Abstract
All extant vertebrates possess an adaptive immune system wherein diverse immune receptors are created and deployed in specialized blood cell lineages. Recent advances in DNA sequencing and developmental resources for basal vertebrates have facilitated numerous comparative analyses that have shed new light on the molecular and cellular bases of immune defense and the mechanisms of immune receptor diversification in the “jawless” vertebrates. With data from these key species in hand, it is becoming possible to infer some general aspects of the early evolution of vertebrate adaptive immunity. All jawed vertebrates assemble their antigen-receptor genes through combinatorial recombination of different “diversity” segments into immunoglobulin or T-cell receptor genes. However, the jawless vertebrates employ an analogous, but independently-derived set of immune receptors in order to recognize and bind antigens: the variable lymphocyte receptors (VLRs). The means by which this locus generates receptor diversity and achieves antigen specificity is of considerable interest because these mechanisms represent a completely independent strategy for building a large immune repertoire. Therefore, studies of the VLR system are providing insight into the fundamental principles and evolutionary potential of adaptive immune recognition systems. Here we review and synthesize the wealth of data that have been generated towards understanding the evolution of the adaptive immune system in the jawless vertebrates.
Keywords: Adaptive immune system, Evolution, Jawless vertebrates
1. Introduction
The elimination and neutralization of structurally diverse viruses, bacteria and parasites by metazoan immune systems requires a corresponding diversity of immune receptor molecules. In general, adaptive immune systems must provide clonally diverse anticipatory and self-tolerant repertoires that can be deployed by lymphocyte cell lineages. Each lymphocyte bears a unique cell surface receptor in order to recognize and respond to antigenic determinants of diverse and evolving pathogens in the ongoing struggle for survival. Until recently, the molecular details of adaptive immunity only had been described for the jawed vertebrates (gnathostomes). All jawed vertebrates, including the most phylogenetically divergent extant representatives, such as sharks, are equipped with similar adaptive immune systems that employ VDJ recombination to achieve combinatorial diversity of immunoglobulins (Igs) and T cell receptors (TCRs) [1]. In developing lymphocytes, the recombination activating genes (RAG1 and RAG2) mediate recombination of variable region segments by random selection of each of V, D and J segments into the maturing immune receptor loci. In addition, the Activation-Induced Deaminase gene (AID) encodes an enzyme that facilitates gene conversion and somatic hypermutation of immunoglobulin loci in higher vertebrates, and also is required for class switch recombination. Recent findings in the cyclostomes (lamprey and hagfish) have revealed that jawless vertebrates possess an alternative form of immune receptor system that evolved independently of the Igs and utilizes a RAG-independent strategy to generate receptor variants that recognize and facilitate elimination of pathogens [2;3]. This system undergoes somatic rearrangements of leucine-rich repeat cassettes in the variable lymphocyte receptor (VLR) locus in order to generate immune receptor diversity that is presumably comparable to that of Igs in jawed vertebrates. Two similar VLR genes (VLRA and VLRB) have been identified in jawless vertebrates. Both VLR genes undergo somatic diversification and probably contribute to immune defense system, but the nature of somatic rearrangement and immune functions of the VLRB gene have been studied in greater detail. Two AID-APOBEC family member genes (CDA1 and CDA2) were identified in lamprey and have been implicated as being facilitators of these gene conversion-type rearrangements of the VLR loci in lymphocytes [4]. Lymphocytes expressing rearranged VLR molecules can bind bacterial or erythrocyte antigens and subsequently differentiate into VLR antibody-secreting cells that morphologically resemble plasma cells [5]. The binding affinities of VLR to its cognate antigens have been empirically determined to be similar to that of mammalian Igs [6;7]. Thus, it is now clear that two radically different systems evolved in cyclostomes and gnathostomes in which either LRR (leucine rich repeat) or Ig (and TCR) gene fragments are randomly assembled to generate diverse repertoires of lymphocyte receptors and respond to antigenic challenges. This evolutionary scenario raises intriguing questions as to whether these two adaptive immune strategies represent truly convergent evolution or if one was ancestral to and replaced by the other in either the cyclostome or gnathostome stem lineage.
2. Evolution of Adaptive Immune System in Gnathostomes (brief overview)
Although this review primarily focuses on the adaptive immune system in cyclostomes, it is best understood in the context of the gnathostome adaptive immune system, because the gnathostome system is relatively well-characterized and provides an outgroup perspective on the cyclostome condition. The adaptive immune system of gnathostomes was presumably appended to the innate immune system and provided the capability to mount non-self-challenge by inducing diversity in a limited set of specialized molecules and deploying these in lymphocytes. The gnathostome adaptive immune system’s vast repertoire of antigen-specific receptor molecules is generated by somatic gene rearrangement, which is mediated by RAG genes. In addition to Igs, which recognize unprocessed antigens and neutralize them, these antigen receptors include TCRs, which recognize antigen fragments presented by highly polymorphic major histocompatibility complex (MHC) molecules that display both self and non-self antigens on the surface of T-cells. The emergence of the adaptive immune system is believed to have occurred when an Ig superfamily (IgSf) gene of the “variable” type was invaded by a transposable element containing RAG1 and RAG2 genes [8;9].
The adaptive immune system may have evolved in the context of a pre-existing innate immune system because the randomly-derived and subsequently activated lymphocytes exhibit memory of previous encounters with infectious agents and facilitate a more rapid response when these agents reappear. Several studies have revealed that jawless vertebrates, protochordates and even invertebrates have evolved sophisticated RAG-independent strategies in order to recognize and facilitate elimination of pathogens. The range of such molecules, which include the Down syndrome cell adhesion molecule (DSCAM) in Drosophila [10], fibrinogen-related proteins (FREP) in the snail [11], and variable lymphocyte receptors (VLRs) in jawless vertebrates [2], encompass both the immunoglobulin superfamily and leucine-rich repeat (LRR) proteins. Despite their structural diversity, these molecules participate in various types of host defense programs. The ability to deploy structurally diverse antigen recognition molecules is a general feature of metazoan immune systems, but DNA-based somatic diversification as a means to facilitate receptor diversity is a characteristic that is apparently unique to the vertebrates.
3. Description of the Adaptive Immune System in Cyclostomes
3.1. Presence of immune cells
Mononuclear cells that are morphologically similar to vertebrate lymphocytes have been identified in various tissues and in the peripheral blood of sea lamprey and hagfish. Lamprey lymphocytes possess a very electron dense nucleus and relatively little cytoplasm, which contains numerous ribosomes but a paucity of membranous organelles [12]. The lamprey possesses several organs that may contribute to the maturation of developing lymphocytes. One of the primary immunological organs of the adult lamprey is the supraneural body (also known as the fat body or pro-vertebral arch; Fig. 1A). This structure lies dorsal to spinal column and becomes heavily populated with lymphocytes following after immune challenge [13–15]. The supraneural body from hematopoietically stimulated lampreys appears to be histologically-similar to ‘bone marrow’ in higher vertebrates and contains all blood cell lineages and their precursors, including lymphocytes at all stages of maturity [16]. Lymphocytes are also abundant in the lamprey kidney, where large populations are intermingled around the renal tubules (Fig. 1B), a situation very similar to that seen in the kidney of teleosts [17]. A third primary immunological organ is the typhlosole, which lies within an invagination of the gut proper (Fig. 1C). This organ contains a wide range of lympho-hematopoietic cells interspersed with stromal-like tissue and blood sinusoids. The general histological organization and hematopoietic cell composition of the typhlosole are similar to that found in the hematopoietic nets occurring in the intestinal submucosa of the plexiform veins of hagfishes [18] and in numerous organs in cartilaginous and bony fishes that are considered to be morphological and functional equivalents of the bone marrow [19].
Figure 1.
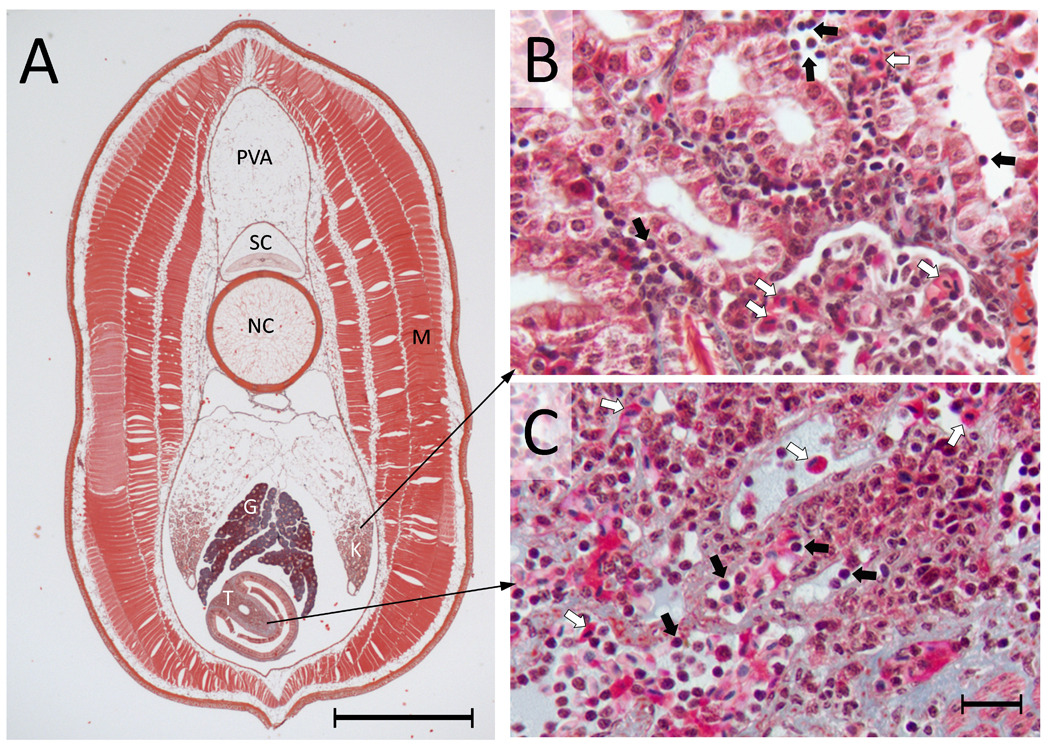
Distribution of cells in primary hematopoietic tissues in larval lamprey. (A) A transverse section from the mid-body region of an ammocoete lamprey (~ 13 cm in length). The 10 µm processed section stained with Masson Trichrome is showing different major internal organs, which include protovertebral arch (PVA), spinal cord (SC), notochord (NC), gonad (G), kidney (K), typhlosole (T) and muscle (M). Scale bar = 1 mm. (B) A magnified view of the kidney showing the distribution of blood cells, including many lymphocytes and erythrocytes (black and white arrows, respectively). Collections of blood cells are seen in and amongst the renal tubules. (C) A magnified view of the typhlosole showing diverse blood cells (lymphocytes and erythrocytes are indicated by black and white arrows, respectively). Scale bar = 10 µm. The structures that are stained light blue are largely extracellular matrix, which is highly abundant in the typhlosole.
3.2. Observation of antibody-like agglutinating activities
As is the case in the gnathostomes, lampreys have the capacity to mount considerable agglutination responses following exposure to heterologous antigens. Serum from the arctic lamprey Lampetra reissneri had been shown to contain natural agglutinins which react, to varying degrees, with the erythrocytes of different species. Upon repeated immunizations with sheep red blood cells, arctic lampreys showed a dramatic increase in specific hemagglutination titers [20]. The induced agglutinins were heat-stable and displayed a high degree of specificity to sheep erythrocytes. Similarly, the sea lamprey, Petromyzon marinus, possesses low titers of agglutinins to the H surface antigen of human “O” cells but subsequent immunization gives rise to a substantial increase in titer of these agglutinins [21]. Furthermore, these agglutinins have been shown to be antigen-specific for the O-type erythrocytes [13;21;22]. In adult P. marinus, the agglutinin-producing cells were found to be abundant within the supraneural body [16], whereas in ammocoetes (larvae) of the arctic lamprey the specific agglutinin producing cells were present in the typhlosole and corresponded morphologically to plasma cells [18]. Thus, the typhlosole and kidney, and supraneural organ are the most likely sites for definitive lymphopoiesis in the larval and adult lamprey, respectively. Alternatively, the general mode of lymphocyte development, differentiation and maturation in lampreys (and cyclostomes, in general) may be different than what is observed in gnathostomes [23].
3.3. Expression of immune related transcription factors in developing lymphocytes
A number of transcription factors that are known to be important in lymphocyte development and early fate specification have presumptive lamprey homologs that also are important in lymphogenesis. The transcription factor Ets is a primary regulator of hematopoiesis in gnathostomes [24] and is expressed in embryonic blood islands and in developing lymphocytes [25;26]. The transcription factor, Bcl11a, which may function as a leukemogenic factor in gnathostomes, also is expressed in lamprey lymphocytes [27]. Other transcription factor genes found as transcripts in lamprey and/or hagfish include: ikaros (zinc finger lymphocyte differentiation factor) [28], blimp-1 (B-cell induced maturation protein, PR domain containing) [29], ubiquitin conjugated enzyme E2A [30], the B-cell homeodomain-containing gene Pax5 [31], and Gata2/3 genes that regulate gene expression in hematopoietic cells. Altogether, expression of numerous transcription factor homologs in both gnathostome and lamprey lymphogenesis suggest that their role in early development of the lymphocyte per se may have been established prior to the divergence of jawless and jawed vertebrates [23].
3.4. Discovery of adaptive immune related genes
Mature cyclostome lymphocytes express many genes that are homologous to those expressed in the mature lymphocytes of higher vertebrates; including genes that have an essential role in lymphocyte proliferation and activation. Sea lamprey lymphocytes express: (1) CD45, which is involved in the regulation of T and B cell stimulation and proliferation [32]; (2) CD9/CD81, which stabilizes the molecular complexes involved in lymphocyte activation, adhesion, migration, and differentiation [33]; (3) BCAP, which serves an adaptor function in signaling and B lymphocyte activation [5;30;33]; (4) CAST, which serves a similar role as BCAP but in T-cell activation [30;33]; (5) CD98, which transports amino acids associated with lymphocyte activation[33]; and (6) SYK protein tyrosine kinase, Src family members, and HS-1 adaptor molecule [30;33], which are lymphocyte activators. Similarly, CD45, a receptor-type protein tyrosine phosphatase that regulates lymphocyte development and activation, has been identified in the lymphocyte population of the hagfish [34]. Other critical genes necessary for the mammalian adaptive immune system have been identified within the lamprey genome, including: (1) CXCR4, a chemokine homing receptor known for mammalian hematopoietic progenitors [35] as well as four additional chemokine receptors [23;33]; (2) two CC chemokines [23]; (3) CXCL12, a chemokine receptor that pairs with CXCR4 and is involved in regulating the precise spatial distribution of lymphoid progenitors in hematopoietic tissues [23]; (4) the chemotactic inflammatory cytokine IL-8 [36]; (5) the IL-17 receptor [5]; (6) CD4-like molecules [37] and (7) Toll-like receptor (TLR) orthologs TLR2a, TLR2b, TLR2c, TLR7 and TLR10 [5], which function in B-cell activation. Cyclostomes also express genes that are similar to those encoding gnathostome adaptive immune receptors, but which lack the diversity of these receptors. A single copy of a TCR-like gene having divergent V and J type sequences and a VpreB-like gene is expressed in lymphocyte-like cells of sea lamprey [38] and a family of paired Ig-like receptor genes encoding transmembrane proteins with activating and inhibitory potential has been isolated from hagfish lymphocytes [37]. Surveys for homologs of genes that are expressed in vertebrate lymphocytes provide evidence for shared regulatory and signaling functions in cyclostome and gnathostome immunity.
3.5. Identification of jawed vertebrate thymus-associated orthologous structures in lamprey
3.5.1. Histological investigation
The search for a thymus in the lamprey was based on the presumption that all higher vertebrates possess and require this structural innovation for immune function. Classical light microscopic studies represented the first attempts to identify a thymus analog in lamprey. These studies revealed numerous lymphocytes and macrophages within structures that are located under the pharyngeal epithelium of ammocoetes [17]. These structures (known as pharyngea lamina propria) appear to be involved in the uptake of foreign materials (particulates) from the pharyngeal cavity [39], but on the basis of more recent observations these clearly were neither a thymus nor a primordial precursor of such an organ [40]. Zurbrigg and Beamish [41] showed that the presumptive lymphocytes of sea lamprey stained positive with Thy-1 (CD90), which is typically associated with the thymus and T-cell reactivity in gnathostomes. They found Thy-1 reactivity in different tissues including typhlosole, opisthonephros, liver, external gill openings in larval lamprey, but were unable to locate a thymus-like structure. Based on all available evidence, the lamprey lacks an identifiable thymus, implying that the thymus, an extremely specialized structure, probably arose as an innovation within the lineage that led to the jawed vertebrates [42].
3.5.2. Molecular investigation
In mammals, T lymphocytes develop from a common lymphocyte progenitor in the bone marrow. These progenitor cells leave the bone marrow and home to the thymus where they undergo development, differentiation and thymic selection. The developing thymocytes are embedded in the interstices of an extensive network of epithelial cells mainly in the cortex of the thymus. The gnathostome transcription factor, Foxn1, occupies a central position in the thymic genetic network and has a role in establishing a functional thymic rudiment. Foxn1 is expressed in the thymus in cartilaginous fishes and all other jawed vertebrates and is considered a key factor in the evolutionary emergence of the vertebrate thymus. A presumed ortholog of the Foxn1 gene (Foxn4L) has been discovered in lamprey, and found to be expressed in epithelial lining of the gill basket in lamprey ammocoetes [23]. In the gnathostome thymus, the differentiation of lymphocyte progenitor cells into the T cell lineage is dependent on the Notch ligand Delta-like 4 (Dll4) [43;44]. Analysis of whole genome sequence data revealed that the lamprey genome contains three distinct Dll4 genes, whereas the teleost fishes possess orthologs of five such genes. One of the lamprey delta-like genes is expressed in a circumscribed region of the epithelium lining the gill basket of ammocoetes, precisely overlapping with expression of the Foxn4L domain, indicating an evolutionarily conserved pattern of co-expression of Foxn4/Foxn4L and Delta-like genes. Moreover, genes for two Notch orthologs (NotchA and NotchB), which transmit Delta-like signals to developing hematopoietic cells in vertebrates [45] have been identified in the lamprey genome and shown to be highly expressed in leukocytes and lymphocytes. However, neither VLRB (a marker of mature lymphocytes) nor CXCR4 (chemokine receptor which is expressed in virtually all leukocytes) were found within the same epithelial structures in histological sections of ammocoetes of Lampetra planeri [30]. The cells expressing VLRB are located largely in the vasculature of gills and gill-associated structures. Importantly, no large aggregations of lymphocytes are localized in any particular site within those tissues. In addition, no VLRB-positive cells were detected within the epithelial structures, corroborating the overall differences between these tissues and thymus [23]. In summary, molecular surveys for thymus-related genes in lamprey provide evidence that the thymic and gill basket epithelium likely share a common developmental origin and that the evolutionary emergence of the thymus may have occurred in the common ancestor of gnathostomes through modification of this epithelium. However, the structures in lamprey are clearly not histologically or functionally equivalent to the gnathostome thymus.
4. Discovery of the Adaptive Immune Receptor of Cyclostomes (VLR)
The first evidence for the existence of the VLR receptors came from a subtractive EST survey of lymphoid cells of immunized ammocoetes, which resulted in the identification of a large number of sequences encoding a particularly complex set of leucine-rich repeat (LRR)-containing molecules [2]. These proteins were named variable lymphocyte receptors because the transcripts were expressed predominantly or exclusively by lymphocytes and each transcript encoded an ectodomain that differed among all other VLR transcripts. The basic structure of the variable lymphocyte receptor is similar to many other LRR-containing proteins and consists of: (1) conserved signal peptide; (2) N-terminal LRR (LRRNT); (3) variable number of diverse LRRs (“diversity region”); (4) connecting peptide followed by C-terminal LRR (LRRCT); (5) conserved C-terminus composed of a threonine/proline-rich stalk (analogous to Ig “constant region”); (6) glycosyl-phosphatidyl-inositol (GPI)-anchor site; and (7) hydrophobic tail. There is no detectable signaling tail. BAC clone analysis revealed that the germline VLR is invariant, lacks the LRR modules that impart the enormous diversity of this molecule, and that the missing LRR modules reside 5’ and 3’ of the transcription start and stop sites. Diversity of the transcribed receptors is thought to occur by subsequent recruitment of LRR cassettes into the diversity region through a novel recombination mechanism that accesses the large banks of flanking LRR cassettes.
4.1. Genomic Structure of VLR Loci in cyclostomes
The germline VLR locus (gVLR) lacks the capacity to encode a functional immune receptor and must undergo rearrangement in order to achieve this functionality [2]. The gVLR core contains the presumptive promoter, transcription initiation and termination sites and the functional start and stop codons, all arranged in the same relative orientation as the mature VLR locus (mVLR) (Fig. 2). However, the region between the start and stop codons does not encode an open reading frame and does not possess cassettes of variable leucine-rich repeats (LRRs) that build the mature locus. Rather, the region that is ultimately populated by LRRs consists of two long (~6kb) intervening regions that are interrupted by a partial LRR-CT module. The LRR minigene cassettes are inserted [46] randomly from elsewhere in the genome between invariant N- terminal and C-terminal LRR cassettes during lymphocyte ontogeny.
Figure 2.
Genomic organization and rearrangement of a mature VLR gene of sea lamprey. The germline VLR (gVLR) configuration with 5’ and 3’-encoding LRR genetic segments (top) contains an additional 13.2 kb of non-coding intervening sequence and lacks the key LRR modules, which are essential to the structure of functional VLR genes. The inserted LRR modules lie both 5’ and 3’ of gVLR. The VLR locus undergoes stepwise assembly (middle) via recombination between short stretches of nucleotide homology found at the junctions of various LRR modules. This process may occur on either (or both) strand(s) during replication and gradually replaces the intervening sequence with all the variable LRR segments. The end product of these recombination events (bottom) is a mature VLR locus capped with an invariant 5’ end of the LRRNT module, an invariant 3’ end of the LRRCT module and a variable number of LRR cassettes (each encoding 24 amino acids) that vary in number and sequence. The illustration is not drawn to scale.
Many, if not all, of the LRRs that are incorporated into the mVLR are found at the upstream and downstream regions that flank the presumptive promoter and core region. These belong to several defined classes. The so-called N-terminal repeats (LRRNT) and C-terminal (LRRCT) repeats are respectively invariant to the 5’ and 3’ regions of the VLR locus that immediately flank the intervening (non-coding) sequence. Other classes of repeats include: LRR1, which always abuts LRRNT, LRRV (variable), LRRVe (a distinct subclass and basically the last LRRV), and CP (“connecting peptide” a truncated LRR), which always abuts LRRCT. Sequencing surveys and reverse transcriptase assays have revealed substantial variation in the length of mVLR molecules, which derives from the incorporation of variable numbers of LRRV cassettes. Thus, a single basic structure can be defined for the mVLR locus: SP-LRRNT-LRR1-LRRV1~9-CP-LRRCT-C terminus.
Rogozin et al. [4] performed a survey of all available sequences from the ~5X lamprey genome sequencing project in order to identify genomic intervals that contribute to the mVLR. This survey searched for matches (>30bp) to a larger number of functional (transcribed) mVLRs and identified 2.2 Mb of sequence fragments (genomic contigs) that contained exact matches to mVLRA molecules and 2.1 Mb that correspond to mVLRB molecules. Understanding how these repeats are distributed in the genome will be the key to understanding the cellular basis of VLR rearrangement/maturation. Specifically this will reveal whether all incorporated cassettes lie in the regions that immediately flank the VLR locus, or if some are incorporated from other chromosomes or distant genomic regions.
The genomic structures of the distinct VLRA and VLRB loci have been characterized for both lamprey and hagfish [2;47] and all loci share the same general modular structure. Notably, this structural conservation contrasts with extensive divergence at the nucleotide sequence level, as might be expected for two lineages that have been evolving independently for >500MY. The most obvious difference among these is between the between gVLRBs. Hagfish gVLRB lacks the 5’ end of the invariant LRRNT module and has only one LRRCT module in contrast to two that have been identified in lamprey. The only notable structural difference in the VLRAs is the presence of an intron in the 5’ UTR of the locus in hagfish that is not seen in lamprey [47] .
4.2. Cellular Basis of Rearrangement
Studies of mVLR molecules, rearrangement intermediates and the lamprey genome have provided several insights into the cellular basis of VLR rearrangements. Notably, these rearrangements are certainly not dependent on the same RAG-mediated mechanism that generates diversity in the immune receptors of jawed vertebrates as no RAG genes have yet been identified in the lamprey genome and patterns of VLR recombination are not consistent with homologous recombination at defined consensus sites [4;48]. The VLR assembly also lacks recombination signal sequences (RSS) that characterize the immunoglobulin variable, diversity and joining segments [30]. Rearrangement intermediates have been isolated from the sea lamprey (Petromyzon marinus) and Japanese lamprey (Lethenteron japonicum) by targeted sequencing of lymphocyte DNAs. These intermediates reveal that LRRs are incorporated onto one and/or both ends (5’ and/or 3’) of the maturing molecule. Incorporated modules show evidence of multiple switches between distinct LRRs that presumably are located in non-continuous segments of the VLR flanking region. The distinct segments that are incorporated into the maturing VLR exhibit short stretches of sequence homology at their boundaries (6–30 nucleotides) [4;48]. In mature lymphocytes, regions of the intervening region that are replaced by LRRs are not observed elsewhere in the genome, arguing against an equal exchange (i.e., cross-over) mechanism. In conjunction with the apparent directionality of LRR incorporation, LRR insertion events likely are mediated by replication, rather than exchange.
This pattern of replication-mediated recombination has been likened to ‘copy-choice’ gene conversion that occurs during mating-type switching in fission yeast [4;48;49]. Copy-choice recombination is, in essence, a targeted version of replication-recombination events that are collectively termed synthesis-dependant strand annealing [50]. Synthesis-dependant strand annealing occurs broadly during DNA repair in yeast [50] and likely, in vertebrates [51]. These recombination mechanisms require invasion of a free (i.e., broken) 3’ end into a non-allelic (donor) region with sequence homology. The homologous sequence acts as a template from which continued synthesis can proceed. Reinvasion and homologous pairing with the parent strand permits resolution of the recombination event without disruption of the donor region. There are, however, distinct differences between VLR recombination and known replication-recombination mechanisms. The most obvious difference between the two is the need for multiple strand invasions, extensions, and terminations that are targeted to different genomic sites (Fig. 2). Moreover, the events are programmed to occur upon maturation of lymphocyte cell populations. Hence, VLR recombination events are seemingly more tightly-regulated (in a developmental sense) and recombinationally-complex than other known forms of replication-mediated exchange.
If, as all evidence seems to indicate, the mVLR is synthesized by a replication-recombination mechanism, then a fuller understanding of the mechanisms underlying VLR diversification will require identification of the molecular players that mediate strand breakage, invasion, synthesis initiation/termination and juxtaposition of the core VLR and imported LRRs. Recently, we have gained an appreciation for the fact that VLR rearrangements are superimposed on a genome that itself undergoes more broad-scale rearrangements much earlier in development [52]. Given that VLR diversification and broad-scale rearrangement result in highly regulated somatic recombinations that are not typical of other vertebrate species, it seems likely that their underlying mechanisms may share some common molecular players.
4.3. Crystal-structure and antigen-binding properties of VLRs
The molecular basis of antigen recognition by antibodies in the vertebrate immune system is well studied and has revealed how the immunoglobulin folds with their CDR (complementarity determining region) loops can form a high-affinity binding site for virtually any antigen it encounters, whether natural or synthetic [53]. How VLR proteins can assess and recognize antigen has also been revealed [54;55], though not as broadly. High sequence variability in the Ig fold is concentrated in CDRs H1, H2, H3, L1, L2, and L3, whereas that of VLRs is confined to the concave surface of each LRR module. The crystal structures of the cyclostome VLRs adopt a solenoid-shaped structure common to LRR family proteins and the backbone structure of the LRR modules between VLRA and VLRB are highly similar [54;56;57] and the sequence variation is concentrated on the concave surface of the protein, which presumably is the site of antigen contact. In general, the length of LRRCT insert is known to differ depending on the size of antigens [7]. Variation in the numbers and sequence of LRRs affects the structure at the concave surface, and modularity that is inherent in the LRR scaffold permits changes in the residues within the this surface without affecting the overall stability of the protein [58]. Recent findings on a VLR-HEL (hen egg lysozyme) protein complex reveal that although lamprey VLR antibodies are fundamentally and structurally different from the antibodies of jawed vertebrates, the soluble monomeric VLRB binds to HEL with similar affinity as that of IgM antibodies, which exist as multimers in the plasma of higher vertebrates[7]. The crystal structure of VLR-HEL protein-antigen complexes reveal that nearly the entire concave surface of lamprey is accessible for antigen binding. The binding surface area by VLR to HEL is similar to the surface buried by Ig-antigen complexes [7]. The VLR binds over the catalytic site of HEL with a loop of LRRCT that penetrates into the carbohydrate-binding cleft. This form of binding is completely distinct from that of mouse VLHH antibodies, which bind to the flatter surface of HEL, but is similar to the heavy chain VH antibodies of camelid and IgNAR of sharks [6;7;59–61]. In vitro mutagenesis of VLRB can increase the binding affinity by altering the electrostatic potential surface, a mechanism which is reminiscent of the effects of somatic hypermutation of Ig antibodies which normally occur in the maturing lymphocytes of jawed vertebrates.
It has recently been shown that VLRA antibodies also are capable of very high-affinity interactions with antigens [6]. The binding ability of VLRB to heterologous RBC and HEL and VLRA to HEL provide further evidence that both VLRs act as adaptive immune receptors in sea lamprey. The extreme conservation of key residues suggests that the few resolved structures are representative of the structures of the entire repertoire of jawless fish VLRs and that these highly stable, modular and relatively small (15–25 kDa) single-chain peptides can bind a broad range of antigenic determinants with high affinity and specificity. Moreover, binding sites can be readily engineered in vitro, and perhaps in vivo [6] for improved binding properties. These findings indicate a functional parallelism between VLR-based and Ig-TCR-based antibodies and support the notion that both classes of antigen receptors were optimized over hundreds of millions of years and have evolved in the context of similar underlying lymphocyte gene networks.
5. Phylogeny of the VLR system
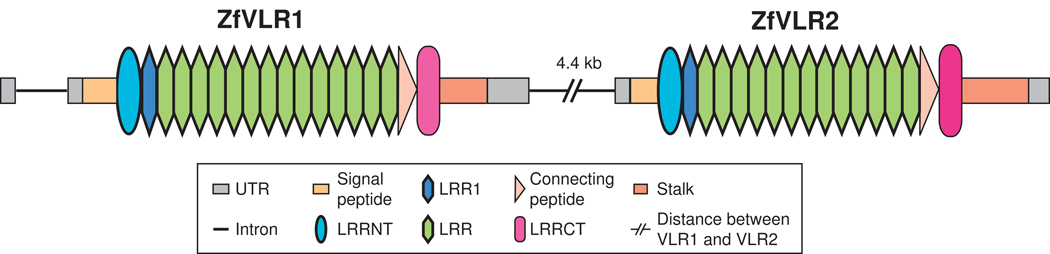
Empirical and in silico experiments have shown that an analogous VLR system capable of recombination is probably not present in urochordates (Ciona), arthropods (Drosophila), amphioxus (Branchiostoma) or mammals. In the teleost fishes (zebrafish, pufferfish), we have identified two adjacent VLR-like sequences that have the same modules as found in cyclostome VLRs (Fig. 3); however, their genomic structures indicate that they cannot undergo rearrangement (i.e., they are fixed in organization and resemble cyclostome mVLR loci). The expression of these genes (named zVLR1 and zVLR2) in zebrafish has been detected by RT-PCR in hematopoietic tissues in adults. Expressed sequence tags (ESTs) have been identified from several tissues for zVLR1 and in situ hybridization has suggested that it is expressed in a number of developing structures, including the blood stem cell compartment (unpublished). The zVLR2 gene also has been identified in zebrafish EST databases but at lower frequencies. It is known that zebrafish possess a well-developed Ig- and TCR-based adaptive immune system [62], but the role of these VLR-like genes in immunity is unknown. On the basis of gene organization and sequence identity, these zebrafish genes appear to be distant homologs of cyclostome VLRs. A more generic inference was put forward by Rogozin et al. [4], who, on the basis of genomic organization and amino acid conservation profiles, suggested that the VLRs share an evolutionary history with the platelet receptor glycoproteins, a class of genes found only in vertebrates. The evolutionary relationship between the cyclostome VLRs and vertebrate LRR-containing loci is not obvious, although remnants of shared ancestry of VLR-like genes have been identified.
Figure 3.
Genomic organization of two VLR-like genes in the zebrafish. The two VLR-like genes separated by 4.4 kb in the zebrafish genome assembly (Accession No. BX569794). Both genes contain all core VLR components identically juxtaposed as in cyclostome mVLRs. However, these genes contain comparatively more LRR cassettes than the average VLR molecule in cyclostomes and lack the ability to undergo genomic diversification. The exact phylogenetic relationship to cyclostome VLRs is unclear, though there is a distinct possibility that they are evolutionarily related at least on the basis of their similar genomic organizations. The functionality of these molecules is largely unknown. Color coding of structural features is presented in the inset and is identical to that in Figure 2. The illustration is not drawn to scale.
6. Distributions and functions of VLR-expressing cells
6.1. Distribution of VLRA+ and VLRB+ lymphocytes
In gnathostomes, B-cells are generated and develop in the bone marrow (or its equivalent), whereas T-cells migrate from distal sites (bone marrow) and undergo development and maturation in the thymus. Recent studies with two-channel FACS-sorted cells by VLRA-and VLRB-specific monoclonal antibodies from four major tissues have shown that VLRA+ and VLRB+ cells belong to discrete lymphocyte populations and exhibit characteristic tissue distribution patterns in lamprey [5]. Although both lymphocyte populations are distributed in the primary lymphoid tissues of lamprey larvae such as blood, kidneys, typhlosole and gill region, the VLRB+ lymphocytes outnumber VLRA+ lymphocytes in all tissues, except the gill region. The VLRB+ lymphocytes are most abundant in the blood and kidneys; whereas, VLRA+ lymphocytes are found primarily in the gills. The distribution pattern is similar in the tissues obtained from both adults and larvae. These results were validated by qPCR with cDNA from flow-sorted cells from all of those tissues from both adult and larvae. The VLRA+ lymphocytes express only VLRA transcripts, whereas the VLRB+ lymphocytes exclusively express VLRB transcripts. The double-negative population did not express VLR transcripts of either type. The relatively high concentration of VLRA+ lymphocytes in the gill region may reflect a developmental origin of these cells or a preferred site of secondary residence [5].
6.2. Differential gene expression by VLRA+ AND VLRB+ cell populations
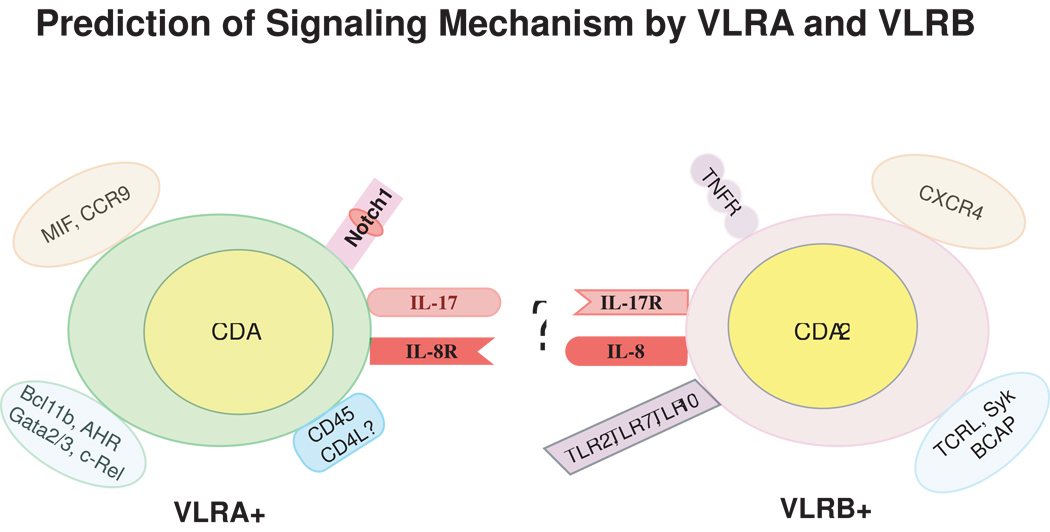
Conventional monoclonal antibody reagents that specifically detect VLRA and VLRB have allowed the isolation of respective lymphocyte populations and subsequent analysis of differential gene expression by respective populations [5]. Lymphocytes from immunized lamprey express homologs of mammalian cytokines and cytokine receptors. The most notable finding is the expression of IL-8R and IL-17 by the VLRA+ lymphocytes but IL-8and IL-17R by the VLRB+ lymphocytes [5] (Fig. 4). The VLRA+ cells expressing IL-17 may attract IL-17R-bearing VLRB+ cells and conversely, VLRB+ cells may utilize IL-8 to attract and engage IL-8R-bearing VLRA+ lymphocytes. This ad hoc hypothesis of cell-cell interaction is based on knowledge of the functions of interleukins and interleukin receptors in gnathostomes, and requires experimental validation. Lamprey lymphocytes also express many chemokines and their receptors. An ortholog for CCR9, which facilitates the first wave of embryonic thymus colonization in mouse, also has been identified in the lamprey genome; however, its expression patterns have not been resolved. CXCR4, another chemokine receptor, which acts as a homing receptor for hematopoietic progenitors, is expressed by VLRB+ cells [5] and is localized in the vasculature of gills [23].
Figure 4.
A conceptual diagram of differential gene expression by VLRA+ and VLRB+ lymphocytes. This illustration highlights the various proteins that were empirically shown [5] to be differentially expressed in flow sorted VLRA+ and VLRB+ lymphocyte populations [5]. These include possible enzymes for genomic rearrangement, signaling molecules, transcription factors, cell surface receptors, and chemokines/cytokines and their cognate receptor genes. The restriction of CDA1 and to VLRA+ and CDA2 to VLRB+ cells implies the potential for their selective involvement in the assembly of the VLRA and VLRB loci during lymphocyte development. It is speculated that the VLRA+ lymphocytes expressing IL-17 may attract IL-17R bearing VLRB+ lymphocytes, and VLRB+ lymphocytes may use IL-8 to attract and engage IL-8R bearing VLRA+ lymphocytes. Likewise, expression of TLR ligands by VLRB+ lymphocytes might trigger the activation of this cell population. Molecules are grouped on the basis of functionality and are not necessarily coexpressed (for example, TCRL, Syk and BCAP are B-cell signaling molecules, whereas Bcl11b, AHR, Gata2/3 and c-REL are T-cell transcription factors, etc).
TLRs regulate the development and persistence of T- and B-cell responses and memory in mammals [63]. Studies have shown that the murine splenic naïve B cells express a variety of TLRs with the exception of TLR5 and TLR8. Induction of robust antibody production and responses to antigenic challenge appears to require direct stimulation of B-cells by TLRs [64][65]. In lamprey, at least three different TLR genes (TLR2, TLR7 and TLR10) have been identified and are expressed by VLRB+ lymphocytes [5]. The expression of TLR ligands by the VLRB lymphocytes suggests that TLRs may facilitate the activation of this population in a manner similar to their role in mammalian B lymphocyte activation. In addition, CD4-like and TCR-like molecules have been reported to be differentially expressed in lamprey lymphocytes [37]. These data suggest that all of these differentially expressed lymphocyte factors probably existed in lymphocyte-like cells prior to the divergence of cyclostomes and gnathostomes. However, it still remains to be seen what role these factors play in VLR function and in lamprey immunity.
6.3. VLRs in humoral and cellular immunity
Experimental evidence suggests compartmentalized roles in humoral (VLRB) and cellular (VLRA) immunity [5]. Immunization of sea lamprey larvae with a single dose of either mouse or human erythrocytes increases VLRB antibody responses, which peaks at around 19 days post-inoculation. However, the hemagglutinin response to erythrocytes was highly variable and depended on antigen dose and individual mouse or human donor. A booster immunization on day-14 results a dramatically increased the specific hemagglutinin response, with a 20-fold amplification. Importantly, when plasma samples were depleted of VLRB by treatment with anti-VLRB coated Sepharose beads, the hemagglutinin activity was almost completely abrogated [66]. This result provides strong evidence that the erythrocytes were indeed agglutinated by VLRB that was produced by the immunized lampreys. The VLRB-bearing lymphocytes are presumed to bind bacterial or erythrocyte antigens and respond by proliferation and differentiation into plasmacytes that secrete multimeric VLRB antibodies specific for protein or carbohydrate epitopes (humoral immunity). Comparative and experimental evidence suggests that the VLRAs remain tethered to the cell surface [5]. This, in combination with experimental evidence that directly supports the idea that VLRA antibodies also are capable of very high-affinity interactions with antigens [6] and the molecular similarity between VLRA lymphocytes and T cells, strongly implies a primary role of VLRA-expressing cells in cellular immunity. These findings provide ample evidence for functional parallelism between VLR-based and Ig-based antibodies and provide further evidence that the origin of adaptive immune system likely predates the common ancestor of all extant vertebrate lineages.
6.4. Lamprey VLRA+ and VLRB+ lymphocytes functionally resemble gnathostome T- and B-cells, respectively
Both VLRA+ and VLRB+ lymphocyte populations responded strongly when larval lampreys were immunized with Bacillus anthracis exosporium by intraperitoneal injection. The most notable functional difference between VLRA+ and VLRB+ populations is that VLRB+ cells secreted their VLRB antibodies in response to immunization [66], whereas the VLRA+ population did not secrete antibodies [5]. Similar results also were observed when the lampreys were immunized with E. coli [5]. Consistent with this observation, 293T human embryonic kidney cells that were transfected with VLRA cDNA produced VLRA proteins that were tethered to the cell surface (i.e., not secreted); whereas transfection of the same cell line with VLRB cDNAs resulted in the secretion of VLRB protein products as multimeric forms [5]. It also is worth noting that a large number of VLRB+ lymphocytes were detected in the blood of both naive and immunized animals, whereas no VLRA+ cells that could bind anthrax spores were detected either before or after immunization. These features raise the possibility that VLRA+ lymphocytes respond to antigenic stimulation in a manner that strongly resembles the T-cell response and that the VLRA proteins, like TCRs in jawed vertebrates are expressed exclusively as transmembrane-bound molecules [5]. Conversely, VLRB+ lymphocytes respond to antigenic stimulation in a manner that more closely resembles the B-cell response. The preferential expression of TLR orthologs by the VLRB+ lymphocytes suggests that TLR ligands may facilitate activation of this population of lymphocytes in a manner similar to that described for mammalian B lymphocytes. Enhanced expression of IL-17 and MIF transcripts by VLRA+ lymphocytes after immune stimulation indicates that VLRA+ lymphocytes may use these pro-inflammatory cytokines to engage the IL-17R-bearing VLRB+ cells in a manner similar to mammalian T-cells. Conversely, upregulation of IL-8 in activated VLRB+ cells suggests that they may use this cytokine to attract IL-8R-bearing VLRA+ lymphocytes [5]. These observations suggest that VLRA+ lymphocytes may be analogous to gnathostome T-cells and VLRB+ lymphocytes analogous to gnathostome B-cells.
7. Conclusions and future prospects
Jawless vertebrates use receptors comprised of variable leucine-rich-repeat modules as counterparts of the immunoglobulin-based receptors (Ig and TCR) that form the basis of the gnathostome antigen recognition system. The discovery that cyclostomes and gnathostomes possess independently-derived immune receptors, which act in the context of very similar underlying lymphocyte genetic programs, suggests that either VLR or Ig, or both evolved in the context of specialized lymphocyte lineages that evolved prior to diversification of the ancestral vertebrate lineage. The question remains open as to whether: (1) VLR evolved in the context of an ancestral Ig expressing system; (2) Ig evolved in the context of an ancestral VLR expressing system; or (3) both systems evolved in parallel, being appended upon the same ancestral system. However the possibility that hagfish are basal to the lamprey-vertebrate split and the observation of VLR-like genes within the zebrafish and fugu genome, suggest the intriguing hypothesis that VLRs represent an ancestral immune receptor molecule.
Major challenges remain in elucidating the evolutionary history of the VLR locus, unraveling the mechanisms underlying its recombinational diversification, and dissecting the mechanisms that regulate its expression and ontogeny. Progress toward sequencing and assembly of an ever-increasing number of genomes from phylogenetically important vertebrate groups (including lamprey and hagfish) should provide broad-scale synteny information that will aid in reconstructing ancestral genomes and may reveal the origin of VLR and the fate of its vertebrate orthologs. Identifying the proteins that interact with the VLR locus at the time of rearrangement will be critical to understanding the precise recombinational mechanisms that give rise to receptor diversity. It is anticipated such studies also will provide insight into broad-scale rearrangements that occur during embryonic development. Dissecting the signaling mechanisms that underlie development and activation of VLRA+ and VLRB+ lymphocytes will be important for deducing whether the lymphocyte-type populations that existed in the ancestral vertebrate lineage gave rise to gnathostome T- and B-cell subsets. Finally, a better understanding of the cellular aspects of the VLR signaling cascade and the mechanisms underlying immunological tolerance are imperative, and will greatly aid in revealing general evolutionary strategies that underlie the emergence of the vertebrate adaptive immune system.
Acknowledgments
We thank Gary Litman of the University of South Florida for critically reviewing this manuscript. The authors are supported by grants from the National Institutes of Health and the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kasahara M, Suzuki T, Pasquier LD. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004 February;25(2):105–111. doi: 10.1016/j.it.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004 July 8;430(6996):174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 3.Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005 December 23;310(5756):1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 4.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007 June;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 5.Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009 June 11;459(7248):796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasumi S, Velikovsky CA, Xu G, Gai SA, Wittrup KD, Flajnik MF, Mariuzza RA, Pancer Z. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci U S A. 2009 August 4;106(31):12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velikovsky CA, Deng L, Tasumi S, Iyer LM, Kerzic MC, Aravind L, Pancer Z, Mariuzza RA. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009 July;16(7):725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci U S A. 1996 September 3;93(18):9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998 August 20;394(6695):744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 10.Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005 September 16;309(5742):1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 11.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004 July 9;305(5681):251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 12.Amemiya CT, Saha NR, Zapata A. Evolution and development of immunological structures in the lamprey. Curr Opin Immunol. 2007 October;19(5):535–541. doi: 10.1016/j.coi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good RA, Finstad J, Litman GW. Immunology. In: Hardesty MW, Potter IC, editors. The Biology of Lampreys. London: Academic Press; 1972. pp. 405–432. [Google Scholar]
- 14.Finstad J, Good RA. THE EVOLUTION OF THE IMMUNE RESPONSE. 3. IMMUNOLOGIC RESPONSES IN THE LAMPREY. J Exp Med. 1964 December 1;120:1151–1168. doi: 10.1084/jem.120.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ardavin CF, Gomariz RP, Barrutia MG, Fonfria J, Zapata AG. The lympho-hemopoietic organs of the anadromous sea lamprey, Petromyzon marinus. A comparative study throughout its life cycle. Acta Zool Stockh. 1984;65:1–15. [Google Scholar]
- 16.Piavis GW, Hiatt JL. Blood cell lineage in the sea lamprey, Petromyzon marinus (Pisces: Petromyzontidae) Copeia. 1971;1971:722–728. [Google Scholar]
- 17.Zapata A, Amemiya CT. Phylogeny of lower vertebrates and their immunological structures. Curr Top Microbiol Immunol. 2000;248:67–107. doi: 10.1007/978-3-642-59674-2_5. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Saito Y, Gotoh H. Vascular architecture and intestinal hematopoietic nests of two cyclostomes, Eptatretus burgeri and ammoncoetes of Entosphenus reissneri: a comparative morphological study. J Morphol. 1981 October;170(1):71–93. doi: 10.1002/jmor.1051700106. [DOI] [PubMed] [Google Scholar]
- 19.Zapata AG, Torroba M, Vicente A, Varas A, Sacedon R, Jimenez E. The relevance of cell microenvironments for the appearance of lympho-haemopoietic tissues in primitive vertebrates. Histol Histopathol. 1995 July;10(3):761–778. [PubMed] [Google Scholar]
- 20.Boffa GA, Fine JM, Drilhon A, Amouch P. Immunoglobulins and transferrin in marine lamprey sera. Nature. 1967 May 13;214(5089):700–702. doi: 10.1038/214700b0. [DOI] [PubMed] [Google Scholar]
- 21.Hagen M, Filosa MF, Youson JH. The immune response in adult sea lamprey (Petromyzon marinus L.): the effect of temperature. Comp Biochem Physiol A. 1985;82(1):207–210. doi: 10.1016/0300-9629(85)90727-3. [DOI] [PubMed] [Google Scholar]
- 22.Pollara B, Litman GW, Finstad J, Howell J, Good RA. The evolution of the immune response. VII. Antibody to human "O" cells and properties of the immunoglobulin in lamprey. J Immunol. 1970 September;105(3):738–745. [PubMed] [Google Scholar]
- 23.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, Diekhoff D, Happe C, Schorpp M, Boehm T. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009 July 10;138(1):186–197. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Oikawa T, Yamada T, Kihara-Negishi F, Sakurai T. Molecular Genetics of Cancer. Trivandrum: Research Signpost; 2005. Ets family of transcription factors in normal hematopoiesis and in leukemogenesis; pp. 119–148. [Google Scholar]
- 25.Remy P, Baltzinger M. The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene. 2000 December 18;19(55):6417–6431. doi: 10.1038/sj.onc.1204044. [DOI] [PubMed] [Google Scholar]
- 26.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007 September;13(3):405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008 January;8(1):9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer WE, O'Huigin C, Tichy H, Terzic J, Saraga-Babic M. Identification of two Ikaros-like transcription factors in lamprey. Scand J Immunol. 2002 February;55(2):162–170. doi: 10.1046/j.1365-3083.2002.01026.x. [DOI] [PubMed] [Google Scholar]
- 29.Hammond KL, Baxendale S, McCauley DW, Ingham PW, Whitfield TT. Expression of patched, prdm1 and engrailed in the lamprey somite reveals conserved responses to Hedgehog signaling. Evol Dev. 2009 January;11(1):27–40. doi: 10.1111/j.1525-142X.2008.00300.x. [DOI] [PubMed] [Google Scholar]
- 30.Uinuk-Ool T, Mayer WE, Sato A, Dongak R, Cooper MD, Klein J. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci U S A. 2002 October 29;99(22):14356–14361. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCauley DW, Bronner-Fraser M. Conservation of Pax gene expression in ectodermal placodes of the lamprey. Gene. 2002 April 3;287(1–2):129–139. doi: 10.1016/s0378-1119(01)00894-0. [DOI] [PubMed] [Google Scholar]
- 32.Uinuk-Ool T, Nikolaidis N, Sato A, Mayer WE, Klein J. Organization, alternative splicing, polymorphism, and phylogenetic position of lamprey CD45 gene. Immunogenetics. 2005 September;57(8):607–617. doi: 10.1007/s00251-005-0019-8. [DOI] [PubMed] [Google Scholar]
- 33.Mayer WE, Uinuk-Ool T, Tichy H, Gartland LA, Klein J, Cooper MD. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc Natl Acad Sci U S A. 2002 October 29;99(22):14350–14355. doi: 10.1073/pnas.212527499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata T, Suzuki T, Ohta Y, Flajnik MF, Kasahara M. The leukocyte common antigen (CD45) of the Pacific hagfish, Eptatretus stoutii: implications for the primordial function of CD45. Immunogenetics. 2002 July;54(4):286–291. doi: 10.1007/s00251-002-0469-1. [DOI] [PubMed] [Google Scholar]
- 35.Ma Q, Jones D, Springer TA. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999 April;10(4):463–471. doi: 10.1016/s1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- 36.Sims-Mourtada JC, Guzman-Rojas L, Rangel R, Nghiem DX, Ullrich SE, Guret C, Cain K, Martinez-Valdez H. In vivo expression of interleukin-8, and regulated on activation, normal, T-cell expressed, and secreted, by human germinal centre B lymphocytes. Immunology. 2003 November;110(3):296–303. doi: 10.1046/j.1365-2567.2003.01745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pancer Z, Mayer WE, Klein J, Cooper MD. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci U S A. 2004 September 7;101(36):13273–13278. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T, Shin I, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005 March 1;174(5):2885–2891. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
- 39.Page M, Rowley AT. A morphological study of pharygeal lymphoid accumulations in larval lampreys. Dev Comp Immunol Suppl. 1982;2:35–40. [Google Scholar]
- 40.Ardavin CF, Zapata A. The pharyngeal lymphoid tissue of lampreys. A morpho-functional equivalent of the vertebrate thymus? Thymus. 1988;11(1):59–65. [PubMed] [Google Scholar]
- 41.Zurbrigg RE, Beamish FWH. Thy-1 immunoreactivity in the larval sea lamprey (Petromyzon marinus L.), a vertebrate without a definitive thymus. Can J Zool. 1995;73:188–197. [Google Scholar]
- 42.Boehm T, Bleul CC. The evolutionary history of lymphoid organs. Nat Immunol. 2007 February;8(2):131–135. doi: 10.1038/ni1435. [DOI] [PubMed] [Google Scholar]
- 43.Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, Macdonald HR, Radtke F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008 October 27;205(11):2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, Zuklys S, Hollander GA, Shima DT, Habu S. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008 October 27;205(11):2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radtke F, Wilson A, Mancini SJ, Macdonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004 March;5(3):247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 46.Snabel V, DeMeews T, Varady M, Nansen P, Bjorn H, Corba J. The sexually linked Mpi locus is presumably involved in imidothiazole resistance in Oesophagostomum dentatum parasites. Parasitol Res. 2000 June;86(6):486–490. doi: 10.1007/s004360050698. [DOI] [PubMed] [Google Scholar]
- 47.Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci U S A. 2005 June 28;102(26):9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagawa F, Kishishita N, Shimizu K, Hirose S, Miyoshi M, Nezu J, Nishimura T, Nishizumi H, Takahashi Y, Hashimoto S, Takeuchi M, Miyajima A, Takemori T, Otsuka AJ, Sakano H. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007 February;8(2):206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 49.d'Alencon E, Petranovic M, Michel B, Noirot P, Aucouturier A, Uzest M, Ehrlich SD. Copy-choice illegitimate DNA recombination revisited. EMBO J. 1994 June 1;13(11):2725–2734. doi: 10.1002/j.1460-2075.1994.tb06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999 June;63(2):349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007 March 1;616(1–2):46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci U S A. 2009 July 7;106(27):11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994 December 4;(6):857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 54.Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, Park BS, Lee H, Yoo OJ, Kasahara M, Lee JO. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007 March 2;282(9):6726–6732. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 55.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008 September 26;321(5897):1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005 July 22;309(5734):581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 57.Istomin AY, Godzik A. Understanding diversity of human innate immunity receptors: analysis of surface features of leucine-rich repeat domains in NLRs and TLRs. BMC Immunol. 2009;10:48. doi: 10.1186/1471-2172-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stumpp MT, Forrer P, Binz HK, Pluckthun A. Designing repeat proteins: modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J Mol Biol. 2003 September 12;332(2):471–487. doi: 10.1016/s0022-2836(03)00897-0. [DOI] [PubMed] [Google Scholar]
- 59.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal Structure of a Shark Single-Domain Antibody V Region in Complex with Lysozyme. Science. 2004 August 19; doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 60.Chan PH, Pardon E, Menzer L, De Genst E, Kumita JR, Christodoulou J, Saerens D, Brans A, Bouillenne F, Archer DB, Robinson CV, Muyldermans S, Matagne A, Redfield C, Wyns L, Dobson CM, Dumoulin M. Engineering a camelid antibody fragment that binds to the active site of human lysozyme and inhibits its conversion into amyloid fibrils. Biochemistry. 2008 October 21;47(42):11041–11054. doi: 10.1021/bi8005797. [DOI] [PubMed] [Google Scholar]
- 61.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, Muyldermans S, Wyns L, Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002 March;11(3):500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- 63.Manicassamy S, Pulendran B. Modulation of adaptive immunity with Toll-like receptors. Semin Immunol. 2009 August;21(4):185–193. doi: 10.1016/j.smim.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gururajan M, Jacob J, Pulendran B. Toll-like receptor expression and responsiveness of distinct murine splenic and mucosal B-cell subsets. PLoS One. 2007;2(9):e863. doi: 10.1371/journal.pone.0000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005 November 17;438(7066):364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 66.Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, Gartland LA, Gartland GL, Boydston JA, Turnbough CL, Jr, Cooper MD. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008 March;9(3):319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]