Abstract
The principal energy generating system in animals is the oxidative phosphorylation (OXPHOS) pathway, which depends on the tight interaction of nuclear and mitochondrial encoded genes to function properly. Mitochondrial genes accumulate substitutions more quickly than nuclear genes, yet the impact of selection on mitochondrial genes is significantly reduced relative to nuclear genes due to the non-recombining nature of the mitochondrial genome and its predicted smaller effective population size. It has therefore been hypothesized that the nuclear encoded genes of the OXPHOS pathway are under strong selective pressure to compensate for the accumulation of deleterious nucleotide substitutions in mitochondrial encoded OXPHOS genes; a process known as compensatory co-adaptation. We evaluated this hypothesis by analyzing nuclear encoded OXPHOS genes for signatures of positive selection as well as evolutionary constraints at amino acid sites. We considered OXPHOS genes of six holometabolous insects and their orthologs from three Nasonia parasitoid wasps; hybrids of which suffer from an increased mortality rate caused by cytonuclear genic incompatibilities. Although nuclear OXPHOS genes are typically highly conserved, we found significant evidence for elevated amino acid divergence in four of the 59 studied nuclear encoded OXPHOS genes. We also found that three of these four genes, as well as six other OXPHOS genes, contain amino acid substitutions between Nasonia species at evolutionarily constrained sites. It is possible that these genes account for the reported incompatibility in Nasonia hybrids and their characterization may lead to a better understanding of the role of positive selection in the genetics of speciation.
Keywords: positive selection, OXPHOS, mitochondria, purifying selection, cytonuclear, compensatory co-adaptation
Introduction
The oxidative phosphorylation pathway (OXPHOS) is the principal energy generating system in animals. Its composition is unique in combining both nuclear and mitochondrial encoded proteins, and the pathway is highly conserved in insects and vertebrates (Porcelli et al., 2007). At the same time there is growing evidence that hybrids between closely related species suffer from a reduction in their OXPHOS pathway efficiency, indicating an accumulation of incompatible substitutions in nuclear and mitochondrial encoded OXPHOS genes between even closely related species (Ellison and Burton, 2006; Ellison et al., 2008; Rand et al., 2004). We set out to assess the hypothesis that positive selection has guided the evolutionary changes found in this otherwise highly conserved pathway.
Mitochondria, in which oxidative phosphorylation occurs, are organelles that are likely derived from a single bacterial endosymbiont ancestor (Gray et al., 1999). While this ancestor is assumed to have contained between 3,000 and 5,000 genes (Boussau et al., 2004), mitochondrial genomes harbor only a fraction of these, typically less than 100 (Ballard and Rand, 2005). The majority of these presumably have been lost because they were no longer required within the eukaryotic environment. The function of others, however, has been maintained through translocation to the nuclear genome or by exaptation of existing nuclear genes for functions within the mitochondria, followed by a deletion of the functionally corresponding mitochondrial genes (Rand et al., 2004, Gross and Bhattacharya, 2009). Over 1,000 nuclear genes have been estimated to encode proteins that are targeted to the mitochondria (Calvo et al., 2006). As a result, mitochondria critically depend in their function on the proper and tight interaction of both nuclear and mitochondrial gene products.
Research on xenomitochondrial cybrid cells (i.e., chimeric cells with a nucleus and mitochondria from two different species) from primate and murid species provided direct evidence for the disruption of mitochondrial functions due to nuclear-cytoplasmic (“cytonuclear”) incompatibility (King and Attardi, 1989; Kenyon and Moraes, 1997; McKenzie et al., 2003). The severity of the disruption was thereby found to positively correlate with the divergence time of the analyzed species. Cytonuclear incompatibility has indeed not only been found in hybrid individuals from crosses between species (i.e., Drosophila, Sackton et al., 2003; Nasonia, Niehuis et al., 2008; Ellison et al., 2008), but also from crosses between diverged populations of the same species (i.e.,Drosophila simulans, Sackton et al., 2003; Tigriopus californicus, Ellison and Burton, 2006, 2008). For this and other reasons, cytonuclear genic incompatibility has been implicated as an important postzygotic barrier to hybridization (Sackton et al., 2003; Ellison and Burton, 2008; Ellison et al., 2008).
The OXPHOS pathway is a prime candidate for cytonuclear genic incompatibilities, given that four of its five complexes (i.e., I, III, IV, and ATPase) are composed of subunits from both the nuclear and mitochondrial genomes (Blier et al., 2001; Rand et al., 2004). In vertebrates as well as insects, all 13 protein coding genes found in the mitochondrial genome are involved in the OXPHOS pathway. This pathway shows reduced ATP generation capacities in interpopulation hybrids of the copepod Tigriopus californicus as well as in interspecific hybrids of Drosophila, and most recently, in those of the parasitoid wasp Nasonia (Sackton et al., 2003; Ellison and Burton, 2006; Ellison et al., 2008). Because ATP is the primary source of energy for cellular processes, these hybrids likely have a reduced fitness due to their deficit of energy. Breeuwer and Werren (1995) found that interspecific hybrid males of Nasonia giraulti and Nasonia vitripennis indeed exhibit significantly increased mortality (i.e., F2 hybrid breakdown) during larval development, a process that has high energetic demands. Niehuis et al. (2008) mapped four loci in the nuclear genome of these hybrids that are incompatible with an allospecific cytoplasm and that account for a major fraction of the observed hybrid mortality. Finally, Ellison et al. (2008) measured OXPHOS enzyme efficiency in Nasonia F2 hybrid males directly and found that they have a significantly reduced efficiency relative to the parental species in each OXPHOS complex except for complex II, which contains only nuclear encoded subunits.
Given that there are far more possibilities for genic incompatibilities to exist between nuclear genes than between nuclear and mitochondrial genes, why do cytonuclear genic incompatibilities seem to play a major role in the early evolution of postzygotic reproductive isolation? One reason could be the fundamentally different way that the nuclear and mitochondrial genomes are processed and transmitted during reproduction. The nuclear genome is, with the notable exception of the sex chromosomes, inherited from both parents with recombination during meiosis, while the mitochondrial genome is usually exclusively maternally inherited and non-recombining (e.g. Hutchinson et al., 1974; Birky, 1995; but see male inheritance in mussels, Zouros et al., 1992). As a consequence, the mitochondrial genome has a theoretically smaller effective population size than the nuclear genome, which significantly reduces the efficacy of natural selection on nucleotide substitutions in the mitochondrial genome (Rand et al., 2004). For this and other reasons (e.g., higher risk of oxidative damage), the mitochondrial genome tends to accumulate nucleotide substitutions at a higher rate than the nuclear genome (Montooth and Rand, 2008). In the genus Nasonia, for example, the substitution rate for mitochondrial genes is, on average, estimated to be approximately 30 times higher than that of nuclear genes (Oliveira et al., 2008).
It has been hypothesized that the accumulation of slightly deleterious nucleotide substitutions in the mitochondrial genome leads to positive selection for compensatory nucleotide substitutions in the nuclear genome to maintain mitochondrial functionality (Rand et al., 2004). If there are nuclear genes of the OXPHOS pathway that compensate for deleterious changes in the mitochondrial encoded OXPHOS genes with which they interact, we would expect to find evidence for positive selection in these nuclear encoded OXPHOS genes. However, to the best of our knowledge there is no study published in which such an analysis has been conducted.
Nasonia is a good model in which to begin the assessment of the role of positive selection in compensating for the accumulation of slightly deleterious nucleotide substitutions in mitochondrial encoded OXPHOS genes for several reasons. First, only the OXPHOS complexes composed of both nuclear and mitochondrial encoded genes show reduced efficiency in (haploid) F2 hybrid males relative to (haploid) males of the parental species (Ellison et al., 2008). Second, we can utilize the newly sequenced genomes of the genus Nasonia (Werren et al., in press) to annotate the nuclear encoded genes of the OXPHOS pathway in N. vitripennis, N. giraulti, and N. longicornis. Third, the incompatibility leading to hybrid breakdown in Nasonia has been mapped to regions of the genome (Niehuis et al., 2008), allowing us to narrow down candidate genes that are potentially involved in the incompatibility.
In comparing the Nasonia nuclear encoded OXPHOS genes with orthologs from other holometabolous insects (Porcelli et al., 2007), we here exploit the evolutionary divergence between these taxa to search for positively selected amino acid sites by analyzing the ratio of nonsynonymous (amino acid replacing) to synonymous (silent) substitutions (ω). Given the high functional constraint on genes that is typically seen in this pathway, we would expect to find high levels of historical purifying selection on the OXPHOS amino acid sequences. In addition, historical compensatory evolution in this pathway is not expected to target all genes, but rather, due to the tight and localized interaction of nuclear and mitochondrial genes, that adaptation has occurred at potentially only a few candidates. Therefore, only a few genes are expected to exhibit such signatures of positive selection, and thus, any signs of adaptation would be interpreted as significant in light of this highly conserved pathway. Further utilizing this divergence, we examine evolutionary constraint on individual amino acid sites in genes of the OXPHOS pathway across the holometabolous insects. We then identify substitutions at these sites within the three Nasonia species that violate this constraint and therefore may impact the protein structure and ultimately the function of the gene. The obtained data enable us to assess the hypothesis that positive selection has guided some of the evolutionary changes in the nuclear genes of the OXPHOS pathway. They also allow us to provide a list of candidate genes that are potentially involved in F2 hybrid breakdown in Nasonia and that will set the stage for future model studies addressing the role of co-adaptation between nuclear and mitochondrial genes for the evolution of intrinsic postzygotic reproductive isolation in this species group.
Methods
Annotation of nuclear encoded OXPHOS genes in Nasonia
For each nuclear encoded gene of the five OXPHOS complexes, we obtained the Drosophila melanogaster protein sequence from the MitoDrome database (D'Elia et al., 2006; http://www2.ba.itb.cnr.it/MitoDrome/). These sequences were used as queries to search the Nasonia genome assembly 1.0 (Werren et al., in press) using the program TBLASTN and applying the SEG filter for low complexity sequences (Altschul et al., 1997). The predicted gene models from the National Center for Biotechnology Information databases (NCBI RefSeq and/or NCBI ab initio) with the highest similarity for a given OXPHOS gene was then manually annotated using the APOLLO genome annotation curation tool (Lewis et al., 2002). Intron-exon boundaries, start and stop sites, and 5′ and 3′ untranslated regions (UTRs) were confirmed or updated based on expressed sequence tags (EST) from the three sequenced Nasonia species (i.e., N. vitripennis, N. giraulti, and N. longicornis; Werren et al., in press). We also considered NCBI-curated EST data from three additional Hymenoptera taxa (i.e., Vespula squamosa, Lysiphelbus testaceipes, and Solenopsis invicta) as well as Swissprot curated protein sequences from the honey bee (Apis mellifera), Drosophila melanogaster, and the red flour beetle (Tribolium castaneum).
The N. vitripennis DNA sequences of annotated OXPHOS genes (Supp I) were used as queries to search the N. giraulti and N. longicornis NCBI trace sequence archives with the program BLASTN, applying an E value cutoff of 10−10 (Altschul et al., 1997). The orthologous DNA sequences from N. giraulti and N. longicornis were manually aligned to the N. vitripennis sequences with the aid of BioEdit (Hall, 1999). In addition, we considered pre-aligned 45-bp long Illumina short-read sequences from N. giraulti and N. longicornis from the Nasonia genome sequencing project (Werren et al., in press). The coordinates of the start and stop codon sites, as well as the intron/exon boundaries of the OXPHOS genes annotated in N. vitripennis were used to trim the corresponding N. giraulti and N. longicornis sequences. All alignments were visually checked for inconsistencies between the trace and the short-read sequences for both N. giraulti and N. longicornis. In cases where short-read and trace sequences were contradictory at a single site, the nucleotide with the majority of support (i.e., present in larger number of short-read/trace sequences) was assigned. If equal support was found for contradictory nucleotides, the nucleotide site was considered ambiguous.
OXPHOS gene sequences from other holometabolous insects
Coding sequences of the nuclear encoded OXPHOS genes from Anopheles gambiae, Aedes aegypti, Drosophila melanogaster, Bombyx mori, Apis mellifera and Tribolium castaneum were obtained from the MitoComp2 database (Porcelli et al., 2007; http://www.mitocomp.uniba.it/). These sequences were translated and aligned to the orthologus amino acid sequences of N. vitripennis, N. giraulti, and N. longicornis with the program CLUSTAL W (Thompson et al., 1994), employing the BLOSUM62 substitution matrix. The alignment of the corresponding coding sequences was deduced from the amino acid sequence alignments. To ensure that all analyses are based on the same amount of sequence information, we excluded gene alignments for which the OXPHOS gene sequence of one or more taxa was missing. If genes had multiple transcripts in a given species, we considered only the transcript with the highest number of conserved nucleotides (to be conservative in subsequent analyses involving tests for positive selection). The exception to this was if multiple transcripts for a gene were found across all studied holometabolous insects, in which case the transcripts were retained in Nasonia.
To ensure comparable data analyses, and to remain conservative in subsequent tests for positive selection, we applied several criteria to remove highly divergent sequences: we manually edited the alignments in BioEdit, removing unalignable N- and C- termini up to the first set of completely conserved amino acid residues. Similarly, if any amino acid residue within the remaining sequence was missing or ambiguous in any species or not reliably aligned across all taxa, that residue was removed from all sequences (unedited alignments are available upon request). Finally the resulting gene alignments were concatenated into a single dataset and the position of each gene was recorded as a code sequence (using a-z, A-P, R-Z, 0 and 1, see Supp. I & II) to facilitate identification of the genes after concatenation. The results reported here are from this concatenation of genes instead of examining them on a simple single gene-by-gene model. This was done because many of these genes are short (e.g., 40% are less than 100 amino acids in length) and thus, parameters such as codon frequencies and neutral substitution rates can have large variances, which can greatly impact the power of a likelihood analysis, leading to erroneous acceptance of the null hypothesis (Anisimova et al., 2001). The concatenation enabled us to estimate these parameters globally to identify patterns in the independent genes and provide us with more statistical power to evaluate the null hypothesis.
Test for positive selection
To assess the impact of positive selection on the evolution of the nuclear encoded OXPHOS genes, we examined the ratio of nonsynonymous (amino acid replacing) substitutions per nonsynonymous site to synonymous (silent) substitutions per synonymous site (ω) with the codon-based maximum-likelihood method (codeml) proposed by Goldman and Yang (1994) and implemented in the software package PAML 4 (Yang, 2007). We performed this analysis on the sequences of the distantly related holometabolous insects to increase the power of the test in order to determine whether historical positive selection has acted on any of the genes in this pathway. We applied the site models M7 and M8, accounting for transitional rate bias (κ estimated) and unequal codon frequencies (F3×4 matrix), and assessed the differences in their likelihood values with the aid of a likelihood ratio test (df = 2). The M7 model assumes a beta distribution of ω values ranging from 0 to 1 (i.e. no positive selection allowed). The M8 model also assumes a beta distribution of ω values, but includes an additional ω class with values > 1 (i.e. positive selection allowed). For both models, we tested a wide range of start parameters for ω (0.01-5.0) and κ (0.5-5.0). Both models require a tree topology, which specifies the phylogenetic relationships between the investigated taxa and which we inferred from the amino acid sequence data in a maximum likelihood framework using the program phyML (Guindon and Gascuel, 2003). We applied three different empirical models of amino acid substitution: JTT (Jones et al., 1992), Dayhoff (Dayhoff et al., 1978), and WAG (Whelan and Goldman, 2001). Substitution rate heterogeneity was approximated using a discrete gamma distribution with four categories. We obtained the same topology for all three substitution models, which proved to be consistent with the phylogeny of the holometabolous insects hypothesized by Savard et al. (2006). The posterior probabilities of individual sites belonging to the class ω > 1 was calculated using the Bayes empirical Bayes approach implemented in PAML (Yang et al., 2005)
Mapping of nuclear-encoded OXPHOS genes
We assessed the possibility that nuclear-encoded OXPHOS genes play a role in cytonuclear genic incompatibility by comparing their position in the Nasonia genome relative to previously identified transmission ratio distorting loci (TRDL; Niehuis et al., 2008). We mapped nuclear encoded OXPHOS genes to chromosomes of Nasonia using positional information from a published high-density linkage map for Nasonia (Niehuis et al., 2009).
Amino acid substitutions in nuclear encoded OXPHOS genes
We used the program MAPP (Stone and Sidow, 2005) to assess the potential negative impact of amino acid replacements in the nuclear encoded genes of the OXPHOS pathway in Nasonia. The program calculates the physicochemical constraints at each amino acid position based on the observed distribution of amino acids across distantly related taxa and then generates an impact score and an associated p value for every potential amino acid substitution. The impact scores are based on the prior distribution (found across many evolutionarily diverged taxa) of physicochemical properties at each amino acid site (i.e. evolutionary constraint), with higher impact scores resulting from greater violations of this constraint (Stone and Sidow, 2005). We again utilized the evolutionary divergence of the holometabolous insects by using MAPP to calculate the evolutionary constraint at each studied amino acid site in the OXPHOS pathway. The substitutions found within Nasonia were then compared with the calculated impact score for each amino acid.
Results
OXPHOS Gene Annotation in Nasonia
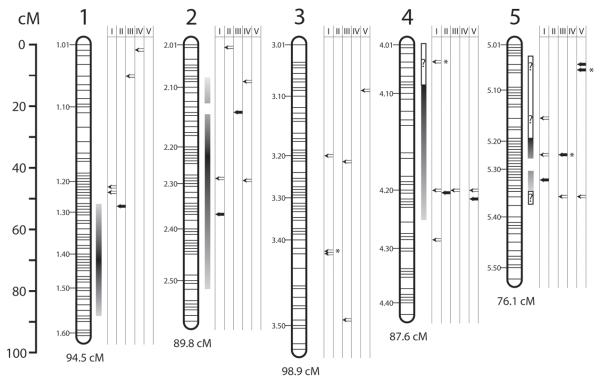
We identified 59 of the 65 OXPHOS pathway subunit genes known from Drosophila (MitoDrome database; D'Elia et al., 2006) in the Nasonia genome (Table 1; Supp. I). It is possible that the remaining genes were not identified because our conservative search strategy parameters did not allow us to reliably identify orthologs that have rapidly diverged. For 9 of the 59 genes (i.e., two of complex I, two of complex III, and five of complex IV), EST data suggested alternate transcripts (Table 1; Supp. I, Werren et al., in press). Utilizing the high-density linkage map for Nasonia (Niehuis et al., 2009), we were able to map 56 of the 59 identified nuclear-encoded OXPHOS genes in the Nasonia genome (Fig. 1, Supp. I).
Table 1.
Annotated nuclear OXPHOS genes across different complexes.
| Number of Genes | |||
|---|---|---|---|
| OXPHOS Complex |
Nasonia 1 | Drosophila | Alternate transcripts2 |
| I | 27 | 31 | 2 (2) |
| II | 4 | 4 | 0 |
| III | 9 | 9 | 0 |
| IV | 6 | 8 | 2 (2) |
| V | 13 | 13 | 4 (2), 1 (4) |
| Total | 59 | 65 | 8 (2), 1 (4) |
Number of Nasonia genes identified is the result of comparisons with Drosophila OXPHOS gene alignments (see Methods).
Number of genes identified from EST data with alternate transcripts in Nasonia, with the number of alternate transcripts shown in parentheses.
Figure 1. Co-localization of OXPHOS genes and cytonuclear incompatibility loci within Nasonia.
Nasonia linkage map adapted from Niehuis et al. (2009). The leftmost scale shows the genetic distance in centimorgans. The horizontal bars in each chromosome (1.01-5.51) represent groups of markers showing no recombination between them, while the space between bars represents recombination. Dark gradient bars to the right of the chromosomes represent incompatibility loci identified by Niehuis et al. (2008); question marks designate locus boundaries that are unclear due to recent expansions of the ends of the linkage groups on the linkage map. The columns to the right of the chromosomes designate the complex (I-V) of each OXPHOS gene.
Empty arrows = genes with amino acid substitutions with non-significant impact scores.
Filled arrows = genes with amino acid substitutions predicted to significantly disrupt protein structure in Nasonia.
stars = genes containing sites that show the signature of positive selection (ω > 1).
Test for Positive Selection
After removing OXPHOS genes for which we were missing sequences in one or more taxa, the concatenated sequence alignment included 53 of the 59 nuclear encoded OXPHOS genes identified in Nasonia, composed of a total of 8,739 codons (Supp. II). The likelihood ratio test indicated a significantly better fit of the M8 sites model, which accounts for positive selection, than the M7 model to the holometabolous insect dataset (−2 log Δ = 8.491, df = 2, p = 0.011). Bayes empirical Bayes revealed four genes (Table 2, Fig 1, Supp. III) containing sites with posterior probability values >80%, corresponding with ω values >1 when accounting for the standard error.
Table 2.
Nuclear OXPHOS genes showing patterns of elevated divergence and amino acid substitutions with potentially disruptive effects on protein structure.
| Gene Name | Complex | Position | P.P. (ω +/− SE)1 |
Amino acid replacement impact (N. vit/ N. gir) 2 |
Residue3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ng | Nl | Nv | Am | Tc | Bm | Ag | Aa | Dm | |||||
| 39 KDA SUBUNIT | 1 | 316 | 0.913 (1.424 +/− 0.252) |
n.s. | P | P | A | I | H | H | Y | L | T |
| 51 KDA SUBUNIT | 1 | 415 | 0.837 (1.355 +/− 0.337) |
n.s. | N | N | S | A | E | R | S | A | K |
| CORE PROTEIN 2 | 3 | 305 | 0.838 (1.356 +/− 0.334) |
n.s. | L | L | L | L | V | A | V | L | V |
| 51 | n.s. | 0.005 / < 0.001 | R | R | H | C | N | N | T | T | N | ||
| 277 | n.s. | 0.010 / 0.680 | Q | Q | E | F | K | V | K | Q | K | ||
| D CHAIN | 5 | 123 | 0.879 (1.390 +/− 0.302) |
n.s. | K | K | R | S | R | Y | K | R | R |
| 89 | n.s. | 0.460 / 0.022 | V | A | A | K | E | A | D | E | E | ||
| 102 | n.s. | 0.455 / 0.029 | T | A | A | A | S | A | S | A | Q | ||
| 30 KDA SUBUNIT | 1 | 166 | n.s. | 0.949 / 0.003 | T | T | A | A | A | A | A | A | A |
| B12 SUBUNIT | 1 | 5 | n.s. | < 0.001 / <0.001 | E | E | K | P | P | P | P | P | P |
| 15 | n.s. | 0.902 / < 0.001 | E | E | A | A | A | A | A | A | G | ||
| CYT B SMALL SUBUNIT | 2 | 47 | n.s. | 0.040 / 0.978 | V | V | L | T | V | V | T | T | V |
| CYT B560 SUBUNIT | 2 | 75 | n.s. | 0.002 / 0.949 | A | A | V | A | A | A | A | A | A |
| 104 | n.s. | < 0.001 / <0.001 | R | R | K | S | A | S | T | S | S | ||
| SULFUR SUBUNIT | 3 | 11 | n.s. | 0.008 / 0.052 | G | G | N | S | S | S | S | S | S |
| 181 | n.s. | 0.109 / 0.022 | V | V | A | P | Y | P | H | H | T | ||
| 188 | n.s. | 0.119 / 0.022 | V | V | I | T | T | L | L | L | L | ||
| E CHAIN | 5 | 2 | n.s. | 0.009 / 0.999 | V | V | I | V | V | V | V | V | V |
| F CHAIN | 5 | 24 | n.s. | < 0.001 / <0.001 | K | K | R | P | A | P | A | P | A |
| 51 | n.s. | 0.949 / 0.003 | T | A | A | A | A | A | A | A | A | ||
| 86 | n.s. | 0.025 / 0.999 | F | F | L | F | F | F | F | F | F | ||
Positions are amino acid sites within each gene of the concatenated dataset (Supp. II).
Posterior probabilities (P.P.) of sites predicted to be subject to positive selection and ω values (both were calculated using the PAML program, Yang, 2007).
Significance values for the impact scores of amino acid substitutions were calculated for each Nasonia species using the program MAPP (Stone and Sidow, 2005). Shaded areas indicate genes suggested by PAML analyses to have putative positively selected sites and disruptive amino acid substitutions.
The residues present at each position for each taxon are abbreviated: Ng = Nasonia giraulti; Nl = Nasonia longicornis; Nv = Nasonia vitripennis; Am = Apis mellifera; Tc = Tribolium castaneum; Bm = Bombyx mori; Ag = Anopheles gambiae; Aa = Aedes aegypti; Dm = Drosophila melanogaster.
When we examined patterns of divergence on a gene-by-gene basis in comparison with the concatenated dataset (using models M7 and M8, results not shown), only one of the four genes previously identified (51 KDA subunit) still showed a significant likelihood ratio test. However, as previously discussed, this is expected given that the power of the likelihood ratio test is greatly decreased when using shorter sequences (Anisimova et al., 2001). Similarly, we also evaluated the effect of divergence time on our results. For this purpose we performed an analysis of ω for each branch of the tree (using the branch model (model=1) in PAML, Supp. IV), which indicated that variation in dS associated with recent vs. deeper divergence times does not explain the significant estimates of dN/dS in our dataset.
Analyses of Amino Acid Substitution Patterns
Of the 56 mapped OXPHOS subunit genes, we found that within Nasonia, 36 genes contain amino acid substitutions. Of these 36 genes, 22 were located within or near the estimated position of previously identified TRDL (Fig. 1; Niehuis et al., 2008). Since the four TRDL had been identified in hybrids of N. giraulti and N. vitripennis, we subsequently restricted our analysis to amino acid substitutions between these two species. This resulted in the examination of 53 amino acid substitutions found in 19 genes. The MAPP analysis assigned a statistically significant impact score to 17 of these amino acid substitutions across 9 of the 19 genes located near TRDL (Table 2, Fig 1, Supp. V).
Discussion
Dobzhansky (1937) and Muller (1942) envisioned the evolution of intrinsic postzygotic reproductive isolation as a consequence of the incompatibility of interacting genes that have diverged between taxa (e.g., Brideau et al., 2006). These interactions are not limited to nuclear genes, but can include mitochondrial genes as well. A prime candidate for the evolution of genic incompatibility between nuclear and mitochondrial genes is the OXPHOS pathway, whose enzyme complexes are composed of both nuclear and mitochondrial encoded gene products. While the pathway itself is highly conserved across taxa in respect to both the number and the function of involved genes, the accumulation of slightly deleterious nucleotide substitutions in the non-recombining mitochondrial genome seems to quickly lead to an array of divergent mitochondrial OXPHOS gene variants (e.g. Oliveira et al., 2008). One possible explanation for the maintenance of these variants, and thus overall OXPHOS functionality, is compensatory selection in nuclear encoded OXPHOS genes, which may also result in the rapid divergence of these particular genes. As a result, genic incompatibilities are expected to quickly develop between nuclear and mitochondrial genes that evolve independently from each other in individuals from different populations and species. Cytonuclear genic incompatibility thus may play a significant role in incipient speciation. It is this aspect that makes the OXPHOS pathway a particularly attractive model to study the co-evolutionary dynamics of nuclear and mitochondrial encoded genes.
Our analysis of the substitution pattern in 53 nuclear encoded OXPHOS genes across nine holometabolous insect species finds that an evolutionary model that accounts for positive selection fits the substitution pattern in the nuclear genes of the OXPHOS pathway significantly better than one that does not. It should be noted that our gene sample is not a random one with respect to the genome in general, in which case we may expect that a large sampling would include a certain proportion reflecting different constraints, i.e., classes of genes each evolving under neutral, positive, and purifying selection. Instead, our sample of genes here is selected with respect to a specific hypothesis regarding selective constraints on OXPHOS genes, which have been shown to be highly conserved (Porcelli et al., 2007). This premise, together with our likelihood modeling, implies that the patterns of elevated divergence are not well explained by simply neutrality or sampling variance (i.e., “chance”), but may represent potential candidates for positively selected genes.
Given this rationale, the observation of even a few genes exhibiting significantly elevated divergence would be surprising without an a priori hypothesis providing an explanation for diverged genes in a conserved pathway. It is possible that our data are consistent with the hypothesis that nuclear genes of the OXPHOS pathway compensate for the accumulation of slightly deleterious nucleotide substitutions in their mitochondrial counterparts. While this is one potential explanation, it is clearly not the only one that may explain elevated divergence and patterns of positive selection, especially since many of the genes still show strong patterns of purifying selection. Thus, it is clear that further studies are needed to distinguish among these competing alternative hypotheses.
It is also possible that the ω value of 2.44 for sites estimated by PAML to be under positive selection (site class −11; Supp. III) could be considered as a low estimate. First, we are missing six OXPHOS pathway genes in our analysis that are known from Drosophila. While we cannot exclude the possibility that these genes are not present in Nasonia (or in the Nasonia genome assembly), it is possible that we did not recognize them because they are sufficiently divergent from their Drosophila orthologs. We also excluded six genes for which we were missing orthologous sequences in one or more taxa of the dataset other than Nasonia. These genes could likewise be absent from those datasets because they evolve rapidly. Finally, we discarded sequence sections from the alignment that could not be reliably aligned across all taxa, because they seemingly have diverged sufficiently to preclude meaningful analysis. Overall, these measures resulted in a relatively conservative analysis that removed genes that may be undergoing rapid sequence evolution and may be erroneously interpreted as positive selection.
Our analysis resulted in the identification of four candidate genes that show patterns of significantly elevated divergence across holometabolous insects (Fig. 1, Table 2, Supp. V). In addition to the conservative data-trimming measures outlined above, we also expect that this set of genes is further reduced in number given that the estimates of ω are averaged across many lineages because our analysis involved multiple taxa that diverged, in some cases, > 300 million years ago (Krauss et al., 2008). We would not expect the same sites to be selected in all lineages, and thus, the number of sites suggested here by PAML to be under positive selection could conservatively also be considered as a low estimate. It should be noted that these results indicate that positive selection may potentially act on the nuclear OXPHOS genes, but the current methods are not able to determine which lineages are responsible for these elevated divergence estimates. As a result, this analysis provides an initial view of the gene and protein subunit regions that reflect long-term evolutionary changes in the OXPHOS pathway among holometabolous insects.
Due to the breadth of the previous analysis, our MAPP results are likely more informative for comparisons within Nasonia, which indicate amino acid changes that are predicted to have a significant impact on the encoded protein. We found that 53 of the amino acid substitutions in our comparison between N. giraulti and N. vitripennis took place in OXPHOS genes that are within the confidence intervals that Niehuis et al. (2008) provided for the position of incompatibility loci in the nuclear genome of Nasonia (Fig. 1, Table 2, Supp. V). Because these intervals do reflect a large proportion of the overall genomic content, the fact that many of our genes reside within or near these TRDL reflects only a preliminary inspection of their actual link to hybrid incompatibility. Nonetheless, the combination of the present data with those published by Niehuis et al. (2008) does narrow the list of candidate genes for additional hypothesis testing. For example, although N. giraulti and N. vitripennis have split very recently, with an estimated divergence time of approximately one million years (Campbell et al., 1993), they have already accumulated substantial protein changes in the otherwise highly conserved nuclear OXPHOS genes that could potentially result in hybrid cytonuclear incompatibilities.
This substitution pattern is even more impressive given that despite our conservative approach, the MAPP analysis predicts that over 30% of the 53 amino acid substitutions located at TRDL may impact protein structures. The possibility remains that this substitution pattern could be the result of relaxed purifying selection due to reduced effective population sizes, especially for the mitochondrial genome. This could lead to deleterious substitutions in both nuclear and mitochondrial genes that could produce the observed hybrid breakdown in Nasonia analogous to synthetic lethals (Hartman, 2001). However, a reduced effective population size and reduced selection efficacy would be expected to impact all genes similarly (with some stochastic variance aside). Thus, it is not readily obvious why only certain genes would be subjected to relaxed selection while others show strong purifying selection. Nonetheless, further studies that examine within population diversity and recent evolutionary changes in selective constraints at these genes may address this question in distinguishing between competing selection models.
Based on the present results, it appears possible that some of the genes and possibly even single amino acid residues identified here are involved in the observed F2 hybrid breakdown in Nasonia. We speculate that these loci may reflect “speciation gene” or even “speciation site” candidates. The Nasonia model system will allow us to test the phenotypic effects of these amino acid substitutions in more detail through future studies. This is one of the more attractive aspects of our study on Nasonia; that is, identifying genes and specific sites with statistically elevated divergence and potential signatures of positive selection is much more compelling when their functional effects can be further validated in vivo. This combination of statistical and functional approaches is likely to provide new insights into the intimate interactions of mitochondrial and nuclear gene products that form the genetic basis of intrinsic postzygotic reproductive isolation in Nasonia and other species. This may also lead to a better understanding of the functioning of the OXPHOS system and the genetics of speciation.
Supplementary Material
Acknowledgements
We thank Stephen C. Pratt for computational assistance and two anonymous reviewers for valuable feedback that helped to improve the manuscript considerably. This work was supported by NIH 1R21RR024199-01 to J. Gadau.
Footnotes
Supplementary information is available at Heredity's website.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and Power of the Likelihood Ratio Test in Detecting Adaptive Molecular Evolution. Mol. Biol. Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst. 2005;36:621–642. [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mecahnisms and evolution. Proc Natl Acad Sci. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier PU, Dufresne F, Burton RS. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 2001;17:400–406. doi: 10.1016/s0168-9525(01)02338-1. [DOI] [PubMed] [Google Scholar]
- Boussau B, Karlberg EO, Frank AC, Legault B, Andersson SGE. Computational inference of scenarios for α-proteobacterial genome evolution. Proc Natl Acad Sci USA. 2004;101:9722–9727. doi: 10.1073/pnas.0400975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer JAJ, Werren JH. Hybrid breakdown between two haplodiploid species: The role of nuclear and cytoplasmic genes. Evolution. 1995;49:705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Campbell BC, Steffen-Campbell JD, Werren JH. Phylogeny of the Nasonia species complex (Hymenoptera: Pteromalidae) inferred from an rDNA internal transcribed spacer (ITS2) Insect Mol Biol. 1993;2:225–237. doi: 10.1111/j.1365-2583.1994.tb00142.x. [DOI] [PubMed] [Google Scholar]
- Dayhoff M, Schwartz R, Orcutt B. A model of evolutionary change in proteins. In: Dayhoff M, editor. Atlas of protein sequence and structure. Vol. 5. National Biomedical Research Foundation; Washington, D.C.: 1978. pp. 345–352. [Google Scholar]
- D'Elia D, Catalano D, Licciulli F, Turi A, Tripoli G, Porcelli D, et al. The MitoDrome database annotates and compares the OXPHOS nuclear genes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Mitochondrion. 2006;6:252–257. doi: 10.1016/j.mito.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press; 1937. [Google Scholar]
- Ellison CK, Burton RS. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution. 2006;60:1382–1391. [PubMed] [Google Scholar]
- Ellison CK, Burton RS. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution. 2008;62:631–638. doi: 10.1111/j.1558-5646.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Niehuis O, Gadau J. Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J Evol Biol. 2008;21:1844–1851. doi: 10.1111/j.1420-9101.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial Evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D. Mitochondrial and plastid evolution in eukaryotes: an outsiders' perspective. Nature Rev Genet. 2009;10:495–505. doi: 10.1038/nrg2610. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hartman JL, Garvik B, Hartwell L. Principles for the Buffereing of Genetic Variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Hutchinson CA, III, Newbold JE, Potter SS, Edgell MH. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974;251:536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- Jones D, Taylor W, Thornton J. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8::275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by establishment of primate xenomitochondrial cybrids. Proc Natl Acad Sci USA. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Krauss V, Thümmler C, Georgi F, Lehmann J, Stadler PF, Eisenhardt C. Near intron positions are reliable phylgenetic markers: An application to Holometablous insects. Mol Biol Evol. 2008;25:821–830. doi: 10.1093/molbev/msn013. [DOI] [PubMed] [Google Scholar]
- Lewis SE, Searle SMJ, Harris N, Gibson M, Iyer V, Ricter J, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0082. research0082.1–0082.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M, Chiotis M, Pinkert CA, Trounce IA. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol Biol Evol. 2003;20:1117–1124. doi: 10.1093/molbev/msg132. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Rand DM. The spectrum of mitochondrial mutation differs across species. PLoS Biol. 2008;6:e213. doi: 10.1371/journal.pbio.0060213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, speciation, and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–423. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Gibson JD, Rosenberg MS, Pannebakker BA, Koevoets T, Judson AK, et al. Recombination and its impact on the genome of the haplodiploid parasitoid wasp Nasonia. PLoS ONE. 2009 doi: 10.1371/journal.pone.0008597. in press. This paper is sheduled to be published when the Nasonia genome paper is published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae) Mol Biol Evol. 2008;25:2167–2180. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli D, Barsanti P, Pesole G, Caggese C. The nuclear OXPHOS genes in Insecta: a common evolutionary origin, a common cis-regulatory motif, a common destiny for gene duplicates. BMC Evol Biol. 2007;7:215. doi: 10.1186/1471-2148-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Haney RA, Rand DM. Cytonuclear co-adaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003;57:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Pysicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2009 doi: 10.1126/science.1178028. (In Press). This is the Nasonia genome sequencing paper. If accepted, we request that publication of this manuscript be delayed until publication of the genome paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zouros E, Freeman KR, Ball AO, Pogson GH. Direct evidence for extensive paternal mitochondrial DNA inheritance in the marine mussel Mytilus. Nature. 1992;359:412–141. doi: 10.1038/359412a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.