Abstract
The creative use of gnotobiotic animals, coupled with the development of modern metagenomic sequencing platforms and metabolomic profiling of biospecimens, has bestowed new insight into the remarkably intricate interface between the host mammal and its resident microbiota. As mutual benefactors, each partner exhibits evidence of adaptation: the host provides a hospitable habitat, giving consideration to its own species of origin, diet, genotype, geographical location, presence or absence of disease, and use of medications; the microbiota, in turn, configures its constituency, collective genome (microbiome), transcriptome, and metabolome to optimally suit its ecological niche. In this review, we discuss the mechanisms through which the gut microbiota and its host collaborate to regulate lipid metabolism, thereby influencing the metabolic response to nutrient intake and ultimately, the development of obesity and associated diseases such as lipotoxicity. These studies therefore demonstrate that the gut microbiota is an `environmental' influence whose synergistic interdependence with its host strongly suggests that we are in fact `supraorganisms'.
Keywords: gut microbiota, lipid metabolism, obesity
Introduction
During the past ~160 million years mammals have co-evolved with a vast and diverse microbial community that colonizes our cutaneous and mucosal surfaces. Most of these microorganisms reside within our gastrointestinal tracts, and their constituency is determined by host phylogeny and diet [1, 2]. The gut microbiota exhibits profound influence over a large number of host (patho)physiological processes, including vitamin biosynthesis, muscosal immunity, inflammatory bowel disease, and colorectal cancer (reviewed in [3–7]). Over the past five years, animal and human studies have revealed remarkable microbial influences on host metabolism, energy utilization, and storage. In this review, we will explore the interdependent relationship between the host and its microbial community in their collaborative effort to coordinate extraordinarily vast metabolic processes.
The microbial community living within its mammalian host consists of an enormous number of microorganisms (10–100 trillion), reaching a cell number that is potentially an order of magnitude greater than all the eukaryotic cells that comprise the host itself [8]. The collective genomes of these resident microorganisms (termed the microbiome) encode a vast arsenal of gene products, which collectively provide a diverse range of biochemical and metabolic activities that complements the host eukaryotic genome [9]. These activities potentially limit the requirement for the evolution of functions encoded by the host's genome, and hence strongly support the notion that we are in fact `supraorganisms' [10].
For the microbiota to thrive, it must consistently meet the dynamic needs of its individual constituents, the community as a whole, and the host (upon which it is dependent for its existence). Thus, it must constantly adapt to its ecological niche, achieving a balance between what it gives to and receives from a host environment that is likely to vary widely within and among individual hosts, based on common factors such as diet, illness, and the use of antibiotics. The microbiota has a number of mechanisms through which it can adapt to an inconsistent niche. Unlike its host's genome, the microbiome can change, often quite rapidly, though modifications in the configuration of the microbial community. And like its host, the microbiota can modify its transcriptome, metabolome, and proteome. The field of metagenomics has transformed our understanding of the structure and remarkable transformative capacity of the microbiota. The family of pathophysiological states that includes obesity has provided a demonstrative window into the reciprocal relationship between host and its dynamic symbionts.
1. The gut microbiota alters its configuration in obesity
Culture-dependent methods for enumerating the microbiota revealed a relationship between gut microbes and obesity as early as 1981, when changes in the microbiota were recorded in patients undergoing gastric bypass surgery [11]. Surgically-induced obesity in rats (via lesion of the ventromedial hypothalamic nucleus) was also associated with changes in microbial ecology [12]. These studies utilized elegant but incomplete methods to survey the microbiota: culture-based methods reflect a minor proportion of the microbial community. The ability to acquire a census of the microbiota and characterize the microbiome have been revolutionized by utilization of molecular based methods based on the ribosomal 16S sequence. Enumerative/comparative platforms include fluorescence in situ hybridization (FISH), competitive PCR, quantitative PCR, restriction fragment length polymorphism (RFLP) mapping, and denaturing (or temperature) gradient gel electrophoresis (DGGE/TGGE) [13]. High-throughput sequencing has spawned the development of useful phylogenetic microarrays [14], but the most robust method to capture the diversity of the microbiota has become multiplex pyrosequencing [15].
Highly-parallel pyrosequencing technology allows the procurement of a vast amount of sequencing data in single runs. The use of these platforms has been widely applied to two methods for characterizing the microbiome: first, enumeration of the microbiota via sequencing of variable regions of 16S rRNA-encoding genes, which are retrieved by PCR using primers designed against flanking conserved regions [16]. This approach has revealed that while diversely represented at the microbial species level among different humans or among vertebrates, the vast majority of mammalian bacterial communities can be classified within two bacterial divisions, or phyla: the Bacteroidetes and the Firmicutes [1, 2]. A second method that allows characterization of the microbiome is `shotgun' sequencing, which samples the microbiome's composite sequence, without rigorously assigning sequences to a discrete member of the community [9]. A third method of using high-throughput sequencing to characterize the microbiome is through the definition of the transcriptome, a `post-array' platform that will become used widely in the near future [17].
Using the former two approaches, mouse and human models have revealed that the gut microbiota robustly and rapidly shifts its membership and representation at the gene content level in response to host adiposity and nutrient environment. Leptin-deficient (ob/ob) C57Bl/6 mice, which are genetically predisposed to develop severe obesity, harbor a microbiota that possesses a significantly higher percentage of Firmicutes, and a correspondingly lower percentage of Bacteroidetes, than their wild-type littermates [18]. Provision of a high-calorie, high-fat/simple carbohydrate, obesity-inducing `Western' diet to wild-type mice effects analogous shifts in the Firmicutes and Bacteroidetes divisions, with the provocation of a `bloom' in the clade Mollicutes, which belongs to the Firmicutes [19]. The same increase in Firmicutes/Bacteroidetes ratio has also been observed in obese humans, compared to lean controls; furthermore, this representation exhibits plasticity, as humans losing weight exhibit a reduction in this ratio irrespective of the type of diet (fat or carbohydrate restricted [20]). Mono- and dizygotic human twins concordant for obesity reveal closely-related microbiomes with the fractional representation of Bacteroidetes directly correlated with leanness [21]. While many features of these constituencies appear to be plastic, their effects could be long-lasting: a comparative study using FISH revealed that the childhood representation of Bifidobacteria (which belong to the phylum Actinobacteria) and Staphylococcus aureus may inversely and directly, respectively, predict the development of adulthood obesity [22].
Results of shotgun sequencing of the microbiome support the hypothesis that shifts in microbial ecology effect functional shifts in the microbiota that could contribute to the perpetuation of the obese phenotype. Compared to lean wild-type littermates, the metagenome of ob/ob or Western diet-fed wild-type mice is enriched in genes that encode the catabolism of complex polysaccharides, including glycoside hydrolases [19, 23]. Furthermore, the microbiota from ob/ob possesses biochemical evidence that it may contribute to the predisposition to obesity: cecal contents from ob/ob mice are enriched in short-chain fatty acids (SCFAs), the products of fermentation of complex polysaccharides, compared to their wild-type littermates. Transplantation of the microbiota from either conventionally-raised normal chow-fed ob/ob donor mice, or Western diet-fed wild-type mice to GF wild-type C57Bl/6 recipient mice causes a greater increase in adiposity than that caused by transplantation of a microbiota from conventionally-raised wild-type littermate donors that had been fed standard chow [19, 23].
In addition to the effects of host genotype and diet, it is also important to consider the ability of antibiotics to manipulate the microbiota, and how this influences obesity and obesity-related disease. A recent 16S rRNA enumeration of human fecal samples revealed that a five day course of orally-administered ciprofloxacin decreased the diversity of the community, to varying extents among treated subjects. Recovery of the community was evident within four weeks, but some taxa had not re-appeared as remotely as six months after treatment [24]. The effects of antibiotic treatment are important to consider: treatment with a combination of norfloxacin and ampicillin yields improvement in fasting serum glucose and tolerance to an oral glucose challenge in ob/ob and diet-induced obesity mice [25, 26]. Some of these changes may be attributable to reductions in circulating lipopolysaccharide (LPS) levels, which correlate with the development of obesity and insulin resistance [27–29].
The most effective therapy to invoke weight loss for refractory obesity in humans is bariatric surgery [30]. Not surprisingly, 16S rRNA-based enumeration of stool reveals marked shifts in microbial ecology of post-gastric bypass patients: obese subjects exhibit enrichment in hydrogen gas-producing bacteria and hydrogen-consuming methanogenic archeons, which may increase energy availability to the obese host, whereas lean and post-bypass subjects exhibit much lower numbers of these organisms [31–33]. The use of postoperative probiotics to further `stack the deck' in favor of reducing the abundance of hydrogen metabolizing organisms may provide incremental improvement in bariatric surgery patients [34].
2. The gut microbiota shapes the host metabolome
Much like metagenomics, the family of approaches used in metabolomics [35] has also revolutionized our understanding host-microbial relationships [36]. The two general approaches – proton nuclear magnetic resonance (NMR) and mass spectrometry (MS) – are applied to biofluids and extracted tissue to generate high density spectra whose interpretation is dramatically enhanced by computational algorithms and chemical database libraries. Mass spectrometry is coupled to chromatographic separation, either in the vapor (gas chromatography, GC) or liquid (LC) phase, and ionization of parental species is effected by one of several methods, such or electron impact or electrospray ionization (see [37, 38] for review).
Comparative metabolomic analysis of biospecimens derived from GF versus colonized mice, via proton NMR has revealed a large number of metabolites whose modification in the host is attributable to the microbiota. Of note, proton NMR profiling of GF mice [39] reveals that levels of phosphocholine and glycine are higher in the livers of GF mice, as are levels of bile acids in the gut: this latter finding is not surprising since the gut microbiota is responsible for bile acid deconjugation, which allows excretion [40]. The metabolite hippurate (benzoyl glycine), a metabolite of phenylalanine, generated through coordinate reactions encoded by the genomes of gut microbes and host, was found at reduced concentrations in the urine of GF mice [39]. Urinary hippurate may be an important biomarker of hypertension in humans: urinary levels have been reported to be inversely proportional to blood pressure [41].
The metabolic significance of the ability of the microbiome to influence phosphatidylcholine metabolism was demonstrated by the use of proton NMR-based metabolomics [42]. The mouse strain 129S6 is prone to develop an insulin resistant, non-alcoholic fatty liver disease phenotype, particularly when fed a high-fat diet, compared to the Balb/C strain. Unbiased proton NMR profiling revealed that circulating phosphatidylcholine levels were higher in high-fat diet fed Balb/C (relatively insulin sensitive) mice than in 129S6 mice. In addition, high levels of urinary methylamines, which are generated via choline metabolism by microbial enzymes, were detected in high-fat diet fed 129S6 but not Balb/C mice. Therefore, the ability of the microbiome in 129S6, presumably distinct from that in Balb/C, to divert choline from host metabolic pathways could create supraorganismal metabolic cross-talk that mimics a choline-deficient diet's ability to induce a fatty liver, insulin-resistant phenotype. The exploration of the microbial architecture, both at the level of taxonomy and gene content, would be an important mechanistic step to underscore the importance of these findings. These findings have been extended to humans, which showed a relationship between urinary methylamines and community structure [43].
A recently published study employed complementary separation techniques (GC and LC) coupled with diverse ionization platforms to chemically profile plasma from GF and colonized mice [44]. The differences were striking: hundreds of metabolite `features' (which includes metabolites, isotopic variations of metabolites, and a small proportion of artifactual adduct ions) were unique to the GF or colonized state (mostly the latter), and approximately 10% of those features present in both conditions were changed by ≥50% in concentration between conditions. Notable changes were observed in the indolamine pathway (tryptophan and serotonin metabolism) and in phase II (conjugation of sulfate and glycine) drug/metabolite sym-xenobiotic pathways, each set of changes illustrating a very important point: mutualistic metabolite exchange between enzymatic activities encoded within the host genome and those encoded by the microbiome. [45].
3. The gut microbiota promotes increased energy harvest from the gut
The gut microbiota may increase energy absorption from the gut by indirect mechanisms (e.g. gut transit time) or direct mechanisms: (1) increasing glucose uptake, (2) fermenting polysaccharides to SCFAs, and (3) modulating lipid absorption. Whereas glucose and acetate are substrates for lipogenesis, dietary triglycerides provide peripheral organs with fatty acids. In order to accommodate the increased nutrient absorption, colonization of the gut is associated with a dramatic increase in microvascular density in the lamina propria of small intestinal villi [46].
3.1 Modulation of glucose absorption
Sugars are ingested in the form of mono- and disaccharides and more complex carbohydrates. The gut microbiota is important for degradation of polysaccharides, but can also directly affect monosaccharide (e.g. glucose) absorption. Colonization of GF mice leads to a two-fold increase in intestinal glucose absorption [47]. Increased glucose absorption is associated with elevated small intestinal Glut1 expression following colonization of GF mice with Bacteroidetes thetaiotaomicron, a prominent human gut symbiont [48]. The underlying molecular mechanism for microbial regulation of intestinal glucose transporters remains to be identified.
3.2 Fermentation of short chain fatty acids
Many dietary complex polysaccharides escape digestion in the small intestine and are metabolized by the colonic gut microbiota [49]. Although ruminants produce the highest amount of SCFAs in the animal kingdom, the human gut microbiota contributes to significant levels of SCFAs in the portal circulation (~375 μmol/l). Acetate, propionate, and butyrate are the main SCFAs and are commonly found at molar ratios ranging from ~75:15:10 to 40:40:20 [50]. Highly fermentable polysaccharides tend to result in high propionate:acetate ratio, whereas high fiber diets increases the proportion of acetate [49]. Most microbially-produced butyrate, and significant amounts of propionate, are used by the colonocytes as energy substrates and for synthesis of membrane lipids. The remaining propionate is used for gluconeogenesis in the liver, and it has been estimated that ~7% of the synthesized glucose can be traced to colonic propionate [51]. Acetate is used by the liver (humans) and liver and adipose tissue (rodents) as a substrate for lipogenesis. Provided sufficient reducing power in the form of NADPH, the calculated efficiency for acetate for lipogenesis is high (~80%) [49]. As noted above, obese mice [19, 23] exhibit increased capacity for gut luminal SCFA production, suggesting that the gut microbiota may affect lipid metabolism and obesity by increasing substrates for energy metabolism in the liver and peripheral tissues.
3.3 Lipid absorption from the gut
Dietary lipids are absorbed in the proximal small intestine. After a meal, bile acids are released into the duodenum, which facilitates emulsification and absorption of dietary lipids. Bacteria modify the bile acids by deconjugation and hydroxylation in the distal intestine, which affects their hydrophobicity [52, 53]. Thus an altered intestinal bile acid composition may affect lipid absorption. Surprisingly, administration of a lipid bolus to GF and conventionally raised mice did not demonstrate altered lipid absorption in the absence of a gut microbiota (Velagapudi, Hezaveh, Reigstad, Gopalacharyulu, Felin, Yetukuri, Borén, Orešič, and F. B. submitted). This may in part be explained by the ability of the microbiota to increase gut transit time [54, 55], which could compensate for relatively reduced absorption rates compared to GF mice.
4. Microbial modulation of peripheral lipid metabolism
In addition to effects on energy absorption from the gut, the microbiota affects lipid composition and metabolism in serum, liver, and peripheral organs. These effects occur indirectly, through modulation of gut hormones, and directly, e.g. through microbial modification of gut-derived lipids.
4.1 Serum
Triglycerides are packaged and transported in serum as (1) chylomicrons, which transport dietary triglycerides from the intestine; and (2) very low-density lipoproteins (VLDL), which transport hepatic triglycerides from the liver to adipose tissue [56]. GF mice have increased serum triglyceride levels after a short 4 hour fast ([57]. Lipoprotein lipase (LPL) is a key regulator of fatty acid release from triglyceride rich lipoproteins in muscle, heart, and fat, and the enzyme is regulated by nutritional status; fasting reduces and refeeding increases enzyme activity [58, 59]. Increased adipocyte LPL activity leads to increased cellular uptake of fatty acids and may affect lipid storage. Interestingly, the gut microbiota affects LPL activity and lipid clearance, at least in part, by down-regulating intestinal expression of a potent LPL inhibitor, Angiopoietin-like protein 4 (Angptl4; also known as fasting-induced adipose factor), which results in increased LPL activity [47]. Thus it is likely that the gut microbiota affects serum triglyceride levels predominantly by modulating triglyceride clearance, which is supported by our finding that we did not observe any significant differences in the rate of free fatty acid absorption following an oral bolus of olive oil in overnight fasted GF and conventionally raised mice maintained on a normal chow diet [57].
4.2 Liver
The portal vein drains the gut and transports nutrients from the intestine to the liver, which is the primary organ responsible for de novo synthesis of triglycerides. Increased intestinal glucose absorption in colonized mice can stimulate lipogenesis either directly, through the transcription factor carbohydrate response element binding protein (ChREBP) [60], or by increased insulin levels through sterol response element binding protein 1 (SREBP-1) [61]: colonization of GF mice induces hepatic expression of both ChREBP and SREBP-1. In addition, expression of downstream target genes acetyl-CoA carboxylase (Acc1) and fatty acid synthase (Fas), which are rate-limiting enzymes for lipogenesis, are also increased by colonization. The increase in de novo synthesis is reflected by increased levels of liver triglycerides in colonized mice [47].
4.3 Muscle – skeletal and cardiac
The microbiota dynamically shapes the metabolic reactions within the host gut and liver, which is reflected in the circulating blood, and excreted urine. These changes in the metabolite profile may influence the development of obesity and insulin resistance. How can skeletal muscle and heart serve as reporters of these microbial activities? Very little experimentation has yet addressed this question, which is important, because it will allow the gut microbiota to teach us previously unrecognized mechanisms through which metabolic disease is initiated and perpetuated. Skeletal muscle (gastrocnemius) of Western diet-fed GF mice exhibits evidence of increased rates fatty acid oxidation compared to that of colonized mice: in GF mice, levels of AMP, phosphorylated AMP-activated protein kinase (α subunit) levels, and consequently, phosphorylated acetyl-CoA carboxylase levels are increased, leading to increased carnitine palmitoyl transferase activity [57].
Until recently, little was known about the effects of the microbiota on the myocardium. Relative to body size, the heart of the GF mouse or rat is slightly small, compared to colonized animals [62, 63]. Given the ability of the microbiota to influence host metabolism and insulin resistance, plus the profound relationship among insulin resistance, nutrient delivery, myocardial substrate metabolism, and cardiac phenotypes [64–66], it is not surprising that the myocardium and its substrate utilization are influenced by the relationship between the coordinated metabolic activities encoded by the microbiome and those by the host genome. Because the heart is entirely dependent on the gut and liver for nutrient acquisition, processing, and delivery, it serves as an excellent reporter of integrated metabolic homeostasis.
Nutrient deprivation induces biochemical and physiological responses across organ systems, requiring orchestrated and complementary responses. After only 24 hours of nutrient deprivation in mice that had been maintained on standard polysaccharide-rich chow, the fractional representation of Bacteroidetes within the cecal microbiota of mice dramatically increases, with a concomitant decrease in the proportion of Firmicutes present, demonstrating the microbiota's remarkably dynamic plasticity [67]. After a 24h fast, the livers of GF mice mount a diminished ketogenic response, compared to colonized mice, which partially deprives the heart of an important myocardial substrate pool during nutrient deprivation. To maintain cardiac functional capacity, hearts of starved GF mice compensate by increasing glucose oxidation. When GF mice are `forced' to generate ketone bodies by feeding them a very low carbohydrate ketogenic diet, their reduction in myocardial mass is reversed [67]. These studies indicate that gut microbial manipulation influences metabolism among organ systems.
4.4 Adipose tissue
The presence of a gut microbiota does not alter expression of key adipogenic and lipogenic genes in adipose tissue. Instead, conventionally raised mice have decreased LPL activity and increased fat storage due to adipocyte hypertrophy. The increased fat storage is positively correlated to leptin levels, which may indicate a state of relative leptin resistance [47]. In the absence of leptin action overnutrition causes accumulation of triglycerides in nonadipose tissues with resulting lipotoxicity, which may lead to diseases such as type 2 diabetes, cardiomyopathy, and insulin resistance [68].
Obesity and increased adipocyte size are associated with macrophage infiltration and elevated levels of inflammatory markers [69, 70]. As discussed above, a recent study indicates that LPS, which is derived from the outer membrane of Gram negative bacteria, may contribute to increased adipose mass and elevated expression of pro-inflammatory markers in adipose tissue [29]. Thus both increased nutrient absorption and elevated levels of LPS may contribute to increased adipose mass and thus suggest a link between the gut microbiota, lipid accumulation, and inflammation. Further support for this association derives from the increased expression of serum amyloid A 3 (SAA3) in adipose and colonic tissue of conventionally raised mice [71]. Adipose tissue-derived SAA3 confers monocyte chemotactic activity [72] and may play a role in metabolic inflammation associated with obesity and insulin resistance [73].
5. Conclusion and perspective
The tools offered by insightfully-designed metabolic and physiological, metagenomic, and metabolomic studies have exponentially expanded our insight into metabolic host-microbial mutualism, plus how these shared relationships are important players in the development of obesity and obesity-related disease. Critical questions are raised by these findings. Perhaps most importantly, what host factors, in addition to diet, determine the constituency of the microbiota in an individual host? Host species is a critical influence [1, 74], but critical differences in community structure may exist based on ethnicity within a species [43]. Clearly, features of host muscosal nutrient environment and provision [75], as well as host mucosal immunity [76] collaboratively orchestrate a community structure that confers maximal mutual beneficence. It will also be important to advance our ability to identify individual species, or more likely, microbial sub-communities, including those in scant minority, that nonetheless can be robustly correlated to biomolecular signatures within host biological specimens. Resolution of the determinants and consequences of spatio-temporal variations in community structure in a given host will undoubtedly advance our understanding of the mechanisms that establish a habitat within the gut. How does the microbiome change in a given individual over time? How does the constituency of the microbiota in the proximal small intestine differ from that of the colon, and what are the metabolic consequences?
Metagenomic analyses have revealed that there may be functional redundancy within the microbiome [24], and that a `core microbiome' exists in all of us, not necessarily at the level of microbial species, but rather at the level of collective function, as predicted by the assembled genes within the microbiome [21]. The functional capacity of the microbiome is therefore determined by host genetic and environmental factors: the ability to manipulate the microbiome, using probiotics, prebiotics, [77–79] or perhaps genetically, using bacteriophage, could provide a set of immensely powerful reagents that encourage therapeutic adaptation of the core microbiome into a maximally mutualistic partner to our genome in conditions such as inflammatory bowel disease and obesity.
Acknowledgements
The authors express their gratitude to Dr. Jeffrey Gordon for invaluable mentorship and support, Anna Hallén for producing Fig 1. Work in the researcher's laboratory is supported by the Human Frontier of Science Program (RGY64/2008), Swedish Research Council (K2007-65X-20421-01-04), Swedish Foundation for Strategic Research (A3 05:207c), EU-funded ETHERPATHS project (FP7-KBBE-222639, http://www.etherpaths.org/), the Åke Wiberg, Lars Hierta's, Nanna Svartz, Fredrik and Ingrid Thurings Foundations, and a LUA-ALF grant from Västra Götalandsregionen to F.B and NIH-K08DK073282 and a planning grant UL1 RR024992 to P.C.
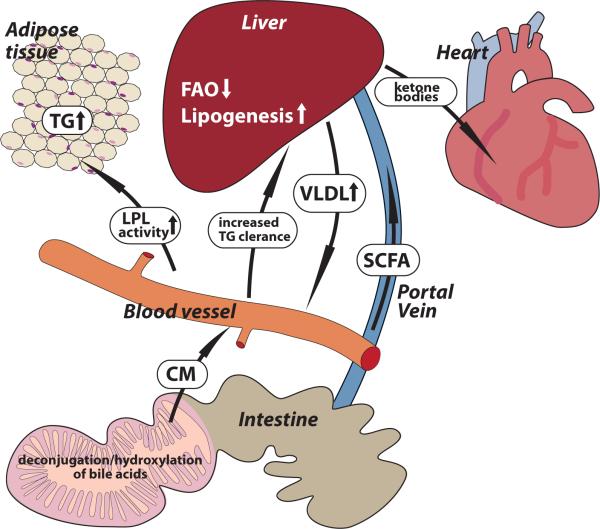
Fig. 1. The gut microbiota affects host lipid metabolism.
The gut microbiota affects lipid uptake and chylomicron formation by modulating bile acid transformations and gut transit time. Furthermore the gut microbiota is instrumental in fermenting complex polysachharides to short chain fatty acids that may act as a lipogenic substrates in the liver. In addition, the gut microbiota suppresses expression of Angiopoietin-like protein 4 (Angptl4) in the intestinal mucosa, which increases LPL mediated triglyceride storage in adipose tissue and reduces serum triglyceride levels. Angptl4 also promote fatty acid oxidation by a yet unidentified receptor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- [4].Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell host & microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O'Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Current opinion in gastroenterology. 2008;24:51–58. doi: 10.1097/MOG.0b013e3282f323f3. [DOI] [PubMed] [Google Scholar]
- [6].Packey CD, Sartor RB. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. Journal of internal medicine. 2008;263:597–606. doi: 10.1111/j.1365-2796.2008.01962.x. [DOI] [PubMed] [Google Scholar]
- [7].Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic Analysis of the Human Distal Gut Microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lederberg J. Infectious History. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- [11].Bjorneklett A, Viddal KO, Midtvedt T, Nygaard K. Intestinal and gastric bypass. Changes in intestinal microecology after surgical treatment of morbid obesity in man. Scand J Gastroenterol. 1981;16:681–687. doi: 10.3109/00365528109182030. [DOI] [PubMed] [Google Scholar]
- [12].Nishizawa Y, Imaizumi T, Tanishita H, Yano I, Kawai Y, Mormii H. Relationship of fat deposition and intestinal microflora in VMH rats. International journal of obesity. 1988;12:103–110. [PubMed] [Google Scholar]
- [13].Deng W, Xi D, Mao H, Wanapat M. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Molecular biology reports. 2008;35:265–274. doi: 10.1007/s11033-007-9079-1. [DOI] [PubMed] [Google Scholar]
- [14].Paliy O, Kenche H, Abernathy F, Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Poretsky RS, Gifford S, Rinta-Kanto J, Vila-Costa M, Moran MA. Analyzing gene expression from marine microbial communities using environmental transcriptomics. J Vis Exp. 2009 doi: 10.3791/1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell host & microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes linked to obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- [21].Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- [23].Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- [24].Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chou CJ, Membrez M, Blancher F. Gut decontamination with norfloxacin and ampicillin enhances insulin sensitivity in mice. Nestle Nutrition workshop series. 2008;62:127–140. doi: 10.1159/000146256. [DOI] [PubMed] [Google Scholar]
- [26].Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Mace K, Chou CJ. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- [27].Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Current pharmaceutical design. 2009;15:1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- [28].Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- [29].Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- [30].Buchwald H, Avidor Y, Braunwald E, Jensen MD, Pories W, Fahrbach K, Schoelles K. Bariatric surgery: a systematic review and meta-analysis. Jama. 2004;292:1724–1737. doi: 10.1001/jama.292.14.1724. [DOI] [PubMed] [Google Scholar]
- [31].Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Nat Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Woodard GA, Encarnacion B, Downey JR, Peraza J, Chong K, Hernandez-Boussard T, Morton JM. Probiotics Improve Outcomes After Rouxen-Y Gastric Bypass Surgery: A Prospective Randomized Trial. J Gastrointest Surg. 2009 doi: 10.1007/s11605-009-0891-x. [DOI] [PubMed] [Google Scholar]
- [35].Blow N. Metabolomics: Biochemistry's new look. Nature. 2008;455:697–700. doi: 10.1038/455697a. [DOI] [PubMed] [Google Scholar]
- [36].Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- [37].Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. Journal of the American College of Cardiology. 2008;52:117–123. doi: 10.1016/j.jacc.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Feng X, Liu X, Luo Q, Liu BF. Mass spectrometry in systems biology: an overview. Mass spectrometry reviews. 2008;27:635–660. doi: 10.1002/mas.20182. [DOI] [PubMed] [Google Scholar]
- [39].Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes E, Nicholson JK. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular systems biology. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wostmann BS. The germfree animal in nutritional studies. Ann Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- [41].Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dumas ME, Wilder SP, Bihoreau MT, Barton RH, Fearnside JF, Argoud K, D'Amato L, Wallis RH, Blancher C, Keun HC, Baunsgaard D, Scott J, Sidelmann UG, Nicholson JK, Gauguier D. Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nat Genet. 2007;39:666–672. doi: 10.1038/ng2026. [DOI] [PubMed] [Google Scholar]
- [46].Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- [49].Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- [50].Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 1997;21:357–365. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- [51].Ford EJ, Simmons HA. Gluconeogenesis from caecal propionate in the horse. Br J Nutr. 1985;53:55–60. doi: 10.1079/bjn19850010. [DOI] [PubMed] [Google Scholar]
- [52].Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- [53].Wostmann BS. The germfree animal in nutritional studies. Annu Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- [54].Abrams GD, Bishop JE. Effect of the normal microbial flora on gastrointestinal motility. Proc Soc Exp Biol Med. 1967;126:301–304. doi: 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- [55].Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Olofsson SO, Asp L, Boren J. The assembly and secretion of apolipoprotein B-containing lipoproteins. Curr Opin Lipidol. 1999;10:341–346. doi: 10.1097/00041433-199908000-00008. [DOI] [PubMed] [Google Scholar]
- [57].Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bergo M, Olivecrona G, Olivecrona T. Forms of lipoprotein lipase in rat tissues: in adipose tissue the proportion of inactive lipase increases on fasting. Biochem J. 1996;313(Pt 3):893–898. doi: 10.1042/bj3130893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Preiss-Landl K, Zimmermann R, Hammerle G, Zechner R. Lipoprotein lipase: the regulation of tissue specific expression and its role in lipid and energy metabolism. Curr Opin Lipidol. 2002;13:471–481. doi: 10.1097/00041433-200210000-00002. [DOI] [PubMed] [Google Scholar]
- [60].Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Towle HC. Glucose and cAMP: adversaries in the regulation of hepatic gene expression. Proc Natl Acad Sci U S A. 2001;98:13476–13478. doi: 10.1073/pnas.251530798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gordon HA, Wostmann BS, Bruckner-Kardoss E. Effects of Microbial Flora on Cardiac Output and Other Elements of Blood Circulation. Proc Soc Exp Biol Med. 1963;114:301–304. doi: 10.3181/00379727-114-28658. [DOI] [PubMed] [Google Scholar]
- [63].Wostmann BS, Bruckner-Kardoss E, Pleasants JR. Oxygen consumption and thyroid hormones in germfree mice fed glucose-amino acid liquid diet. J Nutr. 1982;112:552–559. doi: 10.1093/jn/112.3.552. [DOI] [PubMed] [Google Scholar]
- [64].Abel ED, Litwin SE, Sweeney G. Cardiac remodeling in obesity. Physiol Rev. 2008;88:389–419. doi: 10.1152/physrev.00017.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation. 2007;115:3213–3223. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- [66].Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- [67].Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Unger RH. LIPOTOXIC DISEASES. Annual Review of Medicine. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- [69].Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Reigstad CS, Östergren-Lundén G, Felin J, Bäckhed F. Regulation of Serum Amyloid A3 (SAA3) in Mouse Colonic Epithelium and Adipose Tissue by the Intestinal Microbiota. PLoS ONE. doi: 10.1371/journal.pone.0005842. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Han CY, Subramanian S, Chan CK, Omer M, Chiba T, Wight TN, Chait A. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes. 2007;56:2260–2273. doi: 10.2337/db07-0218. [DOI] [PubMed] [Google Scholar]
- [73].Scheja L, Heese B, Zitzer H, Michael MD, Siesky AM, Pospisil H, Beisiegel U, Seedorf K. Acute-phase serum amyloid A as a marker of insulin resistance in mice. Exp Diabetes Res. 2008;2008:230837. doi: 10.1155/2008/230837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- [77].Martin FP, Wang Y, Sprenger N, Yap IK, Rezzi S, Ramadan Z, Pere-Trepat E, Rochat F, Cherbut C, van Bladeren P, Fay LB, Kochhar S, Lindon JC, Holmes E, Nicholson JK. Top-down systems biology integration of conditional prebiotic modulated transgenomic interactions in a humanized microbiome mouse model. Molecular systems biology. 2008;4:205. doi: 10.1038/msb.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, Kochhar S, Lindon JC, Holmes E, Nicholson JK. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Molecular systems biology. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Molecular systems biology. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]