Abstract
The major bottleneck to the application of high-resolution techniques such as crystallographic X-ray diffraction and spectroscopic analyses to resolve the structure of mammalian membrane proteins has been the ectopic expression and purification of sufficient quantities of non-denatured proteins. This has been especially problematic for members of the major facilitator superfamily, which includes the family of mammalian glucose transporters. A simple and rapid method is described for the purification of milligram quantities of recombinant GLUT1 and GLUT4, two of the most intensively studied GLUT isoforms, after ectopic expression in Pichia pastoris. The proteins obtained were > 95 % pure and exhibited functional transport and ligand-binding activities.
Keywords: Glucose transporter, GLUT1, GLUT4, Membrane protein purification
INTRODUCTION
Membrane proteins are functionally diverse and include receptors for systemic or local messengers as well as transporters and channels that mediate the ability of various hydrophilic substrates to traverse lipid bilayer membranes. Many membrane receptors, channels, and transporters are the targets of pharmacological disease interventions and are thus of intense clinical interest. The determination of the structure of membrane proteins at high resolution can lead to the rational design and development of new drugs with greater efficacy and specificity and that lack undesirable iatrogenic effects.
The major facilitator superfamily (MFS) is the largest superfamily of membrane transport proteins, containing over 5000 members identified in all three taxonomic kingdoms [1]. MFS proteins mediate the membrane transport of a highly diverse set of substrates via both active and passive mechanisms. Most MFS proteins appear to share a common membrane topology with twelve transmembrane alpha helices and are believed to exhibit highly flexible structures [1, 2]. The latter property probably accounts for the extreme difficulty that has been encountered in attempts to obtain three-dimensional crystals of these proteins [3]. Only three members of the MFS have been crystallized to date (lactose permease [4, 5], glycerol-3-P antiporter [6], EmrD multidrug transporter [7]) and none of these transporters is from a eukaryotic source. The principal roadblock to the crystallization of eukaryotic MFS proteins has been the inability to purify large quantities of these proteins in a non-denatured, non-glycosylated form.
The facilitative transport of glucose across the membranes of mammalian cells is mediated by members of the glucose transporter (GLUT) protein family that represents a subset of the MFS [8]. Reflecting their fundamental physiological importance, GLUT proteins are either directly or indirectly involved in a number of human diseases, including type 2 diabetes [9], tumor progression [10], GLUT1 Deficiency syndrome [11], Fanconi-Bickel syndrome [12], and metabolic syndrome associated with highly active antiretroviral therapy [13]. GLUT1 and GLUT4 are perhaps the most intensively studied of the 14 GLUT isoforms expressed in the human. The structure and function of GLUT1 has been studied for several decades using a variety of biochemical approaches including detailed kinetic studies, site-directed and glycosylation-scanning mutagenesis, chemical cross-linking, and many other techniques [14–20]. GLUT1 is expressed at high levels in the human erythrocyte and remains the only GLUT protein that has been purified from its native cell type [21–23]. Unfortunately, attempts to crystallize the purified native protein were unsuccessful, probably due in part to the presence of a heterogeneous N-linked oligosaccharide and to the flexible and insoluble nature of the protein in detergent solutions [22]. GLUT4 is the primary glucose transporter expressed in adipose and muscle tissues and has been the subject of intense investigation [24] because of its rate-limiting role in insulin-stimulated whole-body glucose disposal, a process that is defective in type 2 diabetes [9]. GLUT4 is also a direct cellular target of HIV protease inhibitors, which appears to contribute to the development of metabolic syndrome in patients treated with highly active antiretroviral therapy [25].
Herein we describe a rapid and simple procedure for the purification of milligram quantities of recombinant mammalian glucose transporter proteins that are suitable for functional studies and for crystallization assays.
MATERIALS AND METHODS
GLUT protein expression in Pichia pastoris
Human GLUT1 cDNA isolated from a HepG2 library [26] and Rat GLUT4 cDNA isolated from an adipocyte library [27] were cloned into the expression vector pPICZ B (Invitrogen, Carlsbad, CA).The production of the GLUT1 construct is described here in detail, but similar procedures were used for all four mutant GLUT proteins using appropriate oligonucleotides. PCR was conducted using the pXOV GLUT1 [16] vector as a template in three different steps. In all of the reactions, the forward primer was 5’-GATACAGCGGCCGCATGGAGCCCAGCAGCAA containing a Not I restriction site. For the first reaction the reverse primer was 5’-CCGGTGGTGGTGATGATGGTGGTGGTGTCGACCTTCAATCACTTG. The purified DNA product was used as the template for a second amplification using the same forward primer and with the reverse primer 5’-CCGGTGGTGGTGATGATGGTGGTGGTGTCGACCTTCAATCACTTG for 15 cycles, and then a 10-fold molar excess of the primer 5’- TTCTAGATCAGTCCTTGTCCCGGTGGTGGTGATGATG was added for 25 additional cycles. These latter two primers inserted a Factor Xa cleavage site, a tag of 8 histidines, and a stop codon at the C-terminus of the protein followed by a Xba I restriction site. The site of N-linked glycosylation was removed by mutation of Asp45 to a Thr residue. The Not I / Xba I restriction fragment was cloned into version B of the pPICZ vector. P. pastoris electrocompetent cells (strain X-33, Invitrogen, Carlsbad, CA), were prepared using a standard protocol and transformed by electroporation (Gene Pulser Xcell, BioRad, Hercules, CA) with 10 µg of plasmid linearized by BstX I digestion. Multiple copies of the expression cassette (PAOX1 promoter–gene –Zeocin) were integrated into the yeast genome by homologous recombination in a region upstream or downstream of the alcohol oxidase I (AOX1) gene. Transformed cells were selected in YPDS (Yeast extract Peptone Dextrose Sorbitol) with 100 ug/ml of Zeocin as the selection medium. Clones with multiple copies of the expression cassette were selected using 500 – 2000 µg/ml of Zeocin in YPDS, and then protein expression was evaluated by western blot analysis using antibodies against the C-terminal region of GLUT1 [28]. Clones expressing the highest levels of the mutants were grown in a BioFlo 110 Fermentor & Bioreactor with gas mix controller (New Brunswick Scientific, Edison, NJ). Four liters of medium with 3 % of glycerol were inoculated with 250 ml of an overnight culture of cells grown in BMGY (Buffered Glycerol-complex Medium). After the glycerol was completely exhausted from the culture medium (about 80 – 110 g cells/L), the cells were fed for 24 – 30 hours with methanol on demand by monitoring oxygen levels in order to induce GLUT protein expression. Cells were then harvested and washed twice with PBS plus 1 mM EDTA and stored frozen at −80 °C.
GLUT protein solubilization
Forty-four grams of yeast cells were thawed and suspended at 30% w/v slurry to 135 ml (final volume) in breaking buffer (25 mM Na2HPO4/NaH2PO4, pH 7.40), protease inhibitor (PI) mix (0.01 units/ml aprotinin, 1µg/ml leupeptin, 1 µg/ml benzamidine, 1 µg/ml antipain, 5 µg/ml trypsin inhibitor, 1 µg/ml chymostatin, 1 µg/ml pepstatin A, 1 mM PMSF), and 20 U/ml of DNAase I-type II (Sigma, St Louis, MO). Cells were broken by 7 passes through a M-110S Microfluidizer (Microfluidics Co, Newton, MA) equipped with an interaction chamber of 85 µm at 23,000 psi. Before the last pass 2M DTT and 0.1 M EDTA (4 mM and 2 mM final concentrations, respectively) were added and the homogenate was spun down at 4,500 g for 15 min. The supernate was used for the preparation of total stripped membranes. The pH of the supernate was adjusted to 10.5 using KOH, incubated for 15 minutes on ice, and then centrifuged at 200,000 g for 1 hour at 4°C. The pellet was suspended in 50 ml of 25 mM Hepes, 2 mM EDTA, 2 mM DTT. Detergents were tested for GLUT solubilization using the total homogenate and the stripped membrane preparation. Triton X-100, CHAPS, Decylmaltoside (DM) and Dodecylmaltoside (DDM) were used at a concentration of 1 or 2 % in 40 mM imidazole pH 6.80, 50 mM KCl, 5 % glycerol, 1 mM TECP, 0.2 mM EDTA, containing the PI mix. The preparation was incubated for 45 minutes at 4°C in a bath sonicator VWR M75D (VWR Int, Batavia, IL) containing ice and then spun down at 200,000 g at 4°C for 30 minutes. The supernates were analyzed by western blotting using either polyclonal or monoclonal GLUT antibody [29] or poly-histidine antibody (Qiagen, Valencia, CA) and IRDye 680 donkey anti-rabbit or anti-mouse (LI-COR Biosciences, Lincoln, NE) IgG as secondary antibodies. Band intensities were measured using an Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
GLUT purification
For a standard large-scale preparation, 50 ml of yeast total membrane suspension was adjusted to 1% DDM, 40 mM imidazole pH 6.80, 50 mM KCl, 5 % glycerol, 1 mM TECP, 0.2 mM EDTA, PI mix. After the 200,000 g centrifugation step the pH was adjusted to 7.40 and the solubilized membrane protein was loaded onto a HisTrap HP 5 ml column (GE Healthcare, Uppsala, Sweden) in an AKTA FPLC (GE Healthcare, Uppsala, Sweden). The column was washed with 4 column volumes of 100 mM imidazole in loading buffer and the His-tagged GLUT proteins were eluted in 600 mM imidazole, pH 7.20, 50 mM KCl, 10% glycerol, 0.2% DDM, 3 mM TECP, 0.2 mM EDTA. The eluted protein in a total volume of 6 ml was concentrated to 1.5 ml using a Vivaspin 6 30K (Sartorius, Gottingen, Germany) tube and desalted on a 5 ml HiTrap desalting column (GE Healthcare, Uppsala, Sweden) pre-equilibrated with 25 mM Hepes pH 6.80, 50 mM KCl, 10 % glycerol, 0.2 % DDM, 1 mM TECP, 0.2 mM EDTA. After desalting, the protein solution was concentrated to a volume of 0.5 ml and then loaded onto a 24 ml 10 × 30 cm Superdex 200GL column (GE Healthcare, Uppsala, Sweden) equilibrated in the same buffer. The purity of the recombinant proteins was estimated by analysis of aliquots on 10 % SDS-polyacrylamide gels stained with SimplyBlue Safe Stain (Invitrogen, Carlsbad, CA). The identity of the GLUT1 band was verified by western blot analysis and mass spectrometry.
Photo-affinity labeling of aglyco-GLUT1 and aglyco-GLUT4
Fifteen micrograms of purified GLUT protein in 100 µl 25 mM Hepes pH 6.80, 50 mM KCl, 10 % glycerol, 0.2 % DM, 1 mM TECP, 0.2 mM EDTA were incubated with 5 µM of a biotin derivative of a photoaffinity labeling reagent for glucose transporters [30] N-[2-[2-[2-[(N-Biotinyl-caproylamino)-ethoxy)ethoxyl]-4-[2-(trifluoromethyl)-3H-diazirin-3-yl]benzoyl]-1,3-bis(mannopyranosyl-4-yloxy)-2-propylamine (PEG-biotincap-ATB-BMPA) (TRC Inc., Ontario, Canada) and UV irradiated for 1 min at 18 °C in a Rayonet RPR100 (The Southern New England UV Company, Branford, CT) and, in some of the samples, 1 µM cytochalasin B was included to test for inhibition. After photolabeling the protein was separated from the free reagent using a 0.5 ml G-25 Sepharose (GE Healthcare, Uppsala, Sweden) spin column and then subjected to western blot analysis using streptavidin-HRP (Pierce Biotechnology, Rockfold, IL) to detect the photolabeled protein.
Functional Reconstitution of Purified GLUT1 into Proteoliposomes
Proteoliposomes were prepared by gel filtration [31] using Sephadex G-50. 12.5 mg of soybean phospholipids (Avanti Polar Lipids, Alabaster, AL) in 20 mM HEPES, 0.30 M NaCl were dispersed at 22°C in a bath sonicator (VWR M75D, VWR Int, Batavia, IL) and then mixed with 37 mM CHAPS and incubated for 2 hours at 25 °C. 12.5 µg/ml of purified aglyco- GLUT1 was then added to the lipids. Two hundred microliters of the proteoliposome mixture were loaded onto a 2 ml column of pre-spun Sephadex G-50 equilibrated with 20 mM HEPES, 0.15 M NaCl and the centrifugation-filtration was conducted twice in succession. Proteoliposomes (27 µl) were added to 3 µl of uptake solution (0.2 mM L- or D- [3H] glucose, final concentration), incubated for various time periods, and the uptake was then stopped by the addition of 1 µl of 60 mM HgCl2. This mix was filtered using spin columns of 0.6 ml Sephadex G-50 pre-equilibrated with 20 mM HEPES, 0.15 M NaCl, 2 mM HgCl2. Each point is the average of 2 or 3 independent measurements.
Crystallization of aglyco-GLUT4 Δ38
Concentrated protein was used for screening under different crystallization conditions. Precipitant Synergy P64 from Emerald Biosystem and MembFac from Hampton Research were used. The hanging-drop vapor diffusion method was used more frequently but microdialysis was also tested.
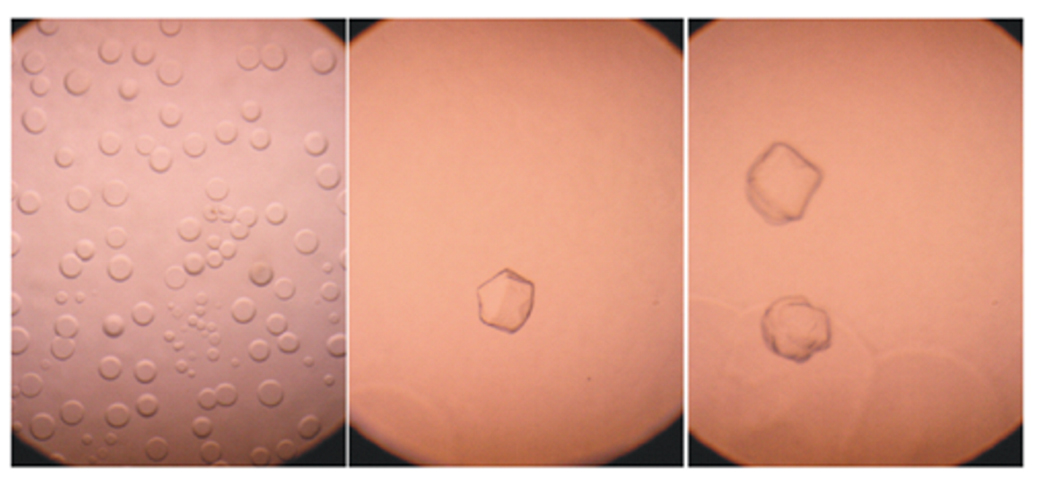
Crystals obtained from different mutants were variable in size, but in general they were small. Aglyco-GLUT4-Δ38 produced crystals after 4 to 5 weeks in 20% PEG400, 15% PEG1000, 0.15 M K2HPO4/NaH2PO4 pH 6.5 as precipitant solution using the hanging-drop method (Fig.6). The size of the crystals was 0.1–.15 mm. Crystals were analyzed in an X-ray detector Mar345dtb (Marresearch-marUSA Inc., Evanston, IL) and produced low resolution diffraction patterns (20 Å).
Figure 6. Aglyco-GLUT4-Δ38 crystals.
Purified aglyco-GLUT4-Δ38 was concentrated to 2 mg/ml and subjected to a variety of crystallization conditions. Protein crystals were observed after 4–5 weeks incubation in 20% PEG400, 15% PEG1000, 0.15 M K2HPO4/NaH2PO4 pH 6.5 using the hanging-drop method (b and c). Crystals were ~ 0.1–0.15 mm in size. On the second and third day phase separation drops were observed (a).
RESULTS
Engineering and expression of mammalian GLUT mutants
GLUT cDNA constructs were designed for expression in the methylotrophic yeast, P. pastoris. This yeast has been successfully used as an expression system for the purification and crystallization of other mammalian membrane proteins, such as voltage-dependent potassium channels [32]. Its major advantage is that it can be grown to an extremely high biomass, thus obviating the need to achieve extremely high levels of overexpression that can result in the aggregation and denaturation of membrane proteins.
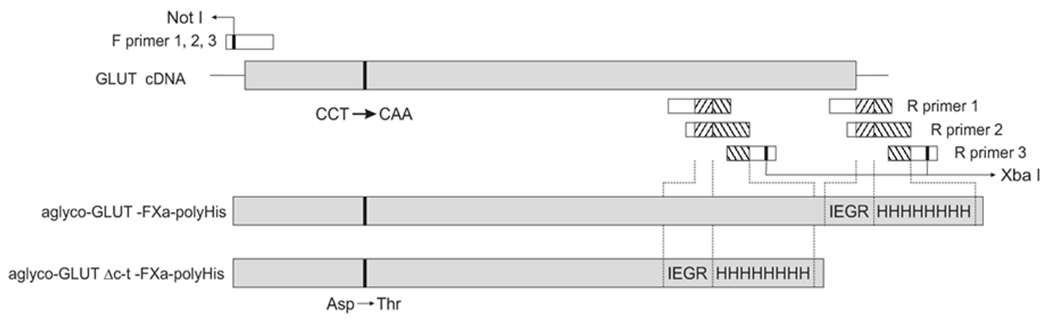
The following human GLUT1 and rat GLUT4 cDNAs were engineered in order to facilitate the large-scale purification of proteins suitable for functional studies or crystallization assays (see Figure 1): 1) aglyco-GLUT1– FXa-polyHis; 2) aglyco-GLUT4– FXa-polyHis,; 3) aglyco-GLUT1 Δ37– FXa-polyHis; and 4) glyco-GLUT4 Δ38 – FXa-polyHis The Δ37 and Δ38 designate the number of amino acids truncated at the C-terminus of the two mutant transporters. The truncation locks the transporters into their inward-facing configurations, which should greatly reduce their flexibility and thus facilitate the formation of 3-dimensional crystals. As an example, the first structure reported for the lactose permease [4] resulted from the crystallization of a C154G mutant, which is locked into the inward-facing conformation, is more resistant to inactivation by heat, and is more stable in solution, properties which probably contributed to its successful crystallization.
Figure 1. Generation of glucose transporter constructs.
For all GLUT constructs the forward primer was the same and included a Not I restriction site. The 3 reverse primers were designed to insert a Factor Xa cleavage site, a tag of 8 histidines, and a stop codon at the C-terminus of the proteins followed by a Xba I restriction site. The glycosylation site was removed by mutation of Asp45 or Asp57 to Thr in GLUT1 or GLUT4, respectively. The Not I / Xba I restriction fragment was cloned into version B of the pPICZ vector (Invitrogen).
All of the constructs were engineered with a factor Xa protease recognition site followed by a sequence of 8 histidine residues at their C-termini in order to remove the tag and to purify the protein by metal-affinity chromatography, respectively. The cDNAs were also subjected to site-directed mutagenesis in order to eliminate the consensus sequence for N-linked glycosylation (N45T or N57T in GLUT1 and GLUT4, respectively). Aglyco GLUT1 is functional and is properly targeted to the cell surface [16]. N-linked oligosaccharides are usually structurally heterogeneous and most likely impede protein crystallization.
Not I / Xba I restriction fragments encoding the GLUT mutants were subcloned into version B of the pPICZ vector and the resulting constructs were introduced into P. pastoris strain X-33 by electroporation, resulting in the integration of the expression cassette by homologous recombination into the yeast genome. Clones with multiple copies of the expression cassette were selected and protein expression was evaluated by western blot analysis using antibodies against the C-terminal region of the two GLUT proteins or poly histidine probe-HRP. Clones that expressed more than 1.5 mg of transporter protein per liter of culture were selected and grown in a BioFlo 110 Fermentor & Bioreactor with gas mix controller. About 80 – 110 g of cells were harvested per liter of culture. The approximate expression level of the mammalian glucose transporters was ~0.1–0.2% of total yeast protein.
Protein Purification
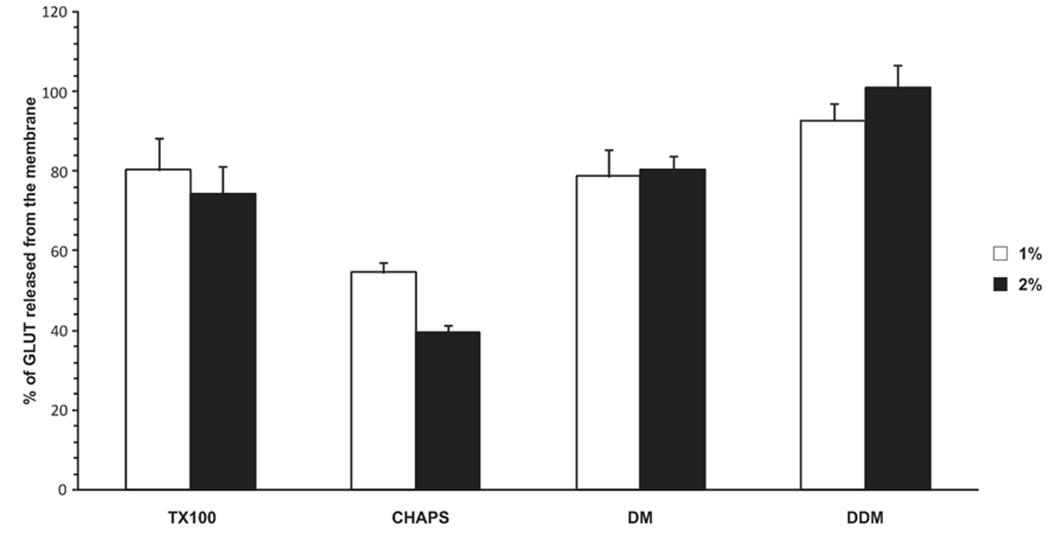
The choice of detergent is a key factor in the successful purification of functional membrane proteins. Recombinant GLUT protein solubilization from P. pastoris membranes was tested using several detergents that have been used for GLUT1 purification from human erythrocytes [19, 21, 22]. Triton X100, CHAPS, decylmaltoside (DM) and dodecylmaltoside (DDM) were used at concentrations of 1 and 2 %. Triton, DM, and DDM released more than 75 % of GLUT protein from the membrane fraction and the 2 % concentrations did not significantly increase the extent of GLUT solubilization compared to 1% (Fig.2). All four GLUT mutants exhibited similar solubilization properties. All of the tested detergents except for CHAPS were effective, but it is known that GLUT1 is irreversibly inactivated in Triton X100 unless the detergent is removed [33]. DDM solubilized almost 100 % of the GLUT proteins and has been used effectively for the purification and crystallization of several membrane proteins, such as the lac permease [4], glycerol-3-P transporter [6] and mammalian voltage-dependent K+-channel [32].
Figure 2. Detergent solubilization of GLUT protein.
Detergents were tested for GLUT solubilization from total yeast membrane preparations. Triton X-100, CHAPS, Decylmaltoside (DM) and Dodecylmaltoside (DDM) were used at 1 or 2 % concentrations in a buffer containing 40 mM imidazole pH 6.80, 50 mM KCl, 5 % glycerol, 1 mM TECP, 0.2 mM EDTA, and a protein inhibitor cocktail. The total membrane preparation was incubated for 45 minutes at 4° C in an ultrasonic bath containing ice and then centrifuged at 200,000 g at 4° C for 15 minutes. The supernatants were analyzed by western blotting using GLUT-specific antibody and IRDye 680 donkey anti-rabbit or anti-mouse (LI-COR Biosciences, Lincoln, NE). GLUT protein levels were measured by Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
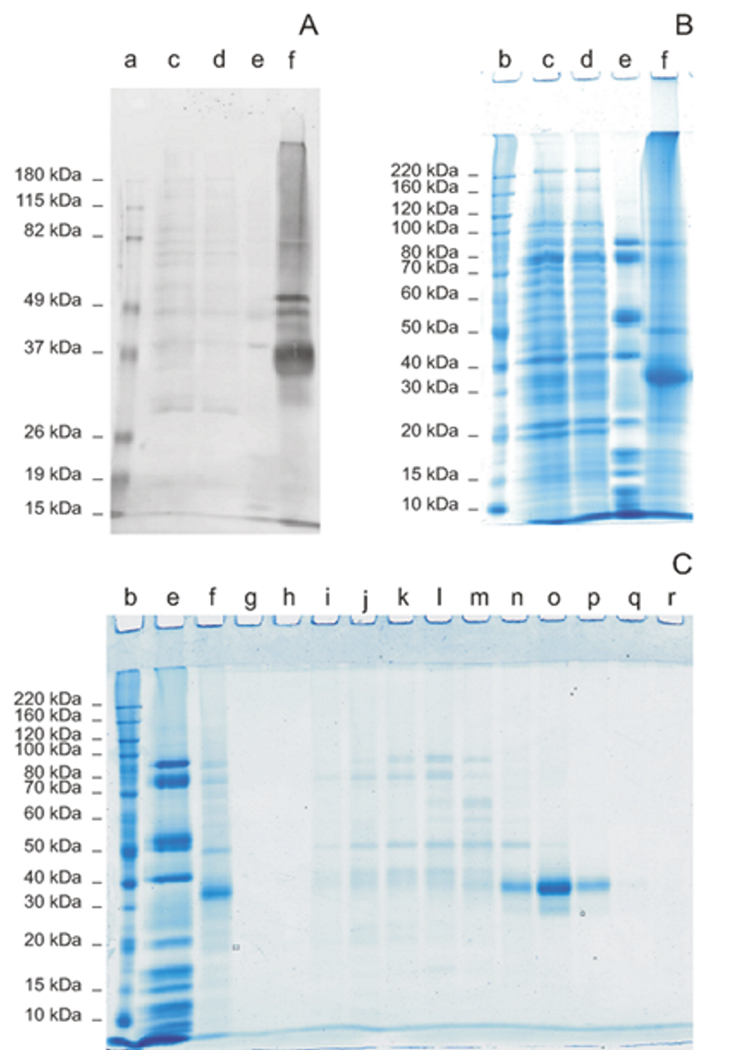
The protein extracted from total membranes of P. pastoris with 1 % DDM was loaded onto a 5 ml Nickel affinity column in an FPLC system. The column was washed with 100 mM imidazole and the recombinant His-tagged proteins were then eluted in 600 mM imidazole. The eluted protein in 6 ml of buffer was concentrated down to 0.5 ml and then subjected to size exclusion chromatography. The purity of the proteins was evaluated on 10 % SDS polyacrylamide gels stained with Coomassie G250. The identity of GLUT1 was confirmed by both western blot analysis and mass spectrometry. Figure 3 illustrates a typical purification of aglyco-GLUT4 Δ38. The nitrocellulose membrane (panel A) was probed using a poly-histidine antibody because of the lack of the C-terminal tail in the recombinant GLUT4 protein, against which all available antibodies for GLUT4 have been raised. The secondary antibody was conjugated to an infrared fluorescent dye that allowed the quantification of protein yield. The Nickel-affinity column was very efficient at enriching the tagged GLUT4 protein, which comprised > 75 % of the total protein eluted in 600 mM imidazole (fig. 3, panels B and C). The use of a size exclusion column permitted the elimination of several contaminating proteins in the imidazole eluate as well as probable oligomeric or aggregated forms of the GLUT protein (fig. 3, panel C). The native oligomeric state of most GLUT proteins is not known, but GLUT1 may exist as a dimer or tetramer in some cell types [19]. However, the functional significance of the oligomerization of GLUT1 is unclear and it appears that GLUT proteins exhibit normal transport activity in the monomeric state [34, 35].
Figure 3. Purification of Aglyco-GLUT4Δ38 expressed in P. pastoris.
Proteins from total membrane preparations of P. pastoris expressing aglyco-GLUT4 Δ38 were solubilized in 1 % DDM and loaded onto a HisTrap HP 5 ml Ni-column. The column was washed with 100 mM imidazole and the His-tagged protein was then eluted in 600 mM imidazole. Panel A, western blot probed using a monoclonal polyhistidine antibody; panel B, Coomassie-G250 stained gel. Lanes a and b: molecular weight markers; c: total membrane solubilized in DDM; d: Ni-affinity column flow through; e: 100 mM imidazole wash; f: 600 mM imidazole elution. The 600 mM imidazole eluted protein was in 6 ml, concentrated to 0.5 ml and subjected to size exclusion chromatography on a Superdex 200GL 10/300 and then analyzed on a Coomassie G250 stained SDS gel: panel C, b: molecular weight markers; e and f: 100 mM and 600 mM imidazole, respectively, from the Ni-affinity column; g to r: fractions from the size exclusion column.
Table 1 outlines the yield at each step of a typical GLUT1 protein purification. GLUT1 represented ~ 0.2 % of the total Pichia protein and after the total membrane preparation, it was enriched to 0.9 %. Greater than 1 mg of GLUT1 protein was obtained per L of culture with an estimated purity of ~ 95%. The greatest loss of GLUT protein occurred due to retention on the Nickel column, most likely representing aggregated or denatured protein. The affinity column step requires that the sample be present in a high-salt buffer in order to reduce non-specific protein binding, and high-salt concentrations induce irreversible aggregation of GLUT proteins. Rapid de-salting of the eluted GLUT proteins was found to be required in order to minimize aggregation.
Table 1.
Recovery of GLUT1 During the Purification Procedure
| GLUT1 protein | Percentage of Recovery |
|
|---|---|---|
| mg/100 g of wet cells | ||
| Total homogenate | 9.37 ± 0.80 | 100% |
| Total Pichia Membranes | 9.25 ± 0.72 | 99 % |
| Detergent Supernatant | 7.40 ± 0.58 | 79 % |
| GLUT1 bounded to the Ni-Column after loaded and washed |
6.3 ± 0.75 | 67 % |
| GLUT1 eluted from Ni-Column | 1.90 ± 0.24 | 20 % |
| GLUT1 recovered from Size Exclusion Column |
1.31 ± 0.28 | 14 % |
Functional Characterization of the protein
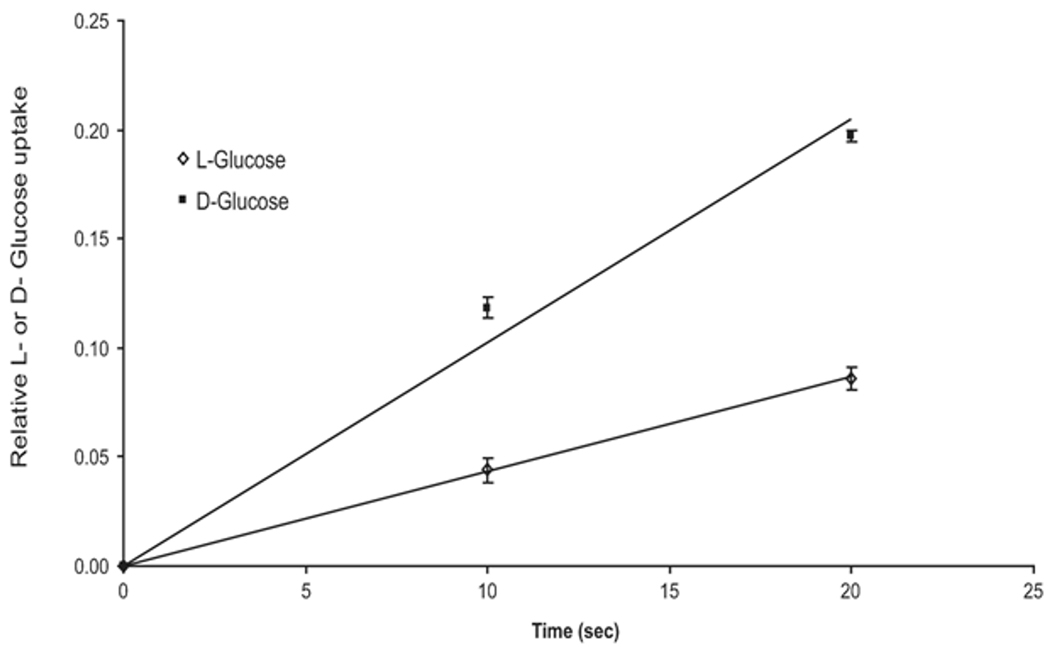
Purified Aglyco-GLUT1 and aglyco-GLUT4 were photolabeled using PEG-biotincap-ATB-BMPA [36], and the photolabeling was inhibited in the presence of cytochalasin B (fig. 4). PEG-biotincap-ATB-BMPA appears to bind to GLUT proteins in their functional exofacial conformations [30], whereas cytochalasin B binds to GLUT proteins only in their cytoplasmic conformations [37]. These results demonstrate that the purified GLUT proteins were present in their two native conformations and were most likely in a functional, non-denatured state. In order to confirm that purified GLUT1 was functional, it was reconstituted into proteoliposomes and the uptake of D- and L-glucose was measured. Figure 5 demonstrates that purified GLUT1 exhibited specific D-glucose transport activity.
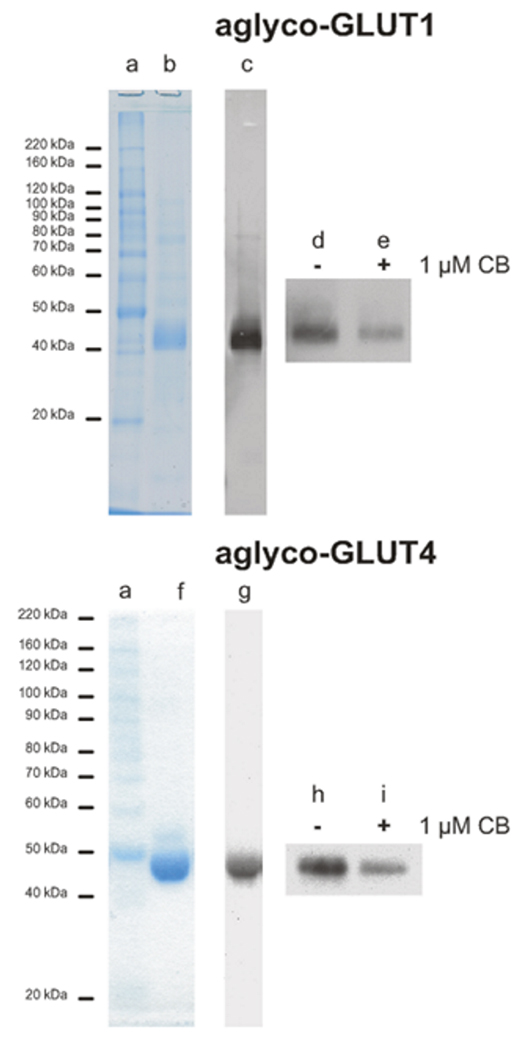
Figure 4. Photolabeling of aglyco-GLUT1 and aglyco-GLUT4 with PEG-biotincap-ATB-BMPA.
Eluted fractions from the Ni-column were desalted by gel filtration and analyzed on 10 % SDS polyacrylamide gels and detected by staining with Coomassie G250 and by western blot analysis using specific antibodies. a: molecular weight markers; b and c: aglyco-GLUT1; f and g: aglyco-GLUT4. Fifteen micrograms of purified protein was incubated in the presence of 4 µM PEG-biotincap-ATB-BMPA without or with1 µM cytochalasin B and then subjected to irradiation for 1 min at 18 °C. The reaction mixes were centrifuged through G-25 Sepharose spin columns and the flow-throughs were loaded onto 10 % SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with streptavidin-HRP.
Figure 5. D-glucose transport activity of purified GLUT1 reconstituted into proteoliposomes.
Proteoliposomes were prepared by gel filtration through Sephadex G-50. 12.5 µg of soybean phospholipids in 20 mM HEPES, 0.30 M NaCl were sonicated, adjusted to 37 mM CHAPS, incubated for 2 hours at 25 °C, and then 12.5 µg/ml of purified aglyco-GLUT1 was added. Two hundred microliters were filtered twice through a 2 ml bed of pre-spun Sephadex G-50 equilibrated with 20 mM HEPES, 0.15 M NaCl. Proteoliposomes (27 ul) were added to 3 µl of uptake solution (0.2 mM L- or D- [3H] glucose, final concentration) and the uptake was quenched by the addition of 1 µl of 60 mM HgCl2 . This mix was filtered through spin columns of 0.6 ml Sephadex G-50 pre-equilibrated with 20 mM HEPES, 0.15 M NaCl, 2 mM HgCl2. Each point is the average of 3 independent measurements.
Crystallization Assays
Attempts have been made to crystallize all four mutant GLUT proteins under a variety of conditions used for membrane proteins. Aglyco-GLUT4 Δ38 was the only mutant that produced crystals of sufficient size to be examined by X-ray diffraction (see Figure 6). The crystals were about ~ 100–150 µm in size but were poorly ordered and produced low resolution (> 20 Å) X-ray patterns. Efforts are currently underway to refine the quality of GLUT protein crystals.
DISCUSSION
Glucose transport was one of the first membrane transport processes to be rigorously explored and has been the subject of intense investigation for more than five decades [38]. Glucose is an essential fuel substance and anabolic substrate for the human and is transported across the plasma membrane of virtually every cell in the body [39–41]. Therefore, it is not surprising that either defects in or abnormal regulation of glucose transporters is associated with a number of rare genetic diseases, such as GLUT1 deficiency syndrome [42] and Fanconi-Bickel syndrome [12] as well as several more common pathological conditions. For example, upregulation of glucose transporter expression is almost invariably associated with oncogenesis [43] reflecting the obligate increase in glycolysis that occurs in tumors [10]. The abnormal insulin-mediated regulation of the GLUT4 isoform is a well-known defect associated with insulin resistant states such as type 2 diabetes [9].
Knowledge of the three dimensional structure at high resolution of glucose transporters will facilitate an understanding of their specific functions and regulation and may also be useful in the rational design of drugs intended to augment or inhibit their activity, as may be beneficial. Unfortunately, homology modeling of the structure of mammalian glucose transporters based on bacterial MFS templates is of extremely limited value due to the lack of sequence similarity [44]. Although the MFS is the largest superfamily of membrane transport proteins with over 5000 known members [45–47] and (www.tcdb.org), the structure of only 3 of these proteins has been determined, and these are all at moderate resolution (3–4 Å) and from bacterial sources. A major barrier to crystallizing mammalian membrane proteins in general has been the inability to purify sufficient quantities of non-denatured, non-glycosylated proteins to conduct crystallization assays. Recently however, the yeast, P. pastoris, has proven to be a useful system to express and purify milligram quantities of mammalian membrane proteins [48]. In this report we describe a relatively simple affinity-tag procedure using P. pastoris for the purification of functional mammalian glucose transporters. A very similar procedure has proven to be highly successful for the purification of several bacterial membrane transporters [4, 6, 49], but we are unaware of any eukaryotic member of the MFS for which it has been successively applied in conjunction with the purification of a protein ectopically expressed in P. pastoris membranes. This procedure should ultimately lead to the successful production of highly ordered protein crystals suitable for high-resolution X-ray diffraction analysis as well as proteins suitable for structural analysis using other biophysical approaches.
ACKNOWLEDGMENTS
We thank Haowey Song from DRTC core facility at Washington University for the mass spectrometry experiments. This work was supported by National Institutes of Health Grant R01 DK43695.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nie Y, Smirnova I, Kasho V, Kaback HR. Energetics of ligand-induced conformational flexibility in the lactose permease of Escherichia coli. J Biol Chem. 2006;281:35779–35784. doi: 10.1074/jbc.M607232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins (Review) Mol Membr Biol. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 4.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 7.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uldry M, Thorens B. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch. 2004;447:480–489. doi: 10.1007/s00424-003-1085-0. [DOI] [PubMed] [Google Scholar]
- 9.Kahn BB. Lilly lecture 1995. Glucose transport: pivotal step in insulin action. Diabetes. 1996;45:1644–1654. doi: 10.2337/diab.45.11.1644. [DOI] [PubMed] [Google Scholar]
- 10.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 11.De Vivo DC, Wang D, Pascual JM, Ho YY. Glucose transporter protein syndromes. Int Rev Neurobiol. 2002;51:259–288. doi: 10.1016/s0074-7742(02)51008-4. [DOI] [PubMed] [Google Scholar]
- 12.Santer R, Schneppenheim R, Dombrowski A, Gotze H, Steinmann B, Schaub J. Fanconi-Bickel syndrome--a congenital defect of the liver-type facilitative glucose transporter. SSIEM Award. Society for the Study of Inborn Errors of Metabolism. J Inherit Metab Dis. 1998;21:191–194. doi: 10.1023/a:1005379013406. [DOI] [PubMed] [Google Scholar]
- 13.Hruz PW, Murata H, Qiu H, Mueckler M. Indinavir induces acute and reversible peripheral insulin resistance in rats. Diabetes. 2002;51:937–942. doi: 10.2337/diabetes.51.4.937. [DOI] [PubMed] [Google Scholar]
- 14.Graybill C, van Hoek AN, Desai D, Carruthers AM, Carruthers A. Ultrastructure of human erythrocyte GLUT1. Biochemistry. 2006;45:8096–8107. doi: 10.1021/bi060398x. [DOI] [PubMed] [Google Scholar]
- 15.Hruz PW, Mueckler MM. Structural analysis of the GLUT1 facilitative glucose transporter (review) Mol Membr Biol. 2001;18:183–193. doi: 10.1080/09687680110072140. [DOI] [PubMed] [Google Scholar]
- 16.Alisio A, Mueckler M. Relative proximity and orientation of helices 4 and 8 of the GLUT1 glucose transporter. J Biol Chem. 2004;279:26540–26545. doi: 10.1074/jbc.M402303200. [DOI] [PubMed] [Google Scholar]
- 17.Mueckler M, Makepeace C. Transmembrane segment 12 of the Glut1 glucose transporter is an outer helix and is not directly involved in the transport mechanism. J Biol Chem. 2006 doi: 10.1074/jbc.M608158200. [DOI] [PubMed] [Google Scholar]
- 18.Mueckler M, Makepeace C. Model of the Exofacial Substrate-Binding Site and Helical Folding of the Human Glut1 Glucose Transporter Based on Scanning Mutagenesis. Biochemistry. 2009 doi: 10.1021/bi900521n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blodgett DM, Graybill C, Carruthers A. Analysis of glucose transporter topology and structural dynamics. J Biol Chem. 2008;283:36416–36424. doi: 10.1074/jbc.M804802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blodgett DM, Carruthers A. Quench-flow analysis reveals multiple phases of GluT1-mediated sugar transport. Biochemistry. 2005;44:2650–2660. doi: 10.1021/bi048247m. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin SA, Baldwin JM, Lienhard GE. Monosaccharide transporter of the human erythrocyte. Characterization of an improved preparation. Biochemistry. 1982;21:3836–3842. doi: 10.1021/bi00259a018. [DOI] [PubMed] [Google Scholar]
- 22.Boulter JM, Wang DN. Purification and characterization of human erythrocyte glucose transporter in decylmaltoside detergent solution. Protein Expr Purif. 2001;22:337–348. doi: 10.1006/prep.2001.1440. [DOI] [PubMed] [Google Scholar]
- 23.Kasahara M, Hinkle PC. Reconstitution and purification of the D-glucose transporter from human erythrocytes. J Biol Chem. 1977;252:7384–7390. [PubMed] [Google Scholar]
- 24.Graham TE, Kahn BB. Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm Metab Res. 2007;39:717–721. doi: 10.1055/s-2007-985879. [DOI] [PubMed] [Google Scholar]
- 25.Murata H, Hruz PW, Mueckler M. The mechanism of insulin resistance caused by HIV protease inhibitor therapy. J Biol Chem. 2000;275:20251–20254. doi: 10.1074/jbc.C000228200. [DOI] [PubMed] [Google Scholar]
- 26.Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, Allard WJ, Lienhard GE, Lodish HF. Sequence and structure of a human glucose transporter. Science. 1985;229:941–945. doi: 10.1126/science.3839598. [DOI] [PubMed] [Google Scholar]
- 27.James DE, Strube M, Mueckler M. Molecular cloning and characterization of an insulin-regulatable glucose transporter. Nature. 1989;338:83–87. doi: 10.1038/338083a0. [DOI] [PubMed] [Google Scholar]
- 28.Mueckler M, Weng W, Kruse M. Glutamine 161 of Glut1 glucose transporter is critical for transport activity and exofacial ligand binding. J Biol Chem. 1994;269:20533–20538. [PubMed] [Google Scholar]
- 29.Marshall BA, Murata H, Hresko RC, Mueckler M. Domains that confer intracellular sequestration of the Glut4 glucose transporter in Xenopus oocytes. J Biol Chem. 1993;268:26193–26199. [PubMed] [Google Scholar]
- 30.Holman GD. Side-specific photolabelling of the hexose transporter. Biochem Soc Trans. 1989;17:438–440. doi: 10.1042/bst0170438. [DOI] [PubMed] [Google Scholar]
- 31.Enkvetchakul D, Jeliazkova I, Bhattacharyya J, Nichols CG. Control of inward rectifier K channel activity by lipid tethering of cytoplasmic domains. J Gen Physiol. 2007;130:329–334. doi: 10.1085/jgp.200709764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:U373–U376. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 33.Zoccoli MA, Baldwin SA, Lienhard GE. The monosaccharide transport system of the human erythrocyte. Solubilization and characterization on the basis of cytochalasin B binding. J Biol Chem. 1978;253:6923–6930. [PubMed] [Google Scholar]
- 34.Kayano T, Burant CF, Fukumoto H, Gould GW, Fan YS, Eddy RL, Byers MG, Shows TB, Seino S, Bell GI. Human facilitative glucose transporters. Isolation, functional characterization, and gene localization of cDNAs encoding an isoform (GLUT5) expressed in small intestine, kidney, muscle, and adipose tissue and an unusual glucose transporter pseudogene-like sequence (GLUT6) J Biol Chem. 1990;265:13276–13282. [PubMed] [Google Scholar]
- 35.Wellner M, Monden I, Keller K. From triple cysteine mutants to the cysteine-less glucose transporter GLUT1: a functional analysis. FEBS Lett. 1995;370:19–22. doi: 10.1016/0014-5793(95)00783-6. [DOI] [PubMed] [Google Scholar]
- 36.Koumanov F, Yang J, Jones AE, Hatanaka Y, Holman GD. Cell-surface biotinylation of GLUT4 using bis-mannose photolabels. Biochem J. 1998;330(Pt 3):1209–1215. doi: 10.1042/bj3301209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldwin JM, Lienhard GE, Baldwin SA. The monosaccharide transport system of the human erythrocyte. Orientation upon reconstitution. Biochim Biophys Acta. 1980;599:699–714. doi: 10.1016/0005-2736(80)90211-4. [DOI] [PubMed] [Google Scholar]
- 38.Widdas WF. Facilitated transfer of hexoses across the human erythrocyte membrane. J Physiol. 1954;125:163–180. doi: 10.1113/jphysiol.1954.sp005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueckler M. Facilitative glucose transporters. Eur J Biochem. 1994;219:713–725. doi: 10.1111/j.1432-1033.1994.tb18550.x. [DOI] [PubMed] [Google Scholar]
- 40.Olson AL, Pessin JE. Structure, function, and regulation of the mammalian facilitative glucose transporter gene family. Annu Rev Nutr. 1996;16:235–256. doi: 10.1146/annurev.nu.16.070196.001315. [DOI] [PubMed] [Google Scholar]
- 41.Zhao FQ, Keating AF. Functional properties and genomics of glucose transporters. Curr Genomics. 2007;8:113–128. doi: 10.2174/138920207780368187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Vivo DC, Leary L, Wang D. Glucose transporter 1 deficiency syndrome and other glycolytic defects. J Child Neurol. 2002;17 Suppl 3:3S15–3S23. discussion 13S24-15. [PubMed] [Google Scholar]
- 43.Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987;235:1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- 44.Lemieux MJ. Eukaryotic major facilitator superfamily transporter modeling based on the prokaryotic GlpT crystal structure. Mol Membr Biol. 2007;24:333–341. doi: 10.1080/09687680701496507. [DOI] [PubMed] [Google Scholar]
- 45.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 48.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 49.Jiang YX, Lee A, Chen JY, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]