Studies of the dynamical properties of proteins using NMR is an emerging field, which has largely been limited to backbone N-H or sidechain methyl motions. It is often, but not always true that changes to dynamic state of the backbone will reflect changes in the sidechains. The NMR techniques for quantifying fast timescale motion involve measuring the longitudinal (R1) and transverse (R2) relaxation rates, as well as the heteronuclear NOE for each amino acid backbone N-H (or sidechain C-H bond vector) in the protein.1–3, 10 These values can then be used to calculate the generalized order parameter (S2), which is a measure of protein flexibility.
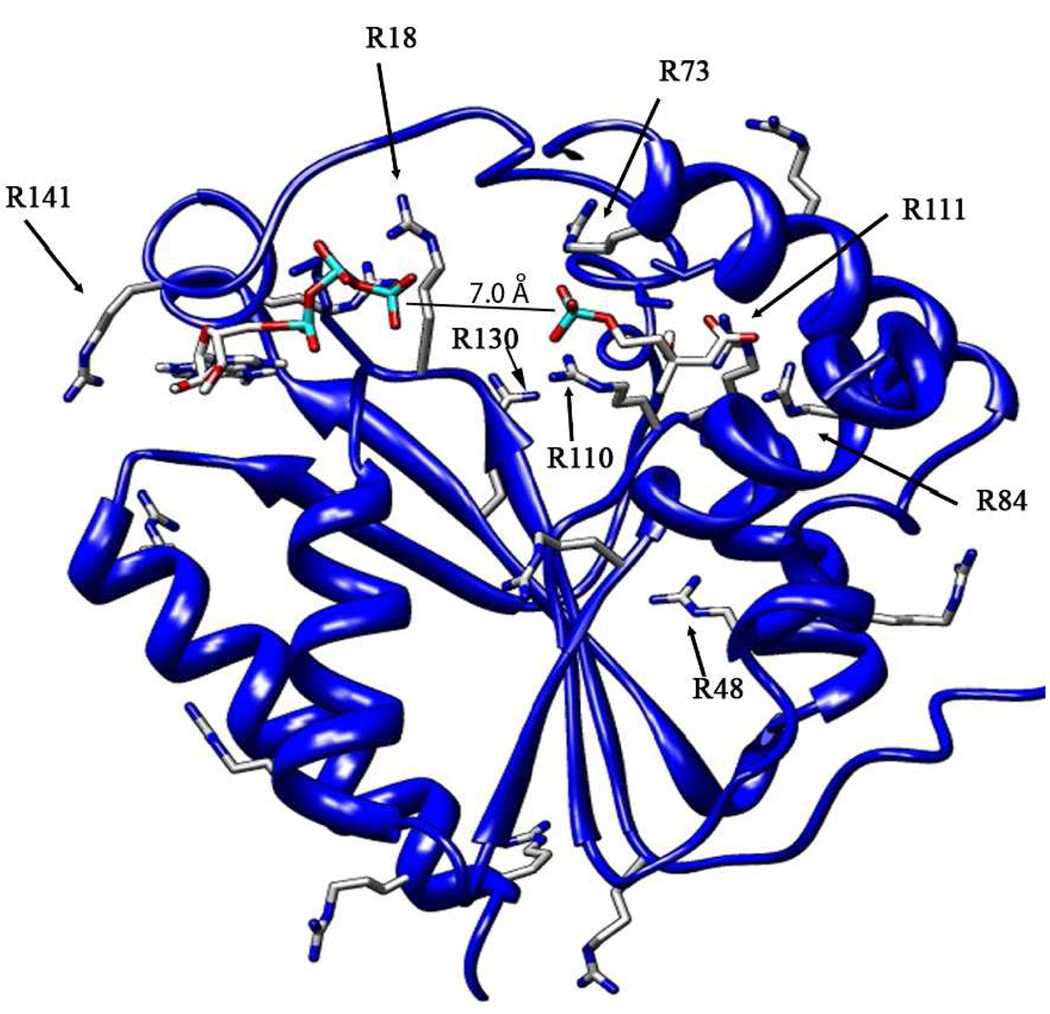
We have recently reported on the ligand induced structural and dynamical changes of human phosphomevalonate kinase (PMK).4 PMK is the fifth enzyme in the mevalonate pathway in humans, and is involved in steroid biosynthesis.5, 6 PMK catalyzes phosphoryl transfer from ATP to mevalonate 5-phosphate (M5P) to form ADP and mevalonate 5-diphosphate. To permit phosphoryl transfer, the substrates are brought close together, resulting in a significant and repulsive buildup of negative charge. To facilitate this difficult task, PMK contains 17 arginines (Figure 1) and 8 lysines, with many in the active site to help neutralize the negative charge on the phosphates. The most important arginines, based on site-directed mutagenesis studies, are R18, R48, R73, R84, R110, R111, R141.7, 8 Here we describe the use of NMR dynamics methods to characterize changes to the mobility of arginine sidechains upon ligand binding. NMR dynamics methods, recently applied to arginine sidechains,10–13 allow study of the role that arginine sidechains play in ligand binding and catalysis. Our studies provide surprising insights into ligand effects on arginine sidechains that are remote from the active site, perhaps due to long-range coulombic attraction.
Figure 1.
Ternary complex of human PMK, with M5P and ATP docked independently into the apo crystal structure (3CH4).9 After docking, structure was optimized using molecular dynamics (50 ps at 300 K). All arginines are shown, and those known to be important for substrate binding or catalysis are labeled. Inter-phosphate distance is 7 Å, so phosphate transfer will require additional domain movement.
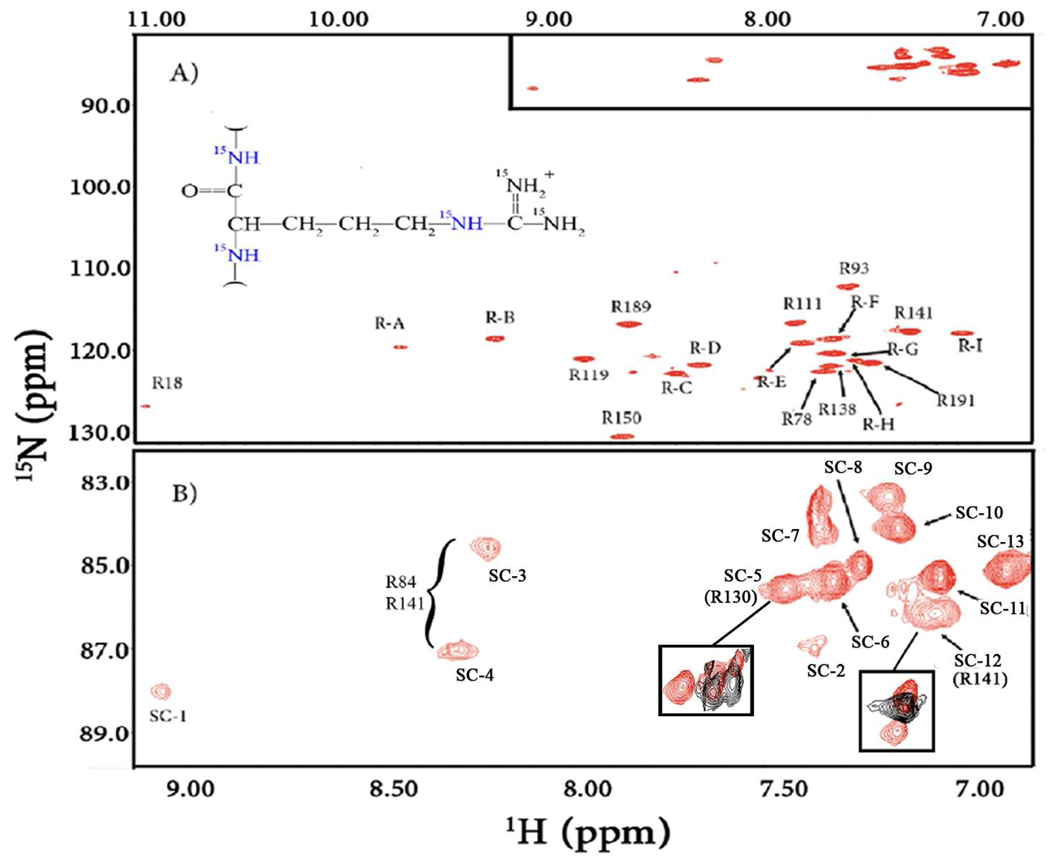
Since PMK is a rather large protein (24 kDa), in terms of NMR studies, samples were prepared with only arginines having the 15N label (Figure. 2A). This allowed us to make sure that every arginine backbone residue was considered, without concern for signal overlap, along with the ε-N-H’s of the sidechain. Other arginine sidechain N-H’s, though labeled, exchange too fast to be observed. As a consequence of PMK having significant resonance overlap in the backbone N-H region, and due to significant relaxation that precluded sidechain assignments via TOCSY experiments, only half of the arginine residues could be assigned. The assigned arginines are labeled accordingly on the spectra, with the rest of the arginine backbone N-H’s being labeled R-(A-I). The ε- N-H’s of the arginine sidechains are shown at the top of Figure 2A and expanded in Figure 2B. As these sidechain N-H’s could not be assigned to their respective backbone N-H’s, they have been labeled SC (1–13); furthermore, four of the 17 residues are apparently exchange broadened.
Figure 2.
1H-15N HSQC spectra of (A) 15N-Arg-PMK with backbone residues labeled and (B) expansion of the arginine sidechain region, with assignment labels. Overlaid expansions in boxes show data for mutation-based assignments (mutated PMK in black). Spectra were of 500 µM PMK (20 mM potassium phosphate, 5 mM DTT, 10% D2O, 10% d6-glycerol), at 298K on a 600 MHz Varian NMR System.
To monitor arginine involvement in the binding of PMK’s substrates, chemical shift titration experiments were performed (Figure S1). Increasing amounts of M5P and MgADP were added to PMK, and arginine backbone and sidechains were monitored using 1H-15N HSQC spectra. The majority of the arginine N-H’s show very small chemical shift changes, indicating little change in chemical environment for residues labeled SC 2, 5–13. For residues labeled SC 1, 2, and 4, the chemical shifts exchange-broaden, indicating some change in environment and motion, due to binding of M5P (Figure S1-C). This experiment was duplicated using MgADP (Figure S1-D), monitoring chemical shift changes as before. Only SC 1, 2, and 4 show large chemical shift changes, suggesting they are involved in binding.
Dynamics experiments were performed as before, where we showed substrate-induced domain movement occurs in PMK.4 Longitudinal relaxation rates (R1) are generally the same for the backbone and sidechains for each particular complex, and are in agreement with previous results4 (Figure S2, Table S1). The transverse relaxation rates (R2) of the backbone N-H’s also correlate with previous studies.4 Sidechains, however, show a decrease in transverse relaxation rates compared to the backbone. This decrease in apo-PMK’s arginine sidechains R2 values is significant; half relative to backbone. The decrease in R2 is small for the M5P/PMK complex, with the other two complexes showing an intermediate decrease. The 1H-15N NOE values for the sidechains vary from residue to residue, but this variability is consistent between complexes.
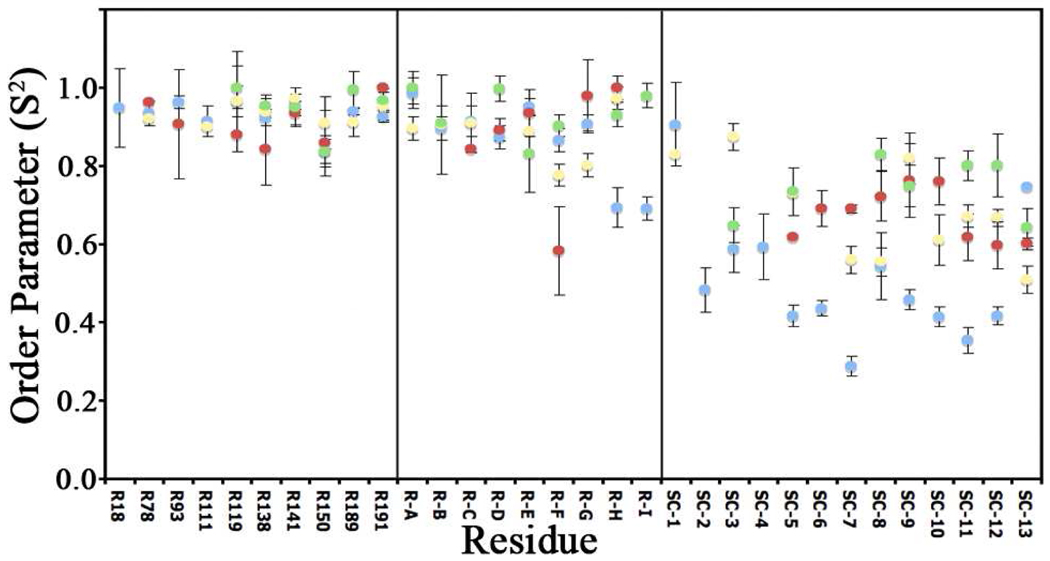
As in previous studies,4 a modelfree analysis2 was used to generate the generalized order parameter, S2. The S2 value measures rigidity on the nsec-psec timescale (where S2 = 1.0 is completely rigid and S2 = 0.0 is completely fluid) for the bond of interest,1–2 in our case arginine backbone and sidechain N-H bond vectors. Modelfree analysis can also differentiate between global and local motions. In our previous work on PMK, we showed that domain movement (global motion) was affected by ligand binding, with little attenuation of local motions. From the sidechain studies presented herein, it was observed that ligand binding causes the arginine ε-N-H’s to increase S2 on average from 0.47 in apo-PMK to 0.64–0.75 in the binary and ternary complexes (Figure 3). This leads to the conclusion that ligand binding causes arginine sidechains to rigidify throughout the protein, even distal to the binding site, perhaps due to long-range coulombic interactions with the substrates. S2 values for the arginine backbones correspond to our previous results4 on fully 15N labeled PMK, where we reported changes to τc due to domain motion, analogous to those in adenylate kinase (Table S1); differences are attributed to different buffer solutions. Surprisingly, backbone dynamics changes were minimal, in stark contrast to sidechain effects.
Figure 3.
Generalized order parameters (S2 values) for all assigned and unassigned arginine backbone and sidechain ε-N-H’s, where blue is apo PMK, red is M5P/PMK complex, yellow is MgADP/PMK complex, and green is the M5P/MgADP/PMK ternary complex. Using site-directed mutagenesis, we have tentatively assigned SC-2 and 3 as being R84 and R111, SC-5 as likely being assigned as R130, and SC-12 probably being R141. Nearly all arginine sidechain residues undergo a significant increase in S2 (i.e. rigidification) upon binding to either ligand. While Arg’s 84 & 111 are close to charged atoms on substrate, Arg130 & 141 are > 10 Å from charged atoms.
It has been noted for studies of sidechain methyl groups that changes to the dynamic state of sidechains may not be well reflected in changes observed in the backbone.14 We also observe quite large differences between backbone and sidechain responses to substrate binding. Whether such dramatic differences between sidechain and backbone dynamics changes are common in arginines (vs. aliphatic sidechains) and/or interactions with highly charged substrates (4 phosphate groups) cannot be concluded until more arginine sidechain dynamics studies are reported on other proteins.
While it is possible that substrate binding simply causes a global rigidification of sidechains in PMK, this seems unlikely. We propose that sidechains rigidify due to coulombic attraction from the highly charged substrate(s); in support of this hypothesis, the ADP-induced S2 effect is abolished in high salt (Figure S8). This must be a long-range effect, since only 4 arginines are within 4 Å and only 6 arginines are within 8 Å, even though ~10 arginine sidechains rigidify (Figure 3).
In summary, we have shown that substrate binding to PMK causes arginine sidechains to transition from a flexible to a rigid state. The magnitude and global nature of this effect is significant. Besides providing mechanistic insights, such studies facilitate docking, by identifying flexible sidechains.
Supplementary Material
Acknowledgement
This research was supported by funding from the American Heart Association (05303072) to D.S.S., NIH DK53766 to H.M.M., and to the Chemical Proteomics Facility at Marquette (NIH-NSF instrumentation grants S 10 RR019012 and CHE-0521323). Thanks also to Garrett Mohr for expressing the PMK mutants.
Footnotes
Supporting Information Available: Detailed procedures and additional NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org
Phosphomevalonate kinase (PMK) catalyzes phosphoryl transfer from ATP to mevalonate 5-phosphate (M5P), on the pathway to synthesizing cholesterol and other isoprenoids. To permit this reaction, its substrates must be brought proximal, which would result in a significant and repulsive buildup of negative charge. To facilitate this difficult task, PMK contains 17 arginines and 8 lysines. But, how this charge neutralization and binding is achieved, from a structural and dynamics perspective, is not known. More broadly, the role of arginine sidechain dynamics in binding charged substrates has not been experimentally defined for any protein, to date.
Herein we report a characterization of changes to the dynamical state of the arginine sidechains in PMK, due to binding its highly charged substrates, ATP and M5P. These studies have been facilitated by the use of arginine-selective labeling, to eliminate spectral overlap. Modelfree analysis indicates that while substrate binding has little effect on arginine backbone dynamics, binding of either substrate leads to significant rigidification of the arginine sidechains throughout the protein, even those that are > 8 angstroms from the binding site. Such a global rigidification of sidechains is unprecedented, and suggests that there are long-range electrostatic interactions of sufficient strength to restrict motion of arginine sidechains on the psec-nsec timescale. It will be interesting to see whether such effects are general for arginine residues in proteins that bind highly charged substrates, once additional studies of arginine sidechain dynamics are reported.
References
- 1.Mandel AM, Akke M, Palmer AG. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 2.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 3.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 4.Olson AL, Yao H, Herdendorf TJ, Miziorko HM, Hannongbua S, Saparpakorn P, Cai S, Sem DS. Proteins. 2009;75:127–138. doi: 10.1002/prot.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellig H, Popjak G. J. Lipid Res. 1961:235–243. [Google Scholar]
- 6.Eyzaguirre J, Valdebenito D, Cardemil E. Arch Biochem Biophys. 2006;454:189–196. doi: 10.1016/j.abb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Herdendorf TJ, Miziorko HM. Biochemistry. 2006;45:3235–3242. doi: 10.1021/bi052231u. [DOI] [PubMed] [Google Scholar]
- 8.Herdendorf TJ, Miziorko HM. Biochemistry. 2007;46:11780–11788. doi: 10.1021/bi701408t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Q, Yan X, Gu S, Liu J, Liang D. Proteins. 2008;73:254–258. doi: 10.1002/prot.22151. [DOI] [PubMed] [Google Scholar]
- 10.Buck M, Boyd J, Redfield C, MacKenzie DA, Jeenes DJ, Archer DB, Dobson CM. Biochemistry. 1995;34:4041–4055. doi: 10.1021/bi00012a023. [DOI] [PubMed] [Google Scholar]
- 11.Cai M, Gong Y, Wen L, Krishnamoorthi R. Biochem. 2002;41:9572. doi: 10.1021/bi0258952. [DOI] [PubMed] [Google Scholar]
- 12.Hsu ST, Cabrita LD, Fucini P, Christodoulou J, Dobson CM. J. Am. Chem. Soc. 2009;131:8366–8367. doi: 10.1021/ja902778n. [DOI] [PubMed] [Google Scholar]
- 13.Trbovic Nikola, Cho Jae-Hyun, Abel Robert, Friesner RA, Rance Mark, Palmer AG. J Am Chem Soc. 2009;21:615–622. doi: 10.1021/ja806475k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igumenova TI, Frederick K, Wand AJ. Chem Rev. 2006;106:1672. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.