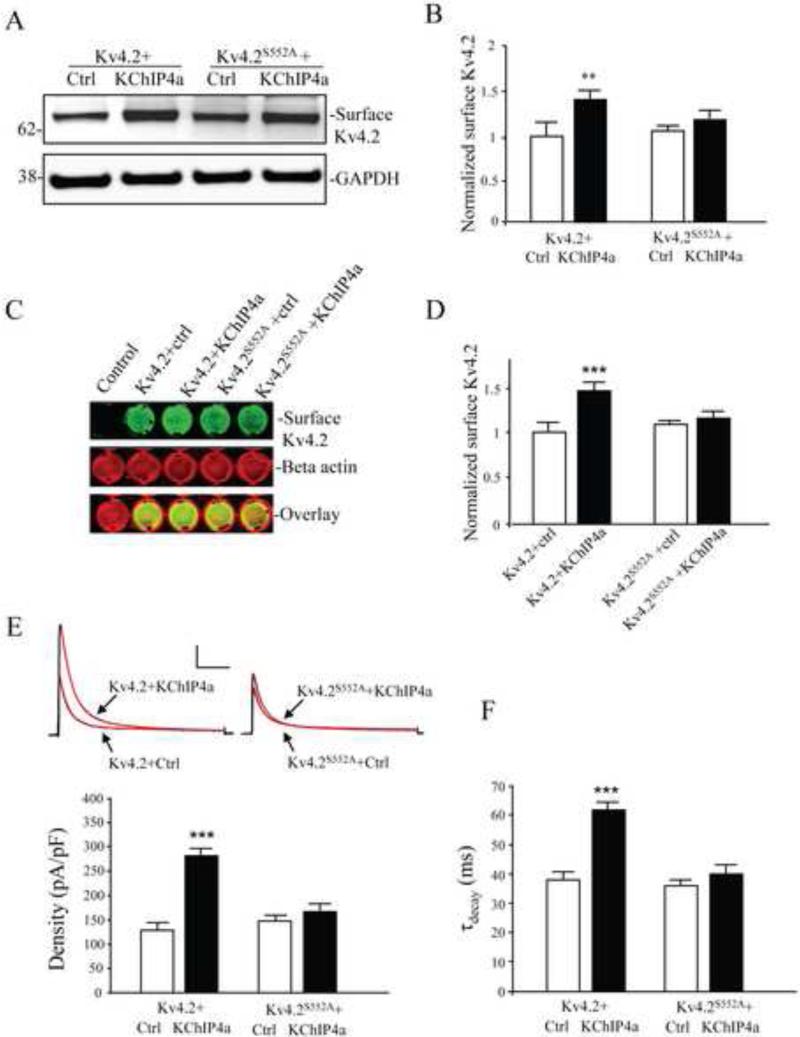
In a biotinylation assay, co-expression of Kv4.2 with KChIP4a (1:1 protein ratio) enhanced surface expression of Kv4.2 approximately 1.5-fold (Figure 1A, B; n = 3; p < 0.05). Surface increase of Kv4.2 by KChIP4a co-expression was also observed using an on-cell western assay (Figure 1C, D; n = 4; p < 0.05). Furthermore, in electrophysiological recordings from HEK293 cells, peak current density of Kv4.2-mediated currents increased after co-expression with KChIP4a (Figure 1E; n = 15; p < 0.05). As found after Kv4.2 co-expression with other KChIPs (Jerng et al., 2004b), enhanced surface expression by KChIP4a co-expression was accompanied by a decrease in the rate of channel inactivation (Figure 1F; n = 15; p < 0.05). Together these data from three different experimental approaches confirm Kv4.2 surface expression enhancement by KChIP4a co-expression in these heterologous expression systems.
Figure 1.
KChIP4a-mediated increase in Kv4.2 surface expression requires PKA phosphorylation of Kv4.2 at site S552. (A) Surface proteins of transfected COS7 cells were labeled with NHS-SS-Biotin and probed with mouse anti-Kv4.2 (1:2000). Cells co-expressing Kv4.2 and KChIP4a showed greater surface expression than for those expressing Kv4.2 alone. Surface expression of the Kv4.2 phospho-mutant S552A (Kv4.2S552A) was not enhanced by KChIP4a co-expression. GAPDH served as a loading control. (B) Pooled data normalized to total Kv4.2 expression shows that KChIP4a co-expression increases surface Kv4.2 expression by ~ 1.5-fold. Error bars represent S.E.M. (C) On-cell western assay of surface Kv4.2 in COS7 cells. The first well of the top row is un-transfected, well-2,3,4 and 5 were transfected with Kv4.2, Kv4.2 with KChIP4a, Kv4.2S552A, and Kv4.2S552A with KChIP4a, respectively. Intensity levels of surface Kv4.2 (green) were significantly increased in wells of Kv4.2 co-expressed with KChIP4a. No changes in fluorescence were observed the wells expressing Kv4.2S552A with or without KChIP4a compared to the wells of Kv4.2 alone. The intensity levels of total beta-actin (red) were not different in each well, demonstrating that equal numbers of cells are found in each well. Increased Kv4.2 but not Kv4.2S552A expression with KChIP4 co-expression is evident in the overlay. (D) Pooled data showing a significant enhancement of surface expression of Kv4.2 with KChIP4a, but not Kv4.2S552A in on-cell western assays. Error bars represent S.E.M. (E) In electrophysiological recordings from HEK293 cells, peak current density of Kv4.2-mediated currents also increased (~2-fold) by co-expression with KChIP4a (n = 15) compared to the Kv4.2 alone (n = 15). Peak current density enhancement by KChIP4a was not found in cells expressing the Kv4.2S552A mutation (n = 10). Red lines on traces are single exponential fits to the decay. Error bars represent S.E.M. Scale bars: 1 nA, 100 ms. (F) KChIP4a co-expression with Kv4.2 delayed the rate of inactivation. No delay was found for KChIP4a co-expression with Kv4.2S552A.
To determine whether S552 phosphorylation by PKA affects Kv4.2 + KChIP4a trafficking, we compared surface expression of both wild type Kv4.2 and the phospho-mutant Kv4.2S552A with and without KChIP4a in COS7 cells. When co-expressed with KChIP4a, surface expression of Kv4.2S552A was not significantly increased compared with wild type Kv4.2+KChIP4a in biotinylation experiments (Figure 1A,B; n = 3; p > 0.1) or on-cell westerns (Figure 1C, D; n = 4; p > 0.1). Peak current density increase and delayed inactivation by KChIP4a were also not observed in electrophysiological recordings from Kv4.2S552A expressing HEK293 cells (Figure 1E,F; n = 25; both p > 0.1). These results indicate that PKA phosphorylation at S552 is necessary for the altered surface expression and inactivation rate of Kv4.2 channels produced by KChIP4a co-expression.
S552 phosphorylation of Kv4.2 is not required for Kv4.2 + KChIP4a interaction
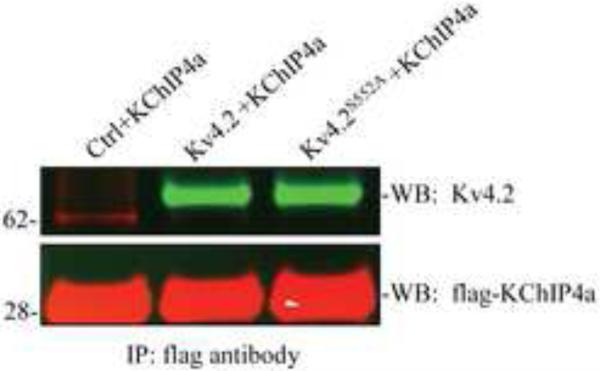
The results in Figure 1, showing the requirement of S552 phosphorylation of Kv4.2 for enhanced surface expression and delayed inactivation kinetics, could simply indicate that phosphorylation is required for the interaction between Kv4.2 and KChIP4a. In fact, the lack of an effect of KChIP4a on A-current inactivation rate in Kv4.2S552A expressing cells (Figure 1F) suggests that surface expressed channels are not in complex with KChIP4a in these recordings. We therefore looked to see if Kv4.2S552A and KChIP4a were physically interacting. To test for this we co-transfected KChIP4a with wild type Kv4.2 or Kv4.2S552A in COS7 cells and performed co-immunoprecipitation experiments. Western blot analysis showed that both Kv4.2 and the Kv4.2S552A mutant could bind to KChIP4a (Figure 2). PKA phosphorylation of Kv4.2 therefore does not appear to be required for Kv4.2-KChIP4a binding but does mediate enhanced surface expression of Kv4.2 through its interaction with KChIP4a.
Figure 2.
PKA S552 is not necessary for the interaction of Kv4.2 with KChIP4a. Co-immunoprecipitation of control, Kv4.2 or Kv4.2S552A with KChIP4a in COS7 cells. Cell lysates were pulled down using an anti-Flag antibody. Western blots were probed with anti-Kv4.2 and anti-Flag antibodies and visualized by anti-rabbit Alexa Fluor 800 secondary (green) and anti-mouse Alexa Fluor 680 secondary (red) antibody. Results demonstrate that both Kv4.2 and the Kv4.2S552A mutant interact with KChIP4a.
S552 phosphorylation is not required for enhanced surface expression by other KChIPs
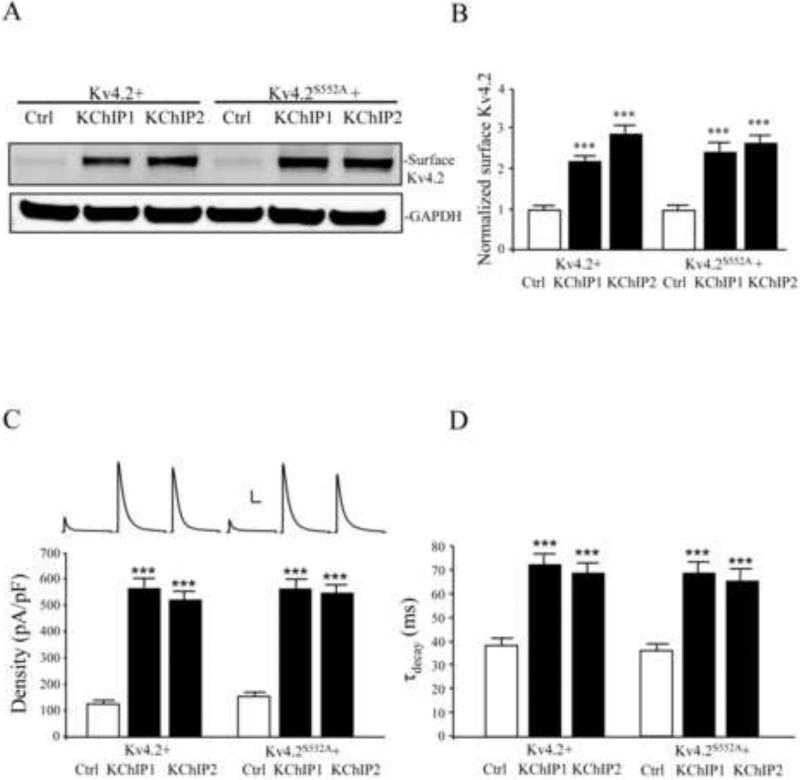
This requirement for PKA phosphorylation at site S552 for enhanced membrane expression is specific for KChIP4a as elevated Kv4.2 expression by neither KChIP1 nor KChIP2 was affected by the Kv4.2S552A mutation (Figure 3). In COS7 cells, biotinylation assays showed that co-expression of either KChIP1 or KChIP2 lead to multi-fold increases in surface expression of both Kv4.2 (Figure 3A,B; n = 3; p < 0.05) and Kv4.2S552A (Figure 3A,B; n = 3; p < 0.05).
Figure 3.
KChIP1 and KChIP2-mediated increases in Kv4.2 surface expression do not require PKA phosphorylation of Kv4.2 at site S552. (A) Surface proteins of transfected COS7 cells were detected using a biotinylation assay. Cells co-expressing Kv4.2 and KChIP1 or KChIP2 showed greater surface expression than for those expressing Kv4.2 alone. Surface expression of the Kv4.2 phosphomutant S552A (Kv4.2S552A) was also enhanced by KChIP1 or KChIP2 co-expression. GAPDH served as a loading control. (B) Pooled data normalized to total protein level of Kv4.2 from three experiments showing significant enhanced membrane expression of Kv4.2 and Kv4.2S552A with KChIP1 or KChIP2 co-expression. Membrane expression levels of Kv4.2 and Kv4.2S552A, in the absence of KChIP1 or KChIP2, were similar. Error bars represent S.E.M. (C) In electrophysiological recordings from HEK293 cells, peak current density of Kv4.2-mediated currents also increased (~4-fold) by co-expression with KChIP1 or KChIP2 (n = 15) compared to the Kv4.2 alone (n = 15). Peak current density enhancement by KChIP1 or KChIP2 was also found in cells expressing the Kv4.2S552A mutation (n = 27). Scale bars: 2 nA, 100 ms. (D) KChIP1 or KChIP2 co-expression with Kv4.2 delayed the rate of inactivation. Delay was also found for KChIP1 or KChIP2 co-expression with Kv4.2S552A.
Electrophysiological recordings in HEK cells produced corresponding results (Figure 3C). Both KChIP1 and KChIP2 produced a similar slowing of macroscopic inactivation as KChIP4a, suggesting a population of surface channels in complex with KChIPs, which do not require S552 phosphorylation of Kv4.2 (Figure 3D). We note also that although KChIP4a does significantly enhance Kv4.2 surface expression (Figure 1), the increase is only half as effective as that found for KChIP 1 or 2 (Figure 3).
PKA activation enhances Kv4.2 surface expression when co-expressed with KChIP4a
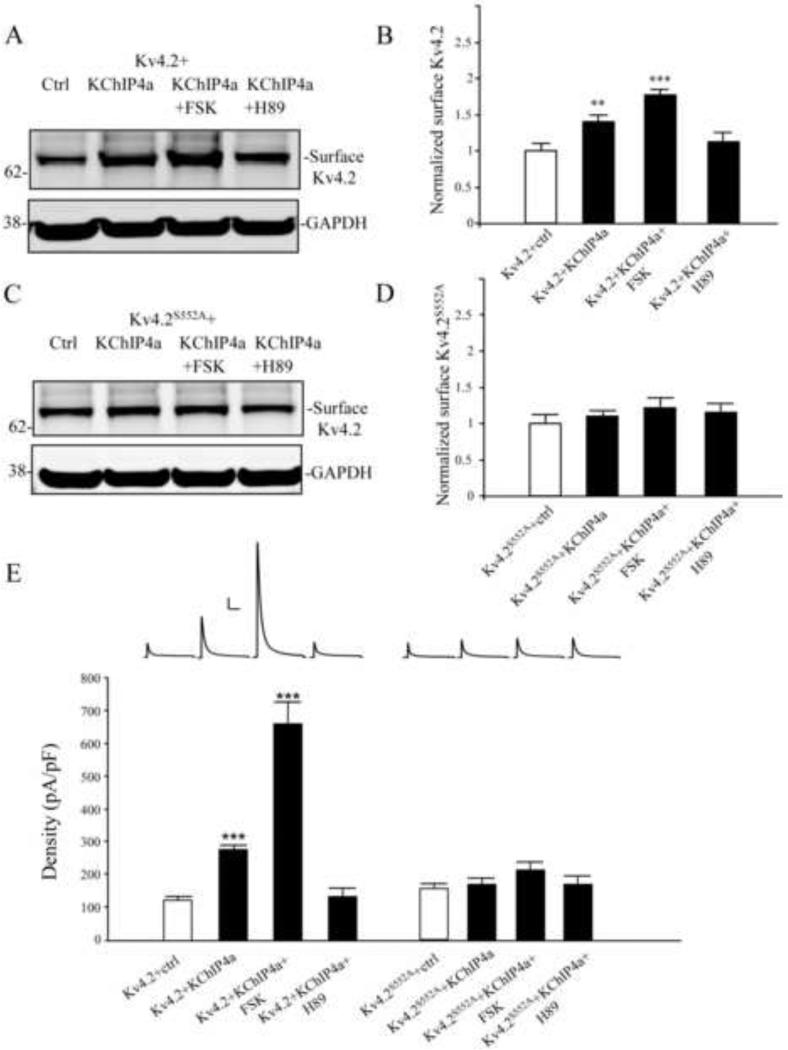
We have previously shown acute PKA activation by forskolin (FSK) to result in an acute decrease in surface expression of EGFP-tagged Kv4.2 (Hammond et al., 2008). However, our present results show S552 PKA phosphorylation to be required for KChIP4a-mediated enhancement of channel surface expression. We therefore predicted that long-term FSK exposure would lead to an increase in surface expression of Kv4.2 in complex with KChIP4a subunits after PKA activation.
To investigate the long-term effect of FSK exposure on the surface expression of Kv4.2 + KChIP4a channel complexes, we performed biotinylation experiments in COS7 cells. While FSK reduced surface expression of Kv4.2 when expressed acutely (10 μM; 15 min; n = 3; p < 0.05, data not shown), long-term (24 h) FSK treatment increased membrane expression levels of Kv4.2 + KChIP4a channels (Figure 4 A, B; n = 3; p < 0.001) as well as Kv4.2 channels expressed in the absence of KChIP4a (Kv4.2+FSK=1.39±0.15 normalized to Kv4.2 + ctrl; n=3; p < 0.05). The FSK effect on Kv4.2 + KChIP4a surface enhancement specifically required PKA phosphorylation as it was blocked by pre-incubation with PKA antagonist H89 (Figure 4 A, B; n = 3; p < 0.001). Enhanced expression after PKA activation required phosphorylation at Kv4.2 site S552 as no increase was found in FSK experiments from cells expressing the Kv4.2S552A mutant (Figure 4 C, D; n = 3; p > 0.1). All biotinylation results after long-term FSK exposure were confirmed in electrophysiological recordings (Figure 4E) including enhanced expression of Kv4.2 in the absence of KChiP4a (Kv4.2+FSK=361.07±14.21 pA/pF; n=19; p < 0.05).
Figure 4.
PKA activation enhances the surface expression of Kv4.2 by KChIP4a (A-B) Biotinylation assay showed surface Kv4.2 were increased about 1.5 fold by co-expressed with KChIP4a in COS7 cells. Long-term treatment with forskolin (10 μM, 24 hr) greatly enhances the surface expression of Kv4.2 compared to Kv4.2 alone, but it was blocked by the PKA inhibitor H89 (10 μM for 1 hr). GAPDH served as a loading control. (C-D) Surface expression of the Kv4.2 phospho-mutant S552A (Kv4.2S552A) was not enhanced either with KChIP4a co-expression or forskolin (10 μM, 24hr). Error bars represent S.E.M. (E) In electrophysiological recording from HEK293 cells, peak current density of Kv4.2-mediated currents were increased about 2 fold by co-expression with KChIP4a (n = 15). Long-term treatment with forskolin (10 μm, 24 hr, n = 10) showed a significant increase (~5-fold) of the peak current density compared to Kv4.2 alone, but it was blocked subsequent application of the PKA inhibitor H89 (10 μm, 1hr, n = 11). Peak current density enhancement by KChIP4a was not found in HEK cells expressing the Kv4.2S552A mutant (n = 10) or forskolin (10 μM, 24hr, n = 10) and H89 (10 μM, 1hr, n = 10) treatment compared to Kv4.2S552A alone. Error bars represent S.E.M. Scale bars: 1 nA, 100 ms.
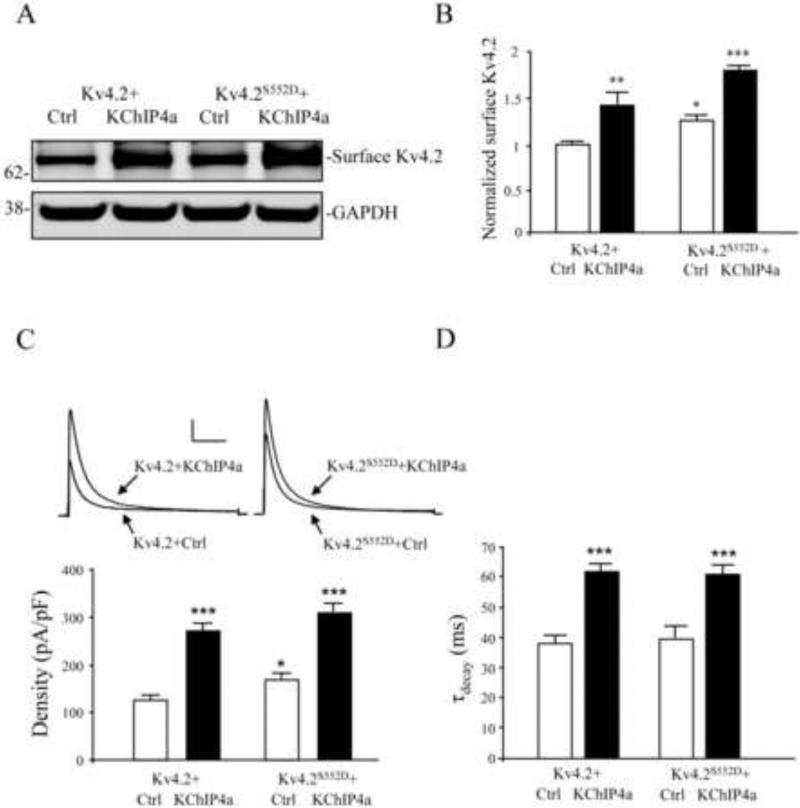
In support of the findings presented thus far, phosphomimetic mutation of Kv4.2 S552 was found, in both biotinylation and electrophysiological recordings, to enhance channel surface expression compared to wild type Kv4.2 (Figure 5 A-C). As expected, increased surface expression of Kv4.2S552D, in the absence of KChIP4a, was not accompanied by a change in the inactivation rate compared to Kv4.2 (Figure 5D). Surface expression of both Kv4.2 and Kv4.2S552D were then further enhanced by KChIP4a co-expression in both biotinylation and electrophysiological recordings (Figure 5A-C). Inactivation time constants were increased for both constructs by KChIP4a (Figure 5D).
Figure 5.
Phosphomimetic mutation of Kv4.2 S552 (Kv4.2S552D) enhances the Kv4.2 channel surface expression. (A-B) Biotinylation results showed that surface expression of Kv4.2S552D is increased compared to Kv4.2 but that surface expression of both are increased with KChIP4a. GAPDH served as a loading control. Error bars represent S.E.M. (C) As with the biotinylation results, electrophysiological recordings from HEK293 cells showed peak current density of Kv4.2S552D-mediated currents was increased compared to Kv4.2 (n = 15). Peak current density was further increased with KChIP4a (n = 18). Scale bars: 1 nA, 100 ms. Error bars represent S.E.M. (D) Inactivation rate was increased for both KChIP4a co-expression with Kv4.2 and Kv4.2S552D. Increased surface expression of Kv4.2S552D was not accompanied by a change in the inactivation rate.
KChIP4a enhancement of Kv4.2 stabilization requires S552 phosphorylation of Kv4.2
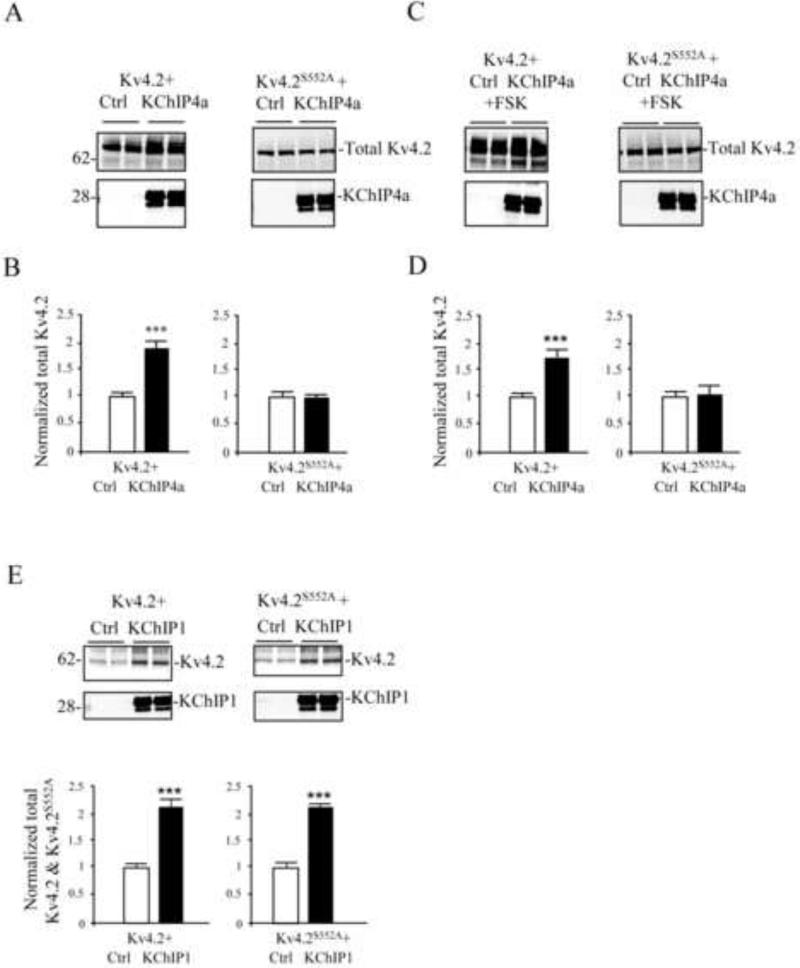
Another reported effect of KChIP co-expression is enhanced Kv4 channel stability (Shibata et al., 2003). We measured the stability of Kv4.2 subunits in stabilization experiments in COS7 cells (Figure 6). Normalized Kv4.2 protein levels were increased ~ 2-fold by 48 h after KChIP4a co-expression (Figure 6 A,B). KChIP4a co-expression thus enhances stabilization of the channel complex, perhaps by preventing the misfolding of Kv4.2 proteins, which would otherwise be targeted for degradation (Shibata et al., 2003).
Figure 6.
KChIP4a-mediated increase in stabilization of Kv4.2 requires PKA phosphorylation of S552. (A) Immunoblots of total protein from COS7 cells, which were co-transfected with Kv4.2 or Kv4.2S552A along with either control vector or KChIP4a for 48 hr. (B) Normalized total protein level of Kv4.2 was increased by co-expression with KChIP4a compared to Kv4.2 expression alone. In contrast, the total level of Kv4.2S552A was not different between KChIP4a and control vector co-expression. (C-D) Chronic forskolin treatment (10 μM, 24 hr) greatly enhanced the stability of total Kv4.2 protein levels when co-expressed with KChIP4a compared with control vector, but no differences were found in the total level of Kv4.2S552A with forskolin treatment between control and KChIP4a co-expression. Error bars represent S.E.M. (E) Enhanced stability of wild type Kv4.2 was found after co-expression with either KChIP isoform. Protein levels of Kv4.2S552A, however, were only increased after KChIP1 expression. KChIP4a but not KChIP1 therefore requires S552 phosphorylation of Kv4.2 to enhance protein stability (n = 4). Error bars represent S.E.M.
To determine if the enhanced stabilization of Kv4.2 by KChIP4a was S552 phosphorylation-dependent, we co-transfected COS7 cells with KChIP4a and either Kv4.2 or Kv4.2S552A. Although KChIP4a co-expression enhanced stability of Kv4.2 (Figure 6A, B; n = 4; p < 0.05), no increased stability was observed for Kv4.2S552A (Figure 6A, B; n = 4; p > 0.05). Chronic FSK exposure (24 h) also enhanced channel complex stability, requiring S552 phosphorylation (Figure 6C, D; n = 4; p < 0.05). The requirement for phosphorylation site S552 for enhanced stability was again specific for KChIP4a as Kv4.2 and Kv4.2S552A protein levels were similarly increased 48 h after KChIP1 co-expression (Figure 6E; n = 4; p < 0.05). KChIP4a therefore both enhances forward trafficking of Kv4.2 and increases the stability of the protein. Both of these effects require S552 phosphorylation of Kv4.2.
A-kinase anchoring proteins anchor PKA to Kv4.2 complexes
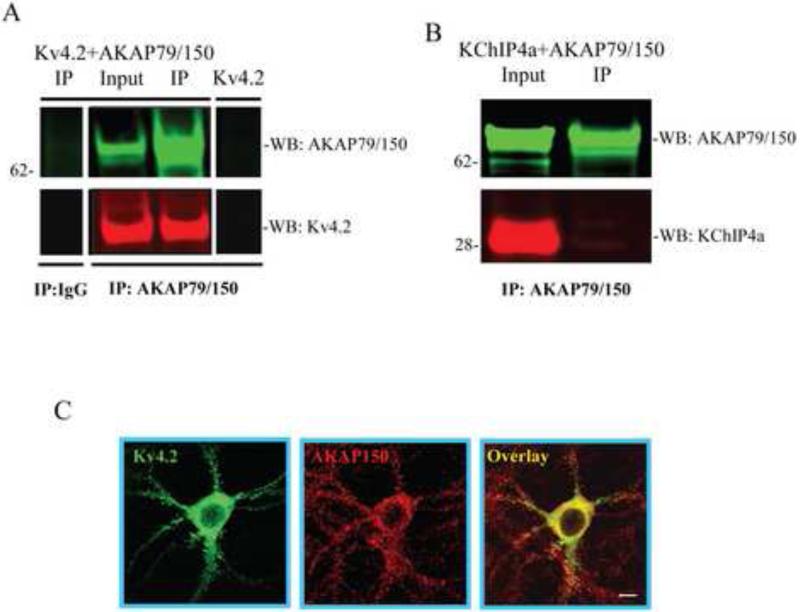
A-kinase anchoring proteins (AKAPs) are a group of proteins that bind to target proteins as well as the regulatory subunit of PKA, allowing for discrete local PKA signaling. Given our results demonstrating the importance of PKA phosphorylation at S552 on Kv4.2 for trafficking, we looked for Kv4.2-AKAP interactions. In co-IP experiments, the AKAP79 antibody pulled down either AKAP79 or Kv4.2 when co-expressed in COS7 cells but not Kv4.2 expressed alone (Figure 7A). No interaction was observed between KChIP4a and AKAP79 in co-IP experiments using the AKAP79 antibody for pulldown (Figure 7B). These results suggest that AKAP79 binds to Kv4.2, but not directly to KChIP4a. Figure 7C shows co-localization of Kv4.2 and AKAP150 (mouse homologue of the human AKAP79) in hippocampal neurons.
Figure 7.
Kv4.2 co-immunoprecipitates with the PKA anchoring protein AKAP79. (A, B) COS7 cells expressing Kv4.2 alone (A, line 4) or coexpressing AKAP79 with either Kv4.2 (A, line1-3) or KChIP4a (B). In co-immunoprecipitation experiments, cell lysates were pulled down using anti-AKAP79 or nonspecific IgG, and probed with anti-Kv4.2 antibody (1:2000) or anti-AKAP79 (1:1000) and anti-Flag antibody (1:6000) to detect KChIP4a, then visualized by anti-mouse Alexa Fluor 680 and anti-rabbit Alexa Fluor 800 secondary antibody, respectively. These results demonstrate that Kv4.2 but not KChIP4a interacts with AKAP79. (C) Images of Kv4.2 and AKAP150 (mouse homologue of AKAP79) in hippocampal showing their co-localization (red, AKAP150; green, Kv4.2; yellow, overlapping signals). Scale bar, 10 μm.
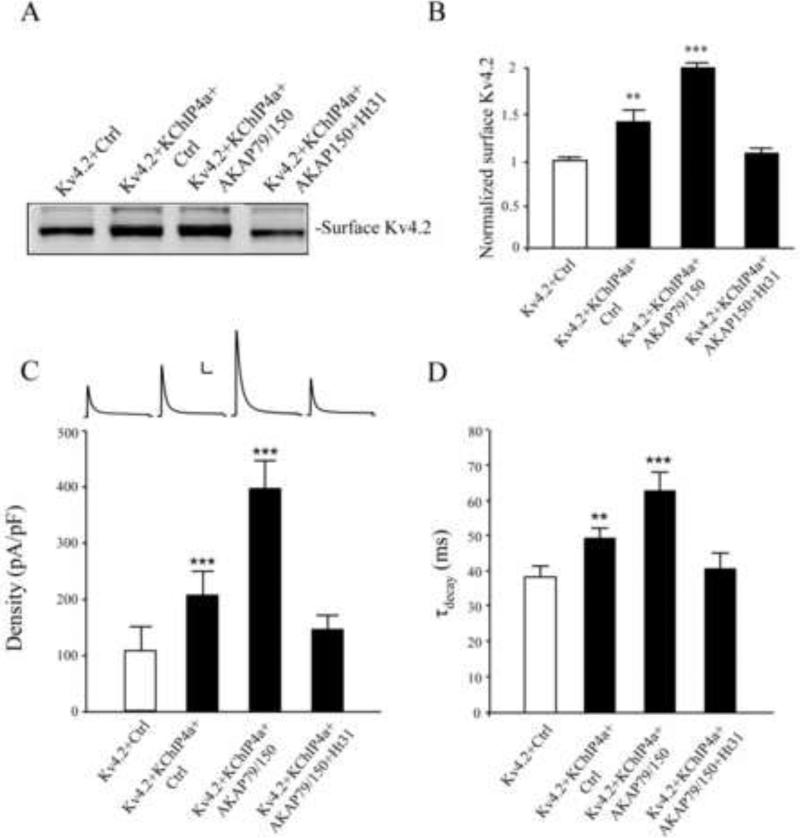
In both biotinylation (n = 3; p < 0.05) and electrophysiology experiments (n = 9-10; p < 0.05), we found AKAP79 to augment Kv4.2+KChIP4a surface expression (Figure 8A-C). To determine AKAP involvement in PKA-mediated surface enhancement, we used the Ht31 inhibitor peptide, which prevents the interaction between RII subunits of PKA and AKAPs. In both biotinylation (Figure 8A, B, n = 3, p > 0.05 compared to control) and electrophysiology experiments (Figure 8C, n = 9, p > 0.05 compared to control), Ht31 brought surface expression/current levels down toward levels observed without KChIP4a co-expression and counteracted the KChIP4a-mediated increase in the time constant of channel inactivation (Figure 8D).
Figure 8.
KChIP4a regulates Kv4.2 trafficking through PKA anchoring proteins. (A-B) Biotinylation results showed COS7 cells expressing Kv4.2 with KChIP4a or with KChIP4a and AKAP79. Kv4.2 surface expression, enhanced by KChIP4a, was further increased by co-expression with AKAP79. The AKAP inhibitor Ht31 (25 μM, 1 hr), which prevents PKA binding to AKAP proteins, blocked the enhanced surface expression of Kv4.2 by AKAP79. Error bars represent S.E.M. (C) These results were mimicked in electrophysiological recordings where co-expression of Kv4.2 with KChIP4a and control vector (n = 9) increased the peak current density (~2 fold) of Kv4.2-mediated current whereas peak current density was further enhanced (~4 fold) by the addition of AKAP79 (n = 10). Both enhancements were again opposed by the PKA anchoring inhibitor Ht31 (25 μM, 1hr, n = 9). Error bars represent S.E.M. Scale bars: 1 nA, 100 ms. (D) KChIP4a-mediated increases in inactivation rate are abolished by Ht31 treatment.
These Ht31 results in HEK cells could indicate a role for endogenous AKAPs in the KChIP4a-mediated effects on current levels (Willoughby et al., 2006). Accordingly, we found Ht31 to reduce peak A-current in electrophysiological recordings from HEK cells expressing Kv4.2+KChIP4a in the absence of AKAP79 (Kv4.2+KChIP4a+Ht31: 211.77±21.63 pA/pF; n=11, p < 0.05 vs Kv4.2+KChIP4a without Ht31: 273.65±13.10 pA/pF; n=15, data not shown). Alternatively, AKAP79-containing channel complexes, requiring PKA for surface expression, could dominate Kv4.2+KChIP4a-only channels when co-expressed with AKAP79, preventing KChIP4a-mediated increase of surface expression. In any case, our results, along with previous observations that both proteins are expressed in hippocampal spines (Kim et al., 2007; Robertson et al., 2009), suggest that AKAPs likely couple PKA to Kv4-channel complexes to dynamically regulate their activity natively in neurons.
Discussion
Here we present results demonstrating a unique, interactive co-regulation of PKA phosphorylation of the Kv4.2 pore-forming α-subunit and the KChIP4a auxiliary subunit. While phosphorylation at site S552 of Kv4.2 is not required for the interaction between Kv4.2 and KChIP4a, it is necessary for the enhanced membrane expression and protein stability conferred by these auxiliary subunits. This co-requirement of Kv4.2 α-subunit phosphorylation and KChIP co-expression for functional changes was also described for PKA modulation of A-type current kinetic properties (Schrader et al., 2002). However, our findings show that the requirement of PKA phosphorylation of Kv4.2 at site 552 for enhanced surface expression and stability is unique to KChIP4a, as other KChIP proteins affected expression levels even when co-expressed with the Kv4.2 PKA phosphorylation mutant S552A. In addition, we identify AKAP79 as a Kv4.2 binding partner and show that AKAPs augment KChiP4a-mediated Kv4.2 surface enhancement. AKAP binding partners allow for compartmentalized PKA signaling to the Kv4.2 macromolecular complex.
Mechanism of enhanced surface expression and stabilization
In the present manuscript, we provide both biochemical and electrophysiological evidence that KChIP4a increases cell surface expression and stability of Kv4.2 channels after heterologous co-expression. These findings are consistent with previous (Jerng et al., 2004b) and present (Figure 3) results observed after co-expression of Kv4 channels with KChIP1 or KChIP2 subunits. Unlike with KChIP1 or KChIP2 co-expression, however, KChIP4a-mediated surface expression and protein stability enhancement required S552 phosphorylation of Kv4.2. To account for these findings we suggest the following working model. If, as evidence suggests (Shibata et al., 2003), KChIP4a tends to be preferentially trapped in the ER, perhaps anchored in the ER membrane by its unique N-terminal transmembrane domain (Jerng and Pfaffinger, 2008), phosphorylation of Kv4.2 at site 552 may be permissive for the trafficking of KChIP4a-containing complexes out of intracellular membranes as was found for the plasma membrane (Hammond et al., 2008). Enhanced stability through KChIP4a binding may occur through a decrease in the incidence of misfolded Kv4.2, which would otherwise be targeted for degradation. Support for this scenario comes from the electrophysiology data presented in Figures 1 and 3. Although Kv4.2 and Kv4.2S552A current densities are similar (Figure 1E, compare “Ctrl” groups), lack of an effect on Kv4.2 S552A inactivation rate by KChIP4a expression (Figure 1F) suggests that channels in complex with KChIP4a are not forward trafficked to the plasma membrane. This is not the case for KChIP1 and KChIP2 where co-expression leads to delayed inactivation rates (Figure 3D).
Comparison with previous reports
In this study we have found that mouse KChIP4a increases the surface expression of Kv4.2 about 1.5-2 fold compared to Kv4.2 alone. These results were confirmed using a range of techniques (biotinylation assays, on-cell westerns and electrophysiological recordings). However, a number of previous reports have found that KChIP4a, unlike other KChIP subunits, does not increase surface expression of Kv4 channels (Holmqvist et al., 2002; Jerng and Pfaffinger, 2008; Liang et al., 2008; Schwenk et al., 2008; Shibata et al., 2003). We tested a number of hypotheses to explain our results showing increased surface expression. First, we noticed that most previous work was performed using Kv4.3 as the pore-forming α-subunit. We therefore tested whether KChIP4a increases the surface expression of Kv4.3 using biotinylation assays after expression in COS7 cells (Supplementary Figure 1). We found KChIP4a increases Kv4.3 surface expression about 1.5 fold compared to Kv4.3 alone (n = 3, p < 0.05), similar to our results using Kv4.2. Next, we examined the peak Kv4.2 current in HEK293 cells co-transfected with human KChIP4a (Supplementary Figure 2). The result is similar to that found for mouse KChIP4a, showing that human KChIP4a also increases the peak current of Kv4.2 about 2-fold (p < 0.05).
One possible explanation for the different findings in the ability for KChIP4a to enhance channel expression levels lies in the recording techniques used, including cell-type differences. Our study used cDNA transfection into mammalian cells (COS7 and HEK, Figure 1 and Supplemental Figure 3) whereas recordings were predominately performed in Xenopus oocytes after cRNA microinjection in previous studies. We note that there is a tendency toward larger currents after KChIP4a expression in one study also using mammalian CHO cells (Holmqvist et al., 2002). Although we demonstrate enhanced surface expression of Kv4.2 with KChIP4a co-expression, the increase is clearly much smaller than that produced by other KChIP subunits (compare Figures 1B and 3B).
Physiological Relevance
PKA activation leads to decreased A-type K+ channel activity in CA1 dendrites through a hyperpolarizing shift in their activation curve, leading to enhanced dendritic excitability (Hoffman and Johnston, 1998). These changes require Kv4 channels to be in complex with KChIP subunits (Schrader et al., 2002). Our present results show that PKA phosphorylation of site S552 of Kv4.2 would also be expected to enhance the expression and trafficking of KChIP4a containing channels. Previously it was observed that S552 is phosphorylated natively in hippocampal neurons (Anderson et al., 2000), that phosphorylation at this site appears to be associated specifically with membrane bound channels (Shibata et al., 2003), and that it is required for activity-dependent internalization (Hammond et al., 2008). Together with our present results showing AKAP interaction with Kv4 channels, these findings suggest a membrane-bound pool of Kv4.2 channel macromolecular complexes whose activity and expression can be locally regulated by PKA through AKAP interaction. This compartmentalized signaling could allow for local regulation of dendritic action potential propagation and initiation. In addition, dynamic regulation of these channels by PKA may help shape Ca2+ signals in microdomains of the dendrite (Cai et al., 2004; Jung et al., 2008), and may play a role in synaptic (Chen et al., 2006; Jung et al., 2008) and intrinsic plasticity (Jung and Hoffman, 2009).
Experimental Methods
Constructs
Mutations of Kv4.2 C-terminal phosphorylation site at S552 (Kv4.2S552A and Kv4.2S552D) were performed using the Quick-Change site-directed mutagenesis kit (Stratagene, San Diego, CA). We mutagenized the serine (S) to Alanine (A) to remove the phosphorylation of PKA site or to Aspartic acid (D) to mimic constitutive phosphorylation at site 552 on construct Kv4.2-GFP (Kv4.2, GFP tagged at C-terminus). Mouse KChIP4a was produced by PCR amplifying mouse cDNA using 5’-GGGAATTCGATGAACTTGGAGGGGCTTG and 5’-GGTCTAGAGATCACATTTTCAAAGAG primers (GenBank accession number AF453243) and cloned into p3xFlag-CMV vector (Sigma, St. Louis, MO). Since there is no satisfactory commercially available antibody against KChIP4a, we used a Flag tag for detection. KChIP1, KChIP2a and human KChIP4a constructs were kindly provided by Dr. Pfaffinger, Baylor College of Medicine. Dr. Covarrubias, Thomas Jefferson University, kindly provided the Kv4.3 construct and the pcDNA-AKAP79 construct was kindly provided by Dr. Dell'Acqua at University of Colorado. All constructs were confirmed by sequencing analysis.
Cell Western Assays
Assays were performed as described by the manufacturer (LI-COR Biosciences application note (2004)). Briefly, 0.2×106 of COS7 cells were plated into 24-well tissue culture plates. The cells were co-transfected with wild type Kv4.2 or Kv4.2S552A with KChIP4a-Flag for 24-48 hr using Fugene 6. COS7 cells were fixed with PBS containing 4% paraformaldehyde. After blocking with Odyssey blocking solution for 1-1.5 hr at RT, cells were incubated with mouse anti-Kv4.2 antibody (1:200, NeuroMab, Davis, CA) in Li-COR blocking buffer at 4°C for over night. Cells were washed and incubated with the secondary antibody IRDye 800 Goat anti-mouse (1:1000, Rockland, Gilbertsville, PA) at 37 °C for 1 hr. After a wash in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min. We used rabbit anti-beta actin antibody (1:1000, sigma) and goat anti-rabbit secondary antibody IRDye 680 (1:800, Invitrogen) to detect actin. The intensity of the 700-nm and 800-nm infrared signal for each well was quantified using the Odyssey infrared imaging system software (LI-COR Biosciences, Lincoln, NE).
Co-immunoprecipitation and Western Blotting
COS7 cells were co-transfected with wild type Kv4.2 or Kv4.2S552A with KChIP4a-flag. To confirm an interaction between AKAP79 and Kv4.2 or KChIP4a, COS7 cells were transfected with Kv4.2 alone as control or co-transfected with AKAP79 and either KChIP4a or Kv4.2. After 24-48 h transfection, cells were lysed in lysis buffer: 150 mM NaCl, 20 mM Tris-HCl, 1% NP40, 0.5% SDS and protease inhibitor mixture (Roche, Indianapolis, IN). Anti-Flag (2μg/500μg protein, Sigma, St. Louis, MO), nonspecific IgG (Invitrogen) or anti-AKAP79 antibody (2ug/500ug protein, Millipore, Bedford, MA) was then added to the centrifuged detergent lysate. The mixture was then incubated and rotated at 4°C for overnight. The antibody-antigen complex was immobilized by adsorption onto 50 μl of immobilized protein A (Pierce, Rockford, IL) and incubated for 2 h at RT. The protein-bead mixtures were washed six times with lysis buffer. The beads were resuspended in reducing SDS sample buffer and analyzed on 10% SDS polyacrylamide gels. The separated proteins were immuonoblotted using Kv4.2 (1:2000, NeuroMab, Davis, CA) or Flag antibody (1: 6000, Sigma) for KChIP4a and AKAP79 antibody (1:1000) and visualized by Alexa Fluor 800 secondary antibody (1:10,000, Rockland, Gilbertsville, PA) and Alexa Fluor 680 secondary antibody (1:10,000, Invitrogen). Immunoreactivity was detected with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, Nebraska). Quantification of results was performed using Odyssey software (LI-COR Biosciences, Lincoln, Nebraska).
Biotinylation assays
Biotinylation assays were performed as previously described (Kim et al., 2007). Briefly, transfected COS7 cells were rinsed with ice-cold PBS, surface protein were biotinylated with 1.5mg/ml sulfo-NHS-SS-biotin reagent (Pierce, Rockford, IL) in PBS for 30 min on ice. Unbound biotin was quenched with cold 50 mM glycine in PBS. Cells were lysed with ice-cold lysis buffer: 150 mM NaCl, 20 mM Tris-HCl, 1% NP40 and protease inhibitor mixture (Roche, Indianapolis, IN), sonicated and centrifuged at 12,000 g for 10 min. Cell lysates were incubated overnight at 4°C with immobilized-Streptavidin agarose beads (Pierce, Rockford, IL), unbound protein was removed from the beads with 5 washes in lysis buffer. The bound proteins were eluted with 2×SDS sample buffer. Surface expressed proteins were separated by electrophoresis on 10% Tris-bis SDS-PAGE (Invitrogen, Carlsbad, CA) and transferred to PVDF membranes. Western blots were probed with the following antibodies: mouse anti-Kv4.2 (1:2000, NeuroMab, Davis, CA), mouse anti-GAPDH (1:1000, Calbiochem, San Diego, CA). Secondary antibodies conjugated to infrared dyes (Rockland Immunochemicals, Gilbertsville, PA) were detected using Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). Quantification of results was performed using Odyssey software.
Stabilization assay
To study the effect of KChIP4a on the stabilization of Kv4.2 protein complexes, we transfected COS7 cells with Kv4.2 or Kv4.2S552A along with KChIP4a, KChIP1 or control (empty vector) for 24 hr, then treated with 10 μM of forskolin (Sigma) for 24 hr at 37°C. After washing with ice-cold PBS, the cells were solubilized in 500 μl of lysis buffer (described above). Lysates were centrifuged for 10 min at 20,000 g rpm at 4°C after shaking for 2 h at 4°C. 20 μl of the supernatant was analyzed by 10% SDS-PAGE. Kv4.2, KChIP4a and KChIP1 were immuonoblotted using Kv4.2 antibody (1: 2000, NeuroMab, Davis, CA), Flag antibody (1: 6000, Sigma) and KChIP1 antibody (1:1000, NeuroMab, Davis, CA), then visualized by anti-rabbit Alexa Fluor 800 secondary antibody (1:10,000) and anti-mouse Alexa Fluor 680 secondary antibody (1:10,000). Detection and quantification was performed using the Odyssey infrared imaging system and its software as described above.
Rat hippocampal neuron culture and immnofluorescence staining
Primary hippocampal neurons cultures were prepared as previously described (Kim et al., 2007). Neurons were infected by a modified sindbis virus expression system (Kim et al., 2004). Briefly, hippocampi from E18-19 rat embryos were triturated after trypsinization and suspension with MEM plus 10% FBS plating media following by plating on Poly-DLysine and Laminin coated glass coverslips (BD, San Jose, CA) in a 24-well plate. After 5-6 hr, the media was replaced with Neurobasal media plus B27 supplements (Invitrogen). Cultures were maintained at 37°C with 10% CO2. Neurons (DIV18-21) were infected with Kv4.2g (Kv4.2-GFP, see (Kim et al., 2005) ) sindbis virus for 1 hr. The next day, neurons were fixed with PBS containing 4% paraformaldehyde, and 0.12 M sucrose for 20 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min, endogenous AKAP150 was immunostained with anti-AKAP150 antibody (1:100, Millipore, Bedford, MA) for 1 h at 37°C, washed with PBS and then incubated with Alexa Fluor 555-conjugated goat anti-rabbit (1:500, Invitrogen) in DPBS with 5% NGS, 0.05% triton X-100 for 1 h at 37°C. Coverslips were mounted onto glass slides with Hard Set mounting medium (Vector Laboratories, Burlingame, CA). Images were captured with Zeiss LSM510 confocal microscope (Carl Zeiss, Germany) using a 63X objective with 1.4 NA.
Electrophysiology
HEK 293 cells were plated on poly-D-lysine/laminin-coated glass coverslips (BD BioCoat) in 12-well dish at a density of 22-24 × 103 cells/well and transfected using the Fugene 6 system (Roche). Cells were transfected with Kv4.2 , Kv4.2S552A or Kv4.2S552D constructs, along with either mouse KChIP4a, KChIP1, KChIP2, AKAP79 or control plasmids. Recordings were made 24-48 hr after transfection. Coverslips containing transfected HEK 293 or COS7 cells were submerged in the recording chamber and exposed to a continuous flow of ACSF consisting of the following (mM): 145 NaCl, 5 KCl, 2 CaCl2, 1.3 MgCl2, 10 glucose, 10 HEPES. ACSF was perfused with 95% O2-5% CO2, and TTX (~0.5 μM, Sigma) was included to block any endogenous sodium current. An infrared differential interference contrast (IR-DIC) videomicroscopy system (Zeiss Instruments, Diagnostic Instruments) was used to visualize cells. Patch pipettes were pulled from thick-walled borosilicate glass (Warner Instruments) to achieve a tip resistance of 2-5 MΩ. Pipettes were filled with an internal solution containing (mM): 20 potassium chloride, 125 potassium gluconate, 10 HEPES, 4 NaCl, 0.5 EGTA, 10 phosphocreatine, 4 ATP, 0.3 TrisGTP. For the PKA inhibition and activation experiments, H89 (10 μM) and FSK (10 μM) was applied to HEK 293 cells in culture 1 h and 24h before and during recordings, respectively. Ht31(25 μM, Promega) inhibitor peptide was applied to cultures 1h before and during recordings in some experiments.
Whole-cell voltage clamp recordings were made using a Multiclamp 700B amplifier (Molecular Devices) and Clampex 10.1 software (Molecular Devices). Signals were digitized at 10 kHz with a Digidata 1440A (Molecular Devices) and filtered at 4 kHz. A-type K+ currents were elicited by delivering a 400 ms prepulse from the holding potential of -60 mV to -120 mV followed by a 500 ms step to +120 mV. This protocol was repeated 20-45 times for each cell to determine the average peak A-current. Leak currents were subtracted online by a P/6 procedure. Only cells with a holding current greater than -100 pA were selected for recordings. Series resistance ranged from 4-11 MΩ, and was monitored throughout the experiments. All recordings were performed at room temperature. Recordings in which the series resistance increased by more than 25% of the initial value were excluded from analysis. No series resistance compensation was employed. Recordings were analyzed using Clampfit 10.1 (Molecular Devices) and Microsoft Excel. To determine A-current density, peak currents were normalized to whole-cell capacitance. To estimate the time constant of channel inactivation (inactivation rate), current traces were fitted with a single exponential function. Statistical significance was evaluated using Student's t-test (unpaired, two tails).
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank Drs. Paul Pfaffinger and Manuel Covarrubias for providing KChIP and Kv4.3 constructs and Dr. Dell'Acqua for kindly providing the AKAP79 construct. We thank Vincent Schram in the NICHD Microscopy & Imaging Core for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Anderson AE, Adams JP, Qian Y, Cook RG, Pfaffinger PJ, Sweatt JD. Kv4.2 phosphorylation by cyclic AMP-dependent protein kinase. J Biol Chem. 2000;275:5337–5346. doi: 10.1074/jbc.275.8.5337. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nature reviews. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD, Weiss JL. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- Cai X, Liang CW, Muralidharan S, Kao JP, Tang CM, Thompson SM. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Chen X, Yuan L, Zhao C, Birnbaum S, Frick A, Jung W, Schwarz T, Sweatt J, Johnston D. Deletion of Kv4.2 Gene Eliminates Dendritic A-Type K+ Current and Enhances Induction of Long-Term Potentiation in Hippocampal CA1 Pyramidal Neurons. Journal of Neuroscience. 2006;26:12143–12151. doi: 10.1523/JNEUROSCI.2667-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Zhou M, Mann S, MacKinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science (New York, N.Y. 2000;289:123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- Hammond RS, Lin L, Sidorov MS, Wikenheiser AM, Hoffman DA. Protein kinase a mediates activity-dependent Kv4.2 channel trafficking. J Neurosci. 2008;28:7513–7519. doi: 10.1523/JNEUROSCI.1951-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DA, Johnston D. Downregulation of transient K+ channels in dendrites of hippocampal CA1 pyramidal neurons by activation of PKA and PKC. J Neurosci. 1998;18:3521–3528. doi: 10.1523/JNEUROSCI.18-10-03521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist MH, Cao J, Hernandez-Pineda R, Jacobson MD, Carroll KI, Sung MA, Betty M, Ge P, Gilbride KJ, Brown ME, Jurman ME, Lawson D, Silos-Santiago I, Xie Y, Covarrubias M, Rhodes KJ, Distefano PS, An WF. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc Natl Acad Sci U S A. 2002;99:1035–1040. doi: 10.1073/pnas.022509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng H, Pfaffinger P, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Molecular and Cellular Neuroscience. 2004a;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ. Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating. J Biol Chem. 2008;283:36046–36059. doi: 10.1074/jbc.M806852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol Cell Neurosci. 2004b;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Jung SC, Hoffman DA. Biphasic somatic A-type K channel downregulation mediates intrinsic plasticity in hippocampal CA1 pyramidal neurons. PLoS One. 2009;4:e6549. doi: 10.1371/journal.pone.0006549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SC, Kim J, Hoffman DA. Rapid, bidirectional remodeling of synaptic NMDA receptor subunit composition by A-type K+ channel activity in hippocampal CA1 pyramidal neurons. Neuron. 2008;60:657–671. doi: 10.1016/j.neuron.2008.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Dittgen T, Nimmerjahn A, Waters J, Pawlak V, Helmchen F, Schlesinger S, Seeburg PH, Osten P. Sindbis vector SINrep(nsP2S726): a tool for rapid heterologous expression with attenuated cytotoxicity in neurons. J Neurosci Methods. 2004;133:81–90. doi: 10.1016/j.jneumeth.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Kim J, Hoffman DA. Potassium channels: newly found players in synaptic plasticity. Neuroscientist. 2008;14:276–286. doi: 10.1177/1073858408315041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung S, Clemens A, Petralia R, Hoffman DA. Regulation of Dendritic Excitability by Activity-Dependent Trafficking of the A-Type K+ Channel Subunit Kv4.2 in Hippocampal Neurons. Neuron. 2007;54:933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wei DS, Hoffman DA. Kv4 potassium channel subunits control action potential repolarization and frequency-dependent broadening in rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;569:41–57. doi: 10.1113/jphysiol.2005.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjilwar K, Strang C, DeRubeis D, Pfaffinger PJ. KChIP3 rescues the functional expression of Shal channel tetramerization mutants. J Biol Chem. 2004;279:54542–54551. doi: 10.1074/jbc.M409721200. [DOI] [PubMed] [Google Scholar]
- Liang P, Wang H, Chen H, Cui Y, Gu L, Chai J, Wang K. Structural insights into KChIP4a modulation of Kv4.3 inactivation. J Biol Chem. 2008 doi: 10.1074/jbc.M807704200. [DOI] [PubMed] [Google Scholar]
- Papazian DM. Potassium channels: some assembly required. Neuron. 1999;23:7–10. doi: 10.1016/s0896-6273(00)80746-1. [DOI] [PubMed] [Google Scholar]
- Robertson HR, Gibson ES, Benke TA, Dell'Acqua ML. Regulation of postsynaptic structure and function by an A-kinase anchoring protein-membrane-associated guanylate kinase scaffolding complex. J Neurosci. 2009;29:7929–7943. doi: 10.1523/JNEUROSCI.6093-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader LA, Anderson AE, Mayne A, Pfaffinger PJ, Sweatt JD. PKA modulation of Kv4.2-encoded A-type potassium channels requires formation of a supramolecular complex. J Neurosci. 2002;22:10123–10133. doi: 10.1523/JNEUROSCI.22-23-10123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Zolles G, Kandias NG, Neubauer I, Kalbacher H, Covarrubias M, Fakler B, Bentrop D. NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. J Biol Chem. 2008;283:18937–18946. doi: 10.1074/jbc.M800976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing S, Roeper J, Pongs O. Kv beta 1 subunit binding specific for shaker-related potassium channel alpha subunits. Neuron. 1996;16:455–463. doi: 10.1016/s0896-6273(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Shibata R, Misonou H, Campomanes CR, Anderson AE, Schrader LA, Doliveira LC, Carroll KI, Sweatt JD, Rhodes KJ, Trimmer JS. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J Biol Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- Willoughby D, Wong W, Schaack J, Scott JD, Cooper DM. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 2006;25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.