Conspectus
Bright, photostable luminescent labels are powerful tools for the imaging of biological events in vitro and in vivo. Semiconductor nanocrystals have emerged as attractive alternatives to commonly used organic lumophores due to their high quantum yields and the spectral tunability that can be achieved through synthetic control. While conventional synthetic methods generally yield high-quality nanocrystals with excellent properties for biological imaging, ligand exchange and biological conjugation are necessary to make nanocrystals biocompatible and biospecific. These steps can result in substantial deterioration of optical characteristic of these nanocrystals. Moreover, the complexity of multistep nanocrystal synthesis, typically requiring inert and anhydrous conditions, prohibits many end users of these lumiphores from generating their own custom materials. We sought to streamline semiconductor nanocrystal synthesis and develop synthetic routes that would be accessible to scientists from all disciplines. In search of such an approach we turned to nucleic acids as a programmable and versatile ligand set, and found that these biomolecules are indeed appropriate for biocompatible semiconductor nanocrystals preparation. In this account we present a summary of our work on nucleic acids-programmed nanocrystal synthesis that has resulted in the successful development of a one-step synthesis of biofunctionalized nanocrystals in aqueous solution.
We first discuss results obtained with nucleotide-capped cadmium and lead chalcogenide-based nanocrystals that served to guide further investigation of polynucleotide-assisted synthesis. We investigate the roles of individual nucleobases and their structures in passivation of the surfaces of nanocrystals and modulating morphology and optical characteristics. We show that nanocrystals’ optical properties and morphologies are highly influenced by nucleic acid structures and sequences, as well as by reaction conditions. Moreover, studies using live cells reveal low toxicity and rapid uptake of DNA-passivated CdS nanocrystals, demonstrating their suitability for bioimaging.
Finally, we describe a new approach that leads to the production of biofunctionalized, DNA-capped nanocrystals in a single step. Chimeric DNA molecules are the enablers of this strategy, providing both a domain for nanocrystals passivation and a domain for biomolecule recognition. Nanocrystals synthesized using this approach possess good spectral characteristics as well as high specificity to cognate DNA, protein, and cancer cell targets. The development of this approach may make nanocrystal lumiphores more readily accessible to those working in the biological sciences.
Introduction
Nanoscale semiconductor crystals exhibit size- and shape-dependent physicochemical properties that are distinct from those of the corresponding bulk solids.1 These unique properties, arising from size-dependent bandgaps, discrete band structures, and confinement of charge carriers enable the diverse applications proposed for semiconductor nanocrystals in the area of physics,2 biology,3, 4 and medicine.5, 6 The application of semiconductor nanocrystals as fluorescent labels for thein situ investigation of cellular processes in life sciences and medicine has in particular received much attention, since nanocrystals have a variety of advantages over organic chromophores. Indeed, emission tunability, brightness, and superior photostabilily have made nanocrystals a promising alternative to conventionally used organic fluorophores for in vitro bioimaging.7
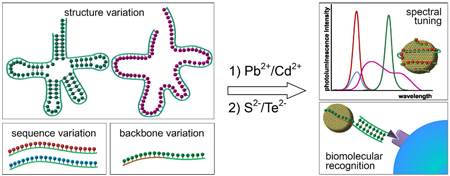
Great strides have been made in the preparation of monodisperse nanocrystals with controlled spectral properties.1, 8 Most preparation methods involve synthesis in organic solvents and post-synthetic modification to render nanocrystals biocompatible and biospecific.9, 10 We sought a “greener”11, simpler, safer, and efficient alternative to the organic routes for biocompatible nanocrystals synthesis, and in this search, turned to biological systems for inspiration. We designed a new approach that would employ a biological template – a nucleic acid molecule - to serve as a modulator of nanocrystals synthesis (Scheme 1).
SCHEME 1. Schematic Representation of Nucleic Acids-Templated Nanocrystals Synthesis.
The role of nucleic acids in living organisms is to store and transfer genetic information – a function seemingly unrelated to semiconductor nanocrystals synthesis. However, a closer examination of the nucleic acids reveals that they have many characteristics advantageous for nanocrystals synthesis: (1) The anionic oxygen of phosphate, the hydroxyl groups of sugar moieties, and nitrogen and oxygen atoms of nucleobases can interact with metal ions that are precursors for nanocrystals, e.g. Cd2+, Pb2+, and Hg2+ 12–14. (2) The three-dimensional structures of naturally occurring and artificial nucleic acids are well defined, and the sizes of many of these biological nanostructures are comparable to that of conventional QDs, which could serve to confine the nanocrystals’ growth and control size. (3) Efficient chemical and biological techniques are currently available for the polynucleotides synthesis15, 16 and can be used to access a wide variety of nucleic acids with desired functionalities. (4) Nucleic acids are water soluble, and if they could be used as ligands, could produce water-soluble nanocrystals that would not need ligand exchange to be compatible with cellular systems. This combination of properties indicated to us that nucleic acids could be effective and functional ligands for nanocrystals.
Nucleic acid monomers as semiconductor nanocrystals ligands
The large amount of data on the synthesis of nanoparticles accumulated in recent years has allowed the attributes of ligands capable of facilitating nanocrystal growth to be defined. Effective ligands for nanoparticle growth must have qualities that allow them to: (i) form a precursor complex with metal ions to control the ion release and nanoparticle growth rate as well as to facilitate heterogeneous nucleation; (ii) terminate the nanoparticle growth and prevent agglomeration once most precursors in the solution are consumed; (iii) passivate nanocrystal surface defects to promote maximal luminescence; and (iv) render nanoparticles soluble in the solvent the synthesis is conducted in.17 To determine whether nucleic acids can indeed serve as ligands for semiconductor NP synthesis and to identify the DNA functional groups essential for this role, we initiated our work in this area with the four natural mononucleotides ATP, GTP, UTP, and CTP, and used these molecules as the sole ligand in the synthesis of PbS and CdS.
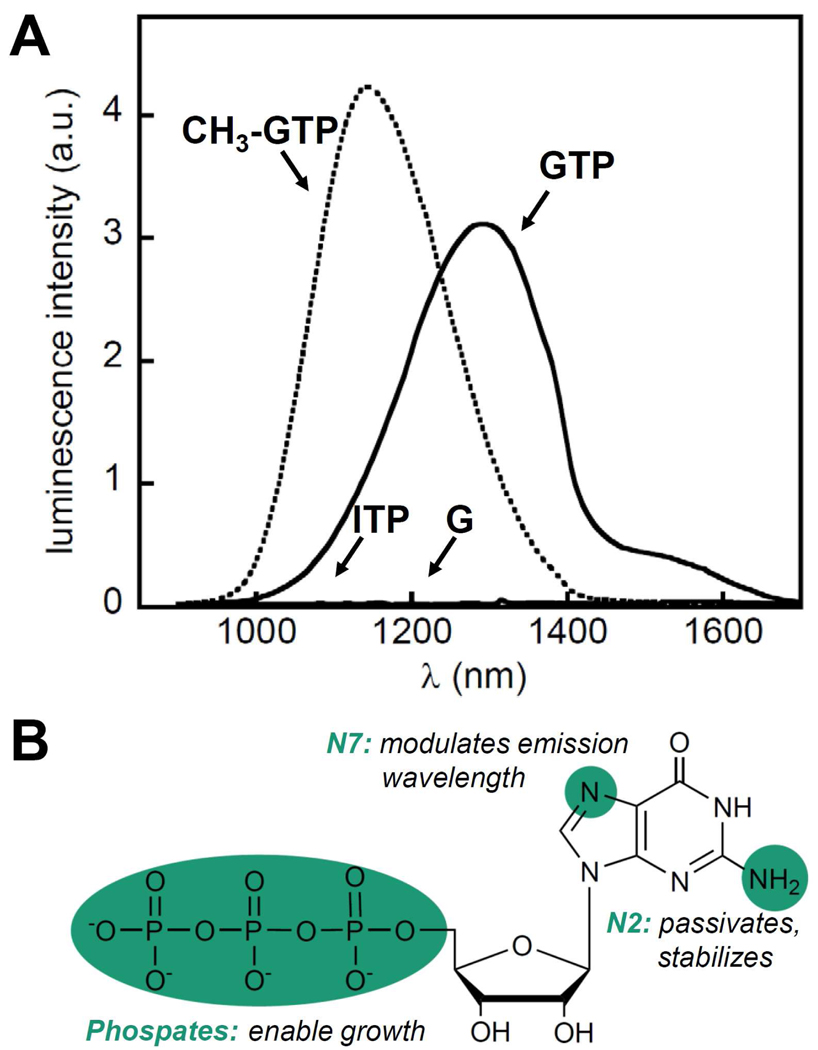
When PbS was synthesized using mononucleotide ligands in aqueous solution, only GTP was found to generate luminescent nanocrystals (Figure 1).18 ATP, CTP, and UTP produced mainly non-soluble bulk materials with little luminescence. Therefore, functional groups unique to the base moiety of GTP appear to play an important role in nanocrystals formation. Previous studies suggest that either the exocyclic N2 or endocyclic N7 of GTP can interact with Pb2+ ions.19
FIGURE 1. Effect of specific chemical functionalities present on GTP on PbS nanocrystals synthesis.
(A) Luminescence spectra obtained when GTP, G, ITP, and 7—CH3-GTP were used for PbS synthesis. (B) Proposed roles of phosphate and base functionalities on GTP in nanoparticle nucleation, growth, termination, stabilization, and passivation.18
To test the role of these moieties, the GTP analogues 7-methyl GTP and inosine triphosphate (ITP, lacks N2) were investigated. When ITP was used instead of GTP, insoluble material was produced instead of nanocrystals. In contrast, methylation of the N7 resulted only in blue shift of the nanocrystals emission maxima relative to GTP (Figure 1), indicating that growth was slowed and smaller particles were formed. FTIR measurements suggested that the exocyclic N2 interacts directly with the semiconductor surface, therefore serving an essential role in nanocrystals surface passivation. Interaction with the N7 was not observed, indicating that this functional group is involved with the growth process but does not serve as a ligand.
The phosphate groups displayed on nucleotides were also shown to be essential for PbS nanocrystals formation. Non-soluble products were generated when G, the nucleoside lacking any phosphate, was used instead of GTP in nanocrystals synthesis, most likely because the negatively charged phosphate groups are necessary to prevent nanocrystal aggregation and to make nanocrystals soluble in water (Figure 1). Moreover, FTIR measurements indicate that the phosphate groups bind to Pb2+ ions initially; however, after the introduction of the S2− source, the exocyclic N2 becomes the dominant binding site.18 Thus, the phosphate groups are important in the early stages of nanocrystals synthesis given that they serve to control the reaction by sequestering lead and feeding it into the reaction mixture with equilibrium control.
When nucleotide-mediated CdS nanocrystals formation was investigated, similar trends with some significant variations were observed.20 The most luminescent CdS nanoparticles were again obtained in the presence of GTP. Materials synthesized with ATP, UTP, and CTP possessed luminescence at least an order of magnitude less than those produced by GTP. Interestingly, unlike in the case of PbS nanocrystals, using 7-methyl GTP had a more substantial effect on CdS nanocrystals properties than the ITP analog. The luminescence intensity of 7-methyl-G-templated nanocrystals was reduced by about an order of magnitude compared to GTP, which suggests that the endocyclic N7 plays a more important role in CdS nanocrystals synthesis than in the case of PbS. However, just as was observed for PbS, the use of GDP, GMP, and G in place of GTP yielded CdS materials with significantly reduced luminescence and solubility. This indicates that the negatively charged triphosphate groups of mononucleotides are critical for CdS synthesis just as they were for PbS. However, for CdS the phosphate groups were found to participate directly in passivation, presumably together with the amino group of the base moiety.21, 22
In general, the studies performed with mononucleotide ligands indicate that the passivation of nanocrystals is dominated by the base moieties, while the phosphate groups prevent their aggregation by maintaining electrostatic repulsion between nanocrystals.
Polymeric nucleic acids as nanocrystal ligands: effects of sequence and structure
Given the observations noted above linking the identities of nucleic acids monomers with nanocrystal properties, it can be envisioned that the virtually infinite number of sequence combinations in a DNA or an RNA strand could make these molecules highly tunable and versatile ligands for nanocrystals synthesis. Sequence determines the functionalities on each polynucleotide strand available for nanocrystals passivation, as well as the number of passivation sites and the spacing between them. Moreover, the secondary and tertiary structure of nucleic acids could affect the nanoparticle growth kinetics by interacting with the nanoparticle surface at different growth stages, leading to the formation of nanoparticles with different sizes or/and shapes.23 This additional degree of freedom could be exploited to produce a greater diversity of nanocrystal products with unique properties.
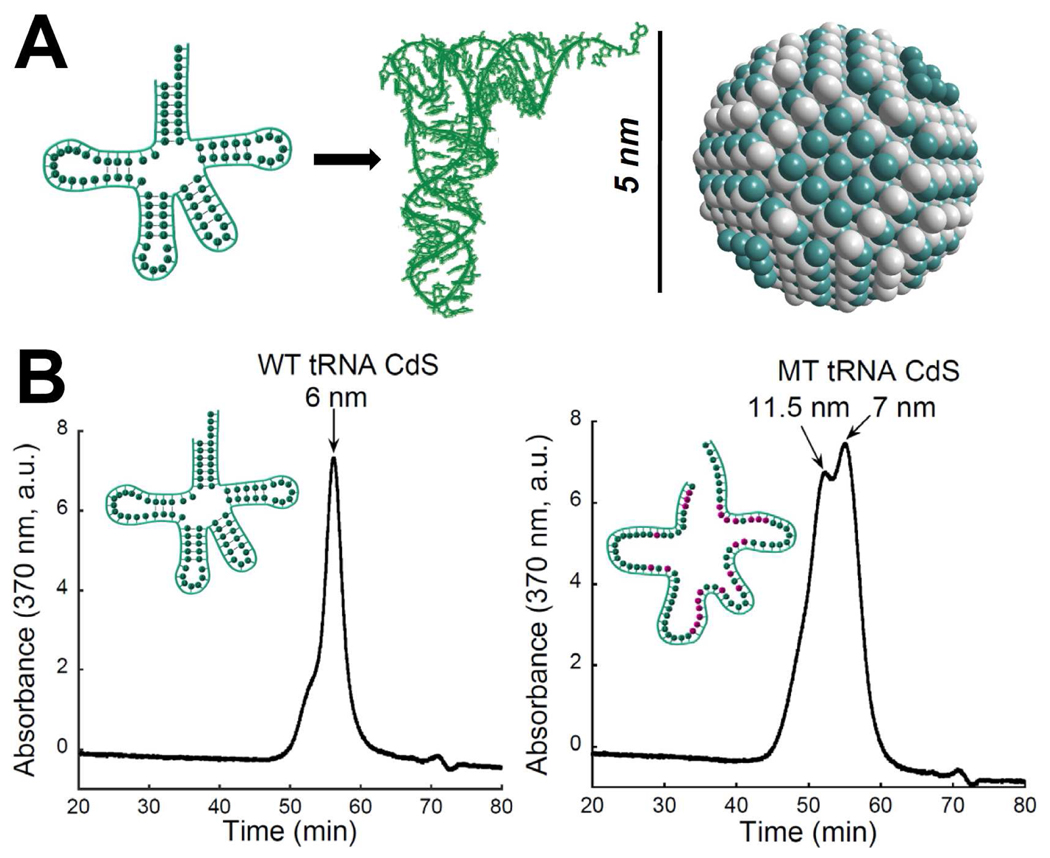
To directly test these ideas and study how nanocrystals were affected by nucleic acids structure, nanocrystal size, morphology, and optical properties were monitored when a biologically active transfer RNA (E. coli tRNALeu) was used as a ligand.24 The wild type (WT) tRNA is a highly structured RNA molecule with a well-defined cloverleaf secondary structure and L-shape tertiary structure.25 The overall size of a tRNA molecule is about 5 nm which is comparable to the size of typical nanocrystals (Figure 2).
FIGURE 2. Transfer RNA as a nanocrystal template and ligand.
(A) Two-dimensional and three-dimensional WT tRNA structures and the same scale view of a 5.4 nm CdS spherical nanocrystal along [1,1,1] axis. (B) Gel filtration chromatography of WT and MT tRNA-CdS complexes.24
To vary the RNA structure and track its influence on nanocrystals formation, we generated an unstructured mutant (MT) tRNA by introducing several structure-perturbing mutations to disrupt the base pairing within all five stems of WT tRNA. Interestingly, the structural properties of CdS nanocrystals made with structured and unstructured RNA were quite distinct. Nanocrystals made with unstructured RNA were significantly larger and more polydisperse than those made with structured RNA. (Figure 2). TEM analysis revealed that WT tRNA produced monodisperse nanocrystals with spherical nanoparticles possessing 4.4 ± 0.4 nm diameters while MT tRNA yielded larger and more irregular nanoparticles with diameters up to 7 nm. Gel filtration chromatography, which reported on the hydrodynamic diameters of the materials, supported this trend (Figure 2). Clearly, the structure of nucleic acids has a strong influence on CdS nanocrystals morphology. Since the unstructured MT RNA molecule is much more flexible compared to WT tRNA, it is likely to adopt various conformations when binding to Cd2+ and CdS nanocrystals’ surface during the initiation and growth stages, which could result in coexistence of several growth regimes and, as a result, nanocrystal sizes.
With clear evidence that nucleic acids structure could modulate the properties of semiconductor nanocrystals, we desired to understand the roles of the sequences of extended unstructured oligomers. Four DNA homooligomers (A20, G20, C20, T20) were studied as ligands in CdS nanocrystals synthesis.26 Unlike mononucleotides,20 all four DNA oligomers produced water-soluble luminescent nanoparticles, demonstrating that these ligands have enhanced activity in nanocrystals growth. Moreover, increased stabilities were observed for nanocrystals synthesized with polynucleotides versus the corresponding mononucleotides most likely because the additive binding of adjacent nucleotides strengthens the overall binding through cooperativity. Interestingly, among the four homooligomer-passivated nanocrystals, those synthesized with T20 exhibit the lowest stabilities (Figure 3). Since thymine lacks an exocyclic amino group, it may be the poorest ligand, even when present in an oligomeric form.
FIGURE 3. Luminescence and stability of DNA-CdS.
(A) Emission spectra for nanocrystals made with DNA and TGA as a co-ligand at different DNA:Cd:TGA ratios. (B) Stability of DNA-CdS nanocrystals obtained with A20, C20, G20, T20 in a phosphate buffer monitored by absorbance at 430 nm.26
Different quantum efficiencies were observed for CdS nanocrystals synthesized with the four 20-mer homoligomers.26 Interestingly, despite lowered stabilities, the pyrimidine oligomers C and T produced CdS nanocrystals that exhibit higher luminescence intensity than those obtained with purine oligomers A and G. Given that the emission observed for these CdS nanocrystals represent that from “trapped states”, this trend may indicate that stronger ligands like A and G promote the formation of trapping sites, leading to higher luminescence intensities for materials with weaker ligands.
Clearly, nucleic acids sequence also influences nanocrystals properties. The effect of polynucleotide functional groups on nanocrystals passivation is enhanced when several of these functionalites act cooperatively in the same chain. The additive affinities of individual units in the oligomeric chains must be considered when extrapolating results obtained with mononucleotides to polynucleotides. This effect has been observed with non-biological ligands,27 and also appears to be operative with DNA ligands, as longer oligonucleotides yield more stable and emissive materials.26 Moreover, the shape and rigidity of oligomeric chains, which is dependent on the sequence, may also affect the binding of nucleic acids to nanocrystal surfaces.28, 29 Others have investigated the dependence of CdS nanocrystals optical properties, size, and dispersity on oligonucleotide composition30 and length31. This work, in combination with ours, indicates that complicated DNA sequences could be designed to “encode” nanocrystals luminescence intensity and wavelength.
Optical properties of DNA-passivated nanocrystals
For bioimaging applications, it is important to be able to prepare nanocrystals with luminescence maxima in a wide spectral range to provide versatility and compatibility with luminophores used for spectral multiplexing in multi-target assays and also to avoid overlap with biological tissues. In addition, individual populations of emitting nanocrystals should possess narrow spectral distributions as well as resistance to photobleaching.
The DNA-passivated CdS nanocrystals originally made exhibit broad luminescence spectra between 500 nm and 700 nm, which is a characteristic of trapped site emission rather than of band gap emission.28, 32–34 The quantum yields of these first-generation materials were low because of the emitting state structure. However, the emission of these materials was improved with the addition of a low molecular weight thiol ligand, thioglycolic acid (TGA).26 TGA molecules were proposed to bind to the exposed to the solvent trap sites on Cd2+-rich CdS nanocrystals surface through the strong Cd2+-thiol interaction, leading to a 3-fold increased luminescence intensity (Figure 3). The binding of the TGA ligand to CdS nanocrystals surface may alter the energies of trap sites and reduce nonradiative decay probability.34
Another parameter that significantly affected nanocrystals properties was the Cd2+ to S2− ratio used for the synthesis. Several studies have reported that a slight excess of Cd2+ is necessary to generate highly luminescent CdS nanocrystals.35, 36 This was also observed for DNA-passivated CdS nanocrystals where nucleotide to Cd2+ to S2− ratio that produced the nanocrystals with the highest luminescence was found to be 3 : 2 : 1.26
Solution pH also affects nanocrystals optical properties. Post-synthetic activation of DNA-passivated CdS nanocrystals by the addition of NaOH and Cd(ClO4)2 resulted in a two-to-three-fold increase in luminescence intensity without a change in emission maxima.37 Nanocrystal luminescence was also increased by performing the synthesis at higher pH. This likely results from hydroxyl groups binding to the trap sites that are unpassivated at lower pHs.20
Overall, limited progress was made with increasing the emission yields of DNA-passivated CdS. Very broad luminescence profiles were consistently obtained, and the quantum yields remained in the single digits despite extensive optimization. Darmatic increases in quantum yield, however, were realized by switching inorganic material. Indeed, the use of CdTe as a material system for DNA-passivated nanocrystals led to much more optimal luminescence characteristics, with PLQY up to 15%.38
One source of this improvement may be the slower kinetics of CdTe formation relative to CdS. CdS nanocrystals form instantly upon mixing of Cd2+-DNA complex with S2− at 25°C, while DNA-passivated CdTe nanocrystals grow over about an hour at 100°C. It is believed that the growth of CdS nanocrystals at low temperature is under kinetic control,39 which results in a rough surface that has various defects. The CdTe nanocrystals, in contrast, grow at a slower rate and elevated temperature closer to the equilibrium of growth and dissolution, which allows the CdTe nanocrystals to develop a more thermodynamically stable surface with fewer defects.40 These materials have therefore constituted the focus of our most recent work,38 which will be described in a subsequent section.
Cytotoxicity and cellular uptake of DNA-passivated CdS nanocrystals
Cytotoxicity has been a major concern associated with nanocrystals for biological applications. For cadmium-based nanocrystals, cytotoxicity can originate from the release of Cd2+ ions from the nanoparticle core, and the generation of reactive oxygen species upon irradiation ofthe nanocrystals.41–43 The stability of the ligand coating covering the nanocrystal surface is a major determinant of cytotoxicity. As mentioned above, DNA-passivated CdS nanocrystals are highly stable in high ionic strength phosphate buffers due to the cooperative binding of polymeric DNA molecules. To directly test their toxicity against living cells, we tested CdS nanocrystals synthesized with four different DNA homopolymers (A20, C20, G20, T20) by incubating HeLa cells with these nanocrystals. Little effect on the cells’ viability was observed, suggesting that only a negligible amount of Cd2+ ions was released from DNA-passivated CdS nanocrystals.26 Nanocrystals prepared by traditional organometallic procedure and solubilized via ligand exchange usually exhibit lower stabilities with increased surface decomposition and Cd2+ release.41,44 It should be noted, however that these materials were not directly compared with DNA-passivated nanocrystals and therefore solid evidence of their relative toxicities is lacking. A comprehensive analysis of the toxicity profiles of the biotemplated materials will be warranted for further biological use.
The cellular uptake and localization of DNA-passivated CdS nanocrystals was also studied.26 Rapid internalization and lysosomal sequestration was observed, which is consistent with previous studies of non-specific cellular uptake of nanocrystals through endocytosis.45 Interestingly, when HeLa cells were treated with TGA-coliganded CdS nanocrystals, less lysosomal trapping and more even distribution within cells were observed. Thus, a surface coating can alter the cellular trafficking of DNA-nanocrystals.
Generation of biofunctionalized DNA-nanocrystals using a one-pot synthesis
The success achieved in using DNA molecules as templates for nanocrystals synthesis prompts an important question: can sequences be designed that will both serve as ligands and as recognition elements? Given that the most powerful applications of nanocrystals can only be realized if the materials can be made specific towards a biological target, this latter issue is of significant interest if the materials are ever to be widely used. As well, the studies performed to understand sequence and structure did not address whether sequences that were used as nanocrystal ligands remained active as binding partners for complementary sequences. The recognition ability of a DNA strand absorbed on the surface could be compromised since its phosphate and nucleobase functionalities are involved in the surface passivation,. To circumvent this issue, we moved towards designing DNA-based ligands that would contain two chemically distinct domains with different roles for nanocrystal liganding and functionalization.
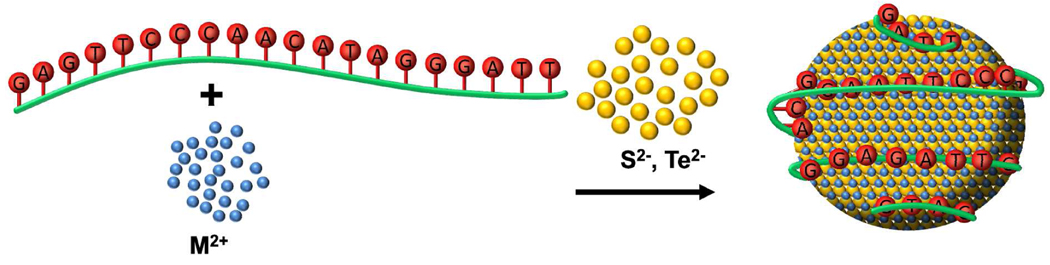
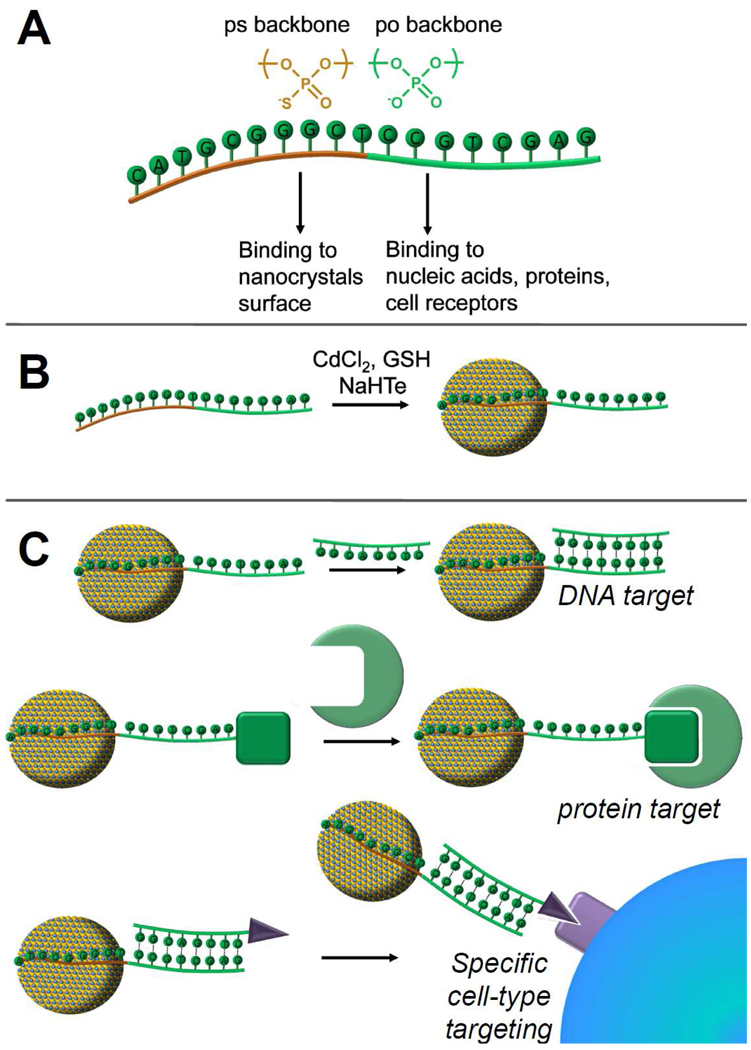
Bi-functional DNA templates with one domain responsible for nanocrystals passivation and the other domain for biorecognition were designed to test whether biofunctionalized nanocrystals could be generated with nucleic acids ligands and affinity domains.38 Phosphorothioate DNA, where the negatively charged oxygen atom on the phosphate backbone is replaced by sulfur, was used as a nanocrystal passivating fragment. Phosphorothioate nucleotides have been shown to possess much higher affinities to Cd2+ ions than phosphate nucleotides.46–48 By combining a phosphorothioate fragment with a phosphate fragment in one DNA strand, the phosphorothioate domain can be expected to bind to the nanocrystals, and the phosphate domain would then be free to interact with various biomolecules and to carry out recognition of a specific target (Figure 4). Thus, biofunctionalized nanocrystals for biotargeting and bioimaging could be obtained in a simple one step aqueous synthesis.
FIGURE 4. DNA-programmed and functionalized CdTe nanocrystals.
(A) Design of chimeric oligonucleotides with ligand (ps) and recognition (po) domains. (B) Schematic representation of one-pot biofunctionalized nanocrystal synthesis and (C) binding to biological targets.
To test this concept, we designed a chimeric DNA template (ps-po) composed of a 10-nucleotide phosphorothioate DNA segment and a 10-nucleotide phosphate DNA segment that are joined together by a 5-nucleotide poly-A linker.38 The ps-po DNA produced monodisperse CdTe nanocrystals ranging from 6.0 to 6.5 nm in size with quantum yields of ~ 15%. To assess the availability of the DNA strands on nanocrystals to bind to biomolecule targets, we first evaluated the hybridization efficiency of each DNA domain with the complementary DNA target. As expected, the po domain possessed much higher hybridization efficiency than the ps domain, which indicates that the ps domain is bound to CdTe nanocrystals more strongly than the po domain.
We then explored the binding of these nanocrystals to protein targets and cancer cells. The thrombin binding aptamer (TBA)49, 50 was introduced as the po domain, and the binding of ps-po-TBA nanocrystals with thrombin was evaluated. A high binding efficiency was observed for the ps-po-TBA-passivated CdTe nanocrystals with thrombin and the binding was confirmed to be sequence-specific. PbS nanocrystals obtained in a similar one-pot procedure for thrombin detection have been reported recently51 indicating that other material systems could be used in such an approach.
We further evaluated the binding of nanocrystals with specific cancer cells by introducing a cancer cell-binding aptamer.52 Specific binding of aptamer-functionalized nanocrystals with CCRF-CEM cells, a leukemia-derived cell line, versus Ramos cells, a lymphoma-derived cell line, was observed. The discrimination of these two cell lines indicates that these biofunctionalized materials could be used for cellular imaging applications where tracking specific cell types is of interest.
These results are most significant because they demonstrate for the first time that functionalized semiconductor nanocrystals can be made in a single step using a ligand source that is available to any researcher who can access a source of synthetic DNA. This advance should be of value to those who desire to use nanocrystals for imaging and biomolecular detection, but have difficulty using commercially available sources of nanocrystals and lack the infrastructure and equipment to use conventional multi-step organometallic synthesis.
Summary
Semiconductor nanocrystals possess optical properties that are superior to traditional organic fluorescent dyes. However, wide adoption of nanocrystals for biological imaging has not yet been realized largely due to the absence of practical synthetic methods for the preparation of biocompatible and nontoxic nanocrystals appended with customized recognition domains. Our nucleic acids-directed synthesis of nanocrystals is conducted in water under mild conditions in the presence of mononucleotides, DNA, or RNA, metal ions, and chalcogenide source. The use of nucleic acids as ligands for nanocrystals synthesis provides a powerful tool for the rational design of nanocrystals via nucleic acid sequence variation. There are many unanswered questions about the structure of the complexes of nanocrystals with nucleic acids, e.g: How many nucleic acid strands are there in the complex? What are their conformation and the mode of interaction with the nanocrystals surface? To answer these and other questions, nanocrystals’ structure and the growth mechanism will be studied in greater detail. Also, the use of in vitro evolution strategies may accelerate the search for optimal nucleotide sequences. The limited search through sequence space for nucleic acids ligands for semiconductor nanocrystals conducted so far has so far yielded sequences that are of great utility for making functional materials, and it is reasonable to expect that a combinatorial approach could propel this research area forward significantly.
Acknowledgements
We wish to acknowledge the Keck Foundation and the National Cancer Institute/National Institute of Health for their generous support of this work. We also thank Edward Sargent and his group at University of Toronto for their participation in the collaborative effort that has helped propel this work forward.
Biographies
Nan Ma received his B.S. degree in Chemistry from Peking University in 2004. He obtained a Ph.D. in Pharmaceutical Sciences at University of Toronto under the guidance of Professor Shana O. Kelley. working on nucleic acids-templated semiconductor nanocrystals in 2009. He is currently a postdoctoral fellow at Stanford Medical School in the research group of Prof. Jianghong Rao.
Grigory Tikhomirov received a B.S. degree in Chemistry from Moscow State University in 2003. He earned his Ph.D. from the University of Alberta in 2008 working with Professor Hicham Fenniri at the National Institute for Nanotechnology. He is currently a postdoctoral fellow in the laboratory of Professor Shana O. Kelly at the University of Toronto.
Shana Kelley is a Professor in the Departments of Biochemistry and Pharmaceutical Sciences at the University of Toronto. Kelley obtained her Ph.D. in 1999 in Chemistry from the California Institute of Technology in Chemistry, and was a NIH Postdoctoral Fellow at the Scripps Research Institute from 1999–2000. Kelley has co-authored over 60 scientific publications, and has developed new biosensing methods, nanomaterials for ultrasensitive biodetection, and novel bioconjugates for cellular studies. Since 2000, Kelley has received a Research Corporation Innovation Award, a Dreyfus New Faculty Award, a National Science Foundation CAREER Award, an Alfred P. Sloan Fellowship, a Camille Dreyfus Teacher-Scholar Award, the Pittsburgh Conference Achievement Award and was named to Technology Review’s list of Top 100 innovators and one of Canada’s Top 40 under 40.
REFERENCES
- 1.Alivisatos AP. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science. 1996;271:933–937. [Google Scholar]
- 2.Mowbray DJ, Skolnick MS. New Physics and Devices Based on Self-Assembled Semiconductor Quantum Dots. J. Phys. D: Appl. Phys. 2005;38:2059–2076. [Google Scholar]
- 3.Gill R, Zayats M, Willner I. Semiconductor Quantum Dots for Bioanalysis. Angew. Chem., Int. Ed. 2008;47:7602–7625. doi: 10.1002/anie.200800169. [DOI] [PubMed] [Google Scholar]
- 4.Chan WCW, Nie SM. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 5.Masumoto Y. Semiconductor Quantum Dots: Physics, Spectroscopy, and Applications. Berlin: Springer; 2002. pp. 457–478. [Google Scholar]
- 6.Klostranec JM, Chan WCW. Quantum Dots in Biological and Biomedical Research: Recent Progress and Present Challenges. Advanced Materials. 2006;18:1953–1964. [Google Scholar]
- 7.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum Dots Versus Organic Dyes as Fluorescent Labels. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 8.Murray CB, Norris DJ, Bawendi MG. Synthesis and Characterization of Nearly Monodisperse CdE (E = sulfur, selenium, tellurium) Semiconductor Nanocrystallites. J. Am. Chem. Soc. 1993;115:8706–8715. [Google Scholar]
- 9.Yu WW, Chang E, Drezek R, Colvin VL. Water-Soluble Quantum Dots for Biomedical Applications. Biochem. Biophys. Res. Commun. 2006;348:781–786. doi: 10.1016/j.bbrc.2006.07.160. [DOI] [PubMed] [Google Scholar]
- 10.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 11.Dahl JA, Maddux BLS, Hutchison JE. Toward Greener Nanosynthesis. Chem. Rev. 2007;107:2228–2269. doi: 10.1021/cr050943k. [DOI] [PubMed] [Google Scholar]
- 12.Martin RB. Nucleoside Sites for Transition-Metal Ion Binding. Acc. Chem. Res. 1985;18:32–38. [Google Scholar]
- 13.Sigel H. Interactions of Metal Ions with Nucleotides and Nucleic-Acids and Their Constituents. Chem. Soc. Rev. 1993;22:255–267. [Google Scholar]
- 14.Hossain Z, Huq F. Studies on the Interaction Between Cd2+ Ions and Nucleobases and Nucleotides. J. Inorg. Biochem. 2002;90:97–105. doi: 10.1016/s0162-0134(02)00411-7. [DOI] [PubMed] [Google Scholar]
- 15.Caruthers MH. Chemical Synthesis of DNA and DNA Analogs. Acc. Chem. Res. 1991;24:278–284. [Google Scholar]
- 16.Wilson C, Keefe AD. Building Oligonucleotide Therapeutics Using Non-Natural Chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Cushing BL, Kolesnichenko VL, O'Connor CJ. Recent Advances in the Liquid-Phase Syntheses of Inorganic Nanoparticles. Chem. Rev. 2004;104:3893–3946. doi: 10.1021/cr030027b. [DOI] [PubMed] [Google Scholar]
- 18.Hinds S, Taft BJ, Levina L, Sukhovatkin V, Dooley CJ, Roy MD, MacNeil DD, Sargent EH, Kelley SO. Nucleotide-Directed Growth of Semiconductor Nanocrystals. J. Am. Chem. Soc. 2006;128:64–65. doi: 10.1021/ja057002+. [DOI] [PubMed] [Google Scholar]
- 19.Sigel H, Da Costa CP, Martin RB. Interactions of Lead(II) with Nucleotides and their Constituents. Coord. Chem. Rev. 2001;219:435–461. [Google Scholar]
- 20.Dooley CJ, Rouge J, Ma N, Invernale M, Kelley SO. Nucleotide-Stabilized Cadmium Sulfide Nanoparticles. J. Mater. Chem. 2007;17:1687–1691. [Google Scholar]
- 21.Green M, Taylor R, Wakefield G. The Synthesis of Luminescent Adenosine Triphosphate Passivated Cadmium Sulfide Nanoparticles. J. Mater. Chem. 2003;13:1859–1861. [Google Scholar]
- 22.Green M, Smith-Boyle D, Harries J, Taylor R. Nucleotide Passivated Cadmium Sulfide Quantum Dots. Chem. Commun. 2005:4830–4832. doi: 10.1039/b506618b. [DOI] [PubMed] [Google Scholar]
- 23.Gugliotti LA, Feldheim DL, Eaton BE. RNA-Mediated Control of Metal Nanoparticle Shape. J. Am. Chem. Soc. 2005;127:17814–17818. doi: 10.1021/ja055039o. [DOI] [PubMed] [Google Scholar]
- 24.Ma N, Dooley CJ, Kelley SO. RNA-Templated Semiconductor Nanocrystals. J. Am. Chem. Soc. 2006;128:12598–12599. doi: 10.1021/ja0638962. [DOI] [PubMed] [Google Scholar]
- 25.Kim S. Transfer RNA: Structure, Properties and Recognition. New York: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 26.Ma N, Yang J, Stewart KM, Kelley SO. DNA-Passivated CdS Nanocrystals: Luminescence, Bioimaging, and Toxicity Profiles. Langmuir. 2007;23:12783–12787. doi: 10.1021/la7017727. [DOI] [PubMed] [Google Scholar]
- 27.Kim S, Bawendi MG. Oligomeric Ligands for Luminescent and Stable Nanocrystal Quantum Dots. J. Am. Chem. Soc. 2003;125:14652–14653. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 28.Mahtab R, Rogers JP, Murphy CJ. Protein-Sized Quantum Dot Luminescence Can Distinguish between "Straight", "Bent", and "Kinked" Oligonucleotides. J. Am. Chem. Soc. 1995;117:9099–9100. [Google Scholar]
- 29.Mahtab R, Rogers JP, Singleton CP, Murphy CJ. Preferential Adsorption of a "Kinked"; DNA to a Neutral Curved Surface: Comparisons to and Implications for Nonspecific DNA–Protein Interactions. J. Am. Chem. Soc. 1996;118:7028–7032. [Google Scholar]
- 30.Bigham SR, Coffer JL. The Influence of Adenine Content on the Properties of Q-Cds Clusters Stabilized by Polynucleotides. Colloids Surf., A. 1995;95:211–219. [Google Scholar]
- 31.Berti L, Alessandrini A, Bellesia M, Facci P. Fine-Tuning Nanoparticle Size by Oligo(guanine)n Templated Synthesis of CdS: An AFM Study. Langmuir. 2007;23:10891–10892. doi: 10.1021/la701867f. [DOI] [PubMed] [Google Scholar]
- 32.Bigham SR, Coffer JL. Thermochemical Passivation of DNA-Stabilized Q-Cadmium Sulfide Nanoparticles. J. Cluster Sci. 2000;11:359–372. [Google Scholar]
- 33.Spanhel L, Haase M, Weller H, Henglein A. Photochemistry of Colloidal Semiconductors. 20. Surface Modification and Stability of Strong Luminescing CdS Particles. J. Am. Chem. Soc. 1987;109:5649–5655. [Google Scholar]
- 34.Chestnoy N, Harris TD, Hull R, Brus LE. Luminescence and Photophysics of CdS Semiconductor Clusters: The Nature of the Emitting Electronic State. J. Phys. Chem. 1986;90:3393–3399. [Google Scholar]
- 35.Coffer JL, Bigham SR, Pinizzotto RF, Yang H. Characterization of Quantum-Confined CdS nanocrystallites stabilized by deoxyribonucleic acid (DNA) Nanotechnology. 1992:69–76. [Google Scholar]
- 36.Celebi S, Erdamar AK, Sennaroglu A, Kurt A, Acar HY. Synthesis and characterization of poly(acrylic acid) stabilized cadmium sulfide quantum dots. J. Phys. Chem. B. 2007;111:12668–12675. doi: 10.1021/jp0739420. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Coffer JL. Effect of Pressure on the Photoluminescence of Polynucleotide-Stabilized Cadmium Sulfide Nanocrystals. Chem. Mater. 1999;11:2326–2330. [Google Scholar]
- 38.Ma N, Sargent EH, Kelley SO. One-Step DNA-Programmed Growth of Luminescent and Biofunctionalized Nanocrystals. Nat. Nanotechnol. 2009 doi: 10.1038/nnano.2008.373. in press. doi:10.1038/nnano.2008.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollingsworth JA, Klimov VI. Semiconductor and Metal Nanocrystals: Synthesis and Electronic and Optical Properties. Vol. Part I. Marcel Dekker, Inc.; 2004. Chapter 1. [Google Scholar]
- 40.Borchert H, Talapin DV, Gaponik N, McGinley C, Adam S, Lobo A, Moller T, Weller H. Relations Between the Photoluminescence Efficiency of CdTe Nanocrystals and Their Surface Properties Revealed by Synchrotron XPS. J. Phys. Chem. B. 2003;107:9662–9668. [Google Scholar]
- 41.Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewinski N, Colvin V, Drezek R. Cytotoxicity of Nanoparticles. Small. 2008;4:26–49. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 43.Lovric J, Cho SJ, Winnik FM, Maysinger D. Unmodified Cadmium Telluride Quantum Dots Induce Reactive Oxygen Species Formation Leading to Multiple Organelle Damage and Cell Death. Chem. Biol. 2005;12:1227–1234. doi: 10.1016/j.chembiol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Cho SJ, Maysinger D, Jain M, Roder B, Hackbarth S, Winnik FM. Long-term Exposure to CdTe Quantum Dots Causes Functional Impairments in Live Cells. Langmuir. 2007;23:1974–1980. doi: 10.1021/la060093j. [DOI] [PubMed] [Google Scholar]
- 45.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term Multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 46.Pecoraro VL, Hermes JD, Cleland WW. Stability Constants of Mg2+ and Cd2+ Complexes of Adenine Nucleotides and Thionucleotides and Rate Constants for Formation and Dissociation of MgATP and MgADP. Biochemistry. 1984;23(22):5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 47.Jiang L, Zhuang J, Ma Y, Yang B, Yang W, Li T. The Coordination Sites of Phosphorothioate OligoG10 with Cd2+ and CdS Nanoparticles. New J. Chem. 2003;27:823–826. [Google Scholar]
- 48.Carla P, Da Costa AOHS. Complex Formation of Divalent Metal Ions with Uridine 5'-O-Thiomonophosphate or Methyl Thiophosphate: Comparison of Complex Stabilities with Those of the Parent Phosphate Ligands. ChemBioChem. 2003;4:593–602. doi: 10.1002/cbic.200200551. [DOI] [PubMed] [Google Scholar]
- 49.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of Single-Stranded DNA Molecules That Bind and Inhibit Human Thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 50.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The Structure of Alpha-Thrombin Inhibited by a 15-Mer Single-Stranded DNA Aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 51.Choi JH, Chen KH, Strano MS. Aptamer-Capped Nanocrystal Quantum Dots: A New Method for Label-Free Protein Detection. J. Am. Chem. Soc. 2006;128:15584–15585. doi: 10.1021/ja066506k. [DOI] [PubMed] [Google Scholar]
- 52.Shangguan D, Tang ZW, Mallikaratchy P, Xiao ZY, Tan WH. Optimization and Modifications of Aptamers Selected From Live Cancer Cell Lines. ChemBioChem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]