Abstract
Insect odorant-binding proteins (OBPs) are small, water-soluble molecules that are thought to transport the hydrophobic odorants to their receptors in the chemosensory neurons. Here we report the identification and molecular characterization of AsteObp1, an Obp1 gene in Anopheles stephensi, a major malaria vector in Asia. We showed that AsteObp1 and Anopheles gambiae Obp1 (AgamObp1) are orthologues. These two genes share similar coding sequences and conserved non-coding sequences (CNSs) that may be involved in their regulation. Transcript of AsteObp1 was observed in larvae and reached a relatively high level in late pupae. Quantitative RT-PCR on female adult chemosensory tissues showed ~900-fold higher expression of AsteObp1 in antennae than in maxillary palp and proboscis. The amount of AsteObp1 in female legs was approximately 15-fold lower than that of maxillary palp and proboscis. The level of AsteObp1 transcript was 7 and 85-fold higher in females than in males in the antennae, and maxillary palp and proboscis, respectively. Moreover, AsteObp1 level was reduced by approximately 20-fold in maxillary palp and proboscis 24 h after a bloodmeal. Our results indicate that AsteObp1 likely functions in female olfactory response and it may also be involved in blood-feeding behaviour.
Keywords: antennae, blood-feeding, comparative genomics, olfaction
Introduction
Mosquito behaviours are mediated by both internal and external factors and olfactory cues are undoubtedly the most important external stimuli that affect behaviours such as host-seeking, oviposition, and sugar-feeding (Takken & Knols, 1999). With the availability of the genome assembly of An. gambiae and Ae. aegypti (Holt et al., 2002; Nene et al., 2007) and through the advent of behavioural, physiological, and molecular studies (e.g., Takken & Knols, 1999; Takken et al., 2001; Meijerink et al., 2001; Dekker et al., 2002, Justice et al., 2003; Xu et al., 2003; Hallem et al., 2004; Biessmann et al., 2005; Zhou et al., 2008; Sengul & Tu, 2008), our understanding of the basis for the sense of smell in mosquitoes has been greatly improved in recent years.
In insects, the proteins that are known to be involved in processing of olfactory cues include odorant-binding proteins (OBPs; Vogt & Riddiford, 1981), odorant receptors (ORs; Clyne et al., 1999; Vosshall et al., 1999), and odorant-degrading enzymes (ODEs; Vogt & Riddiford, 1981; Vogt et al., 1999; Ishida & Leal, 2002). Among these, OBPs are believed to bind the odorant molecule and transport it through the aqueous sensillar lymph to the receptors on chemosensory neurons. OBPs are small (15-17 kDa), water-soluble, extracellular proteins present in the sensillum lymph of the sensilla. Most insect OBPs share a hallmark feature of six cysteines, whose relative positions are conserved. A D. melanogaster OBP mutant, named lush, shows abnormalities in the perception of the aggregation pheromone, 11-cis-vaccenyl acetate (VA) (Xu et al., 2005). Moreover, it has been demonstrated that LUSH is required to stimulate the VA-sensitive receptorto elicit a respond to VA (Ha & Smith, 2006). This response is mediated by the pheromone-induced conformational shifts in the LUSH protein (Laughlin et al., 2008).
In Diptera, the genomes of the D. melanogaster, An. gambiae, and Ae. aegypti encode 51, 57, and 66 Obp genes, respectively (Galindo & Smith, 2001; Hekmat-Scafe et al., 2002; Vogt, 2002; Xu et al., 2003; Zhou et al., 2008). Among OBPs in An. gambiae, AgamObp1 was found at high levels in female antennae, although the expression was observed in headless bodies as well (Biessmann et al., 2002; Li et al., 2005). Moreover, AgamObp1 transcript level was reduced after blood-feeding by less than 2-fold in female heads (Justice et al., 2003; Biessmann et al., 2005), which may indicate a possible role of this gene in blood-feeding behaviour of female mosquitoes.
In this study, we cloned, mapped, and characterized the Obp1 gene in An. stephensi (henceforth referred as AsteObp1), an important malaria vector in Asia. Comparisons with Obp1 gene and protein sequences in other mosquitoes revealed conserved gene structure, potential regulatory sequences, and conserved secondary structure. We also determined the expression profiles of AsteObp1 using qualitative and quantitative RT-PCR. Our results indicate that AsteObp1 likely functions in female olfactory response and it may also be involved in blood-feeding behaviour.
Results
Identification and chromosomal mapping of OBP1 in An. stephensi
Obp1 gene (accession number: AY146721; Xu et al., 2003) and Obp17 gene (accession number: AY146723; Xu et al., 2003) in An. gambiae were previously annotated as two genes that are likely derived through gene duplications since they are found in close proximity to each other on the 2R chromosome, sharing similarity in their flanking sequences as well (Xu et al., 2003). A later study also determined different contigs that have identical sequences to predicted OBP1 (Li et al., 2005). Currently, the updated Ensembl and Vectorbase annotation shows only a single gene with two possible transcripts in An. gambiae: Q8I8TO_ANOGA (AY146721) and OBP17 (AY146723). Q8I8TO_ANOGA codes for the AgamOBP1 peptide (AF437884; Biessmann et al., 2002). We used the gene sequence of AgamObp1 to construct an Obp1 gene probe to screen a bacterial artificial chromosome (BAC) library derived from An. stephensi genomic DNA. Three positive clones were identified from a total of 9216 clones screened, which represents ~ 5-fold coverage of the genome, assuming that the genome size of An. stephensi is approximately 240 Mbp (Rai and Black, 1999). One positive BAC clone was sequenced, which produced 2 contigs. A single An. stephensi Obp1 gene (AsteObp1, FJ410801) was identified in the 98.5 kb assembled contig sequence (GQ250942).
Furthermore, we mapped AsteObp1 gene to the region corresponding to 17A of the chromosomal arm 2R of the ovarian chromosomes of An. stephensi by fluorescent in situ hybridization analysis (Fig. 1). This region is homologous to An. gambiae 14E of chromosomal arm 2R, the region in which AgamObp1 gene resides (http://www. ensembl.org/Anopheles_gambiae/index.html; Sharakhova et al., 2006). Our chromosomal mapping and sequence analysis (discussed below) indicate that the AsteObp1 gene is located at a single locus in the An. stephensi genome and AsteObp1 and AgamObp1 are probably orthologues.
Figure 1.
Fluorescent in situ hybridization performed on the polytene chromosomes of Anopheles stephensi. The An. stephensi odorant-binding protein 1 gene (AsteObp1) is labelled with Cy5 (blue).
Comparison of the Obp1 genomic regions in An. gambiae and An. stephensi
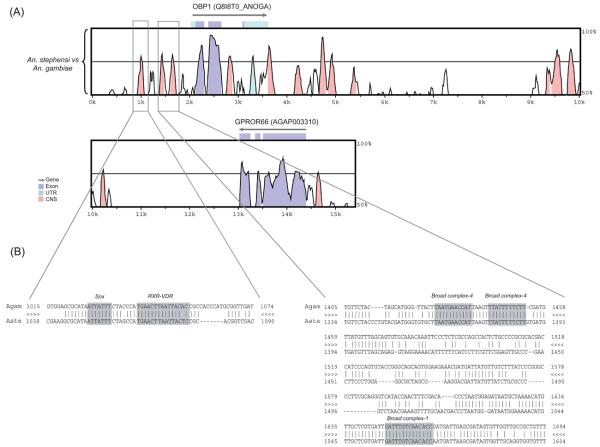
Fig. 2A shows the VISTA display of the sequence alignment between An. gambiae and An. stephensi in the genomic regions including the Obp1 gene and nearby sequences. The coding sequences of the Obp1 genes as well as the order of neighboring genes are conserved between the two species. For example, a homologue of the An. gambiae G-protein coupled odorant receptor GPROr66 gene, which is next to the AgamObp1 gene, is also identified approximately 10 kb downstream of the AsteObp1. We named this gene as AsteGPROr66 (FJ410800).
Figure 2.
Pairwise comparison between Anopheles gambiae and An. stephensi genomic sequences including the odorant-binding protein 1 (Obp1) gene and the nearby sequences. (A) VISTA plot that shows the alignment between An. gambiae and An. stephensi. The x-axis shows the relative position of the An. gambiae genomic DNA used as the reference genome in the alignment. The y-axis shows the % identity between the compared species. Arrows above the plots correspond to genes or coding sequences annotated in the An. gambiae genome according to the Ensembl database. The cut-off for sequence conservation is 75% over 100bp. The coding sequence portion of the third exon of the Obp1 gene is only 18bp long, which is why it does not show a conserved peak. The boxed regions correspond to the conserved noncoding sequences that were further investigated for transcription factor binding sites. Coding regions are shown in purple, untranslated regions are in light blue, and conserved non-coding sequences are shown in pink. CNS, conserved noncoding sequences. UTR, untranslated region. (B) VISTA alignment between An. gambiae and An. stephensi corresponding up to 1 kb upstream of the Obp1 transcription start site (boxed regions). Potential TFBS that are conserved for the two species are highlighted in the alignment. Three sites for two Broad complex isoforms are shown. The first and second Broad complex-4 sites are reverse complementary.
Pairwise alignment revealed high level of conservation in the coding regions of Obp1 gene, ranging between 77% to 94% identity at the nucleotide level. Additionally, there are a few conserved non-coding sequences (CNSs) at the 5′ and 3′ flanking regions as well as in an intron (Fig. 2A, pink peaks). These regions exhibited 71% to 87% identity between the two species. In order to determine whether these regions contain any transcription factor binding sites (TFBSs), we used ConSite prediction tool (Sandelin et al., 2004) and found significant hits for C2H2-type zinc finger (Broad-complex isoforms 1 and 4), a nuclear receptor factor (RXR-VDR) and HMG (Sox) binding sites ~1.1 kb upstream of the Obp1 gene. These sites are conserved between An. gambiae and An. stephensi (Fig. 2B). Interestingly, the conserved segment (the pink peak, Fig. 2A) in the second intron of Obp1 also contained TFBSs for an ecdysone-induced transcription factor E74A and another nuclear receptor factor (CFI-USP) (data not shown). Other highly conserved motifs have also been observed in the pairwise alignment that did not show any significant hits for a known transcription factor in the database.
Experimental characterization of AsteObp1 gene structure
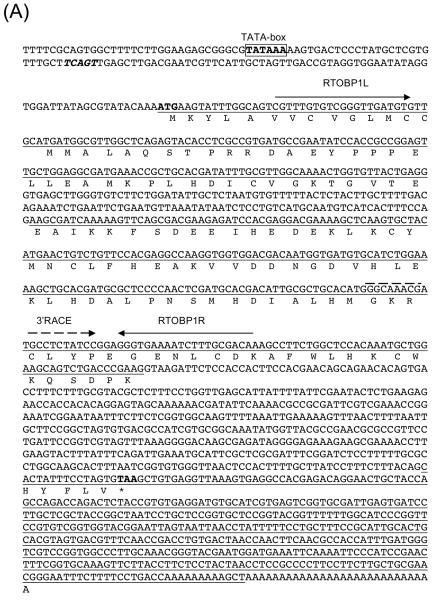
The AgamObp1 gene model served as a good reference for the annotation of the AsteObp1 coding sequences. The alignment shown in Fig. 2A suggests a good conservation in the coding sequences of AsteObp1 and AgamObp1 genes. The observed conservation between AgamObp1 and AsteObp1 genes also helped the identification of a potential TATA-box and the transcription start site. A TCAGT sequence (Fig. 3A, boldface and italicized), which is identical to the arthropod initiator consensus (Cherbas & Cherbas, 1993), is observed 69 nucleotides upstream of the translation initiation codon, ATG. A consensus for TATA-box is located 101 bp upstream of the ATG as well (Fig. 3A; boldface and boxed).
Figure 3.
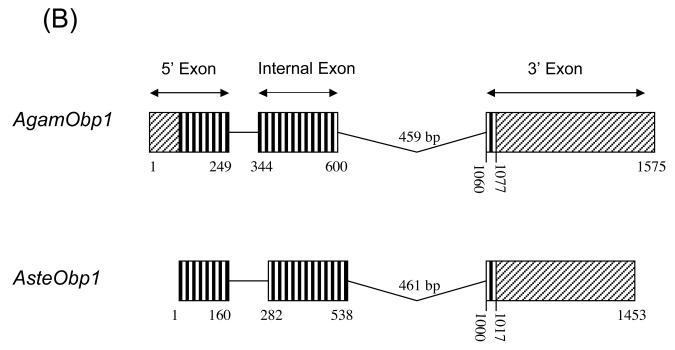
Sequence and structure of anopheline odorant-binding protein 1 (Obp1) genes. (A) Anopheles stephensi Obp1 genomic and cDNA sequences. The AsteObp1 gene contains two introns among the three underlined exons. The deduced amino acid sequence of AsteOBP1 is shown below the coding regions. Start (ATG) and stop (TAA) codons are shown in bold. A putative initiator sequence, TCAGT, similar to the arthropod initiator consensus is in bold and italicized. The consensus for TATA-box, TATAAA, is shown in bold and boxed. The primers used in RT-PCR and RACE reactions are shown by arrows above the corresponding sequences. (B) AsteObp1 gene structure is compared with that of AgamObp1. 5′ exons, internal exons, and 3′ exons are indicated. Boxes with vertical lines indicate open reading frames (ORFs) and they are connected with lines that indicate the introns. The 5′ and 3′ untranslated regions (UTRs) are shown in boxes with diagonal lines. Numbers indicate the relative positions of UTRs, ORFs and intronic regions.
The 3′ end of the AsteObp1 transcript was determined by 3′ RACE, which produced a cDNA product with an expected poly(dA) tail, a 422 bp untranslated region (UTR) and a removed intron. Reverse transcription polymerase chain reaction (RT-PCR) was also performed using primers spanning the first predicted intron (Fig. 3A). Comparing sequences of the RT-PCR and RACE products and the BAC clone confirmed the existence the first and second introns of 121 bp and 461 bp in length, respectively. Overall, we characterized the entire open reading frame (ORF) of AsteObp1 interrupted by two introns as shown in Fig. 3A. The coding region is 432 bp long encoding a 144 amino acid peptide. The overall gene structure of AsteObp1 is highly similar to that of AgamObp1 (Fig. 3B), which is annotated in GenBank (AY146721; Xu et al., 2003).
Expression profile of AsteObp1
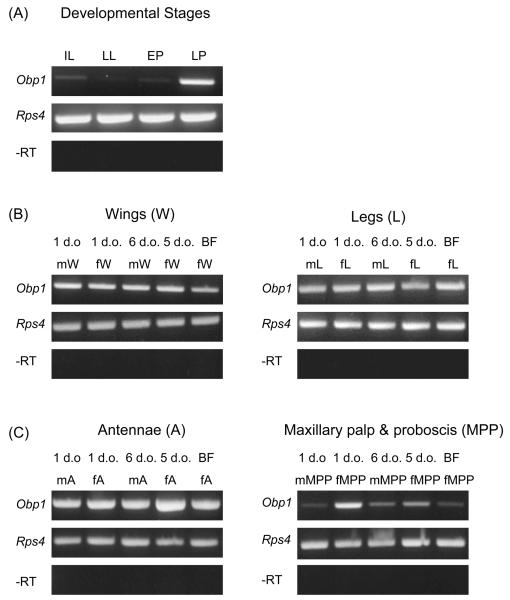
The expression profile of AsteObp1 was initially examined by non-quantitative RT-PCR. We found a higher level of AsteObp1 expression in late pupae compared to larvae or early pupae (Fig. 4A). Either weak expression or nearly no expression was observed in the first instar larvae, late larvae, and early pupae. We also performed RT-PCR using different olfactory and gustatory tissues from 1-day-old and 5 to 6-days-old adults of both sexes as well as females 24 h after a bloodmeal. Broad distribution of AsteObp1 gene expression in most of the chemosensory tissues of mosquitoes was observed (Fig. 4B, 4C). AsteObp1 was expressed in mosquito head tissues such as antennae, maxillary palp and proboscis, and mosquito body parts such as legs and wings. AsteObp1 expression appeared to be reduced in maxillary palp and proboscis in females after a bloodmeal (Fig. 4C). Either very weak or no expression was observed in body samples devoid of head and appendages (not shown).
Figure 4.
Nonquantitative RT-PCR showing expression of Anopheles stephensi odorant-binding protein 1 gene (AsteObp1) in larvae and pupae (A), and chemosensory tissues of adult An. stephensi (B, C). Lanes are as follows: first instar larvae (IL), late larvae (LL), early pupae (EP), late pupae (LP); adult male wings (mW), adult female wings (fW), blood-fed female wings (BFfW); adult male legs (mL), adult female legs (fL), blood-fed female legs (BFfL); adult male antenna (mA), adult female antenna (fA), blood-fed female antenna (BFfA); adult male maxillary palp and proboscis (mMPP), adult female maxillary palp and proboscis (fMPP), blood-fed female maxillary palp and proboscis (BFfMPP). The Rps4 internal control gene is used as the positive control. Minus reverse transcription (−RT) products (negative control without the use of reverse transcriptase) are also shown for each cDNA samples. d.o., days old. All RT-PCR reactions were performed with 35 cycles of amplification.
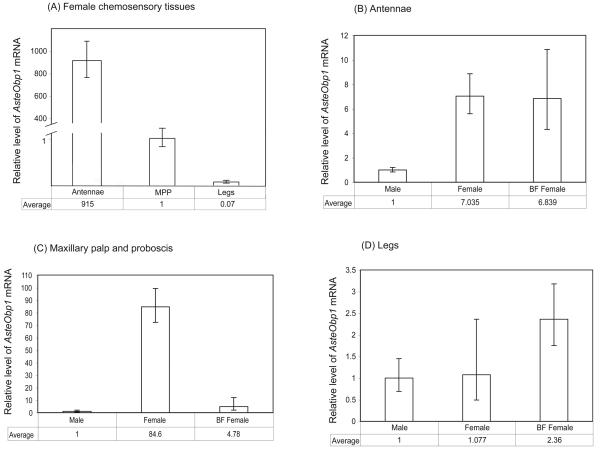
Furthermore, quantitative real-time PCR was performed to compare the AsteObp1 mRNA levels in the antennae, maxillary palp and proboscis, and legs from adult females. We observed a significantly higher (~900-fold) expression in female antennae compared to that of the maxillary palp and proboscis (Fig. 5; Table 1). Very low amount of AsteObp1 was observed in female legs, approximately 15-fold lower than that of in maxillary palp and proboscis. These results clearly show that AsteObp1 is much more abundantly expressed in the primary olfactory organ, the antennae, of the An. stephensi mosquitoes compared to the other chemosensory tissues. Although the expression level in maxillary palp and proboscis was low, the level was significant and likely biologically relevant.
Figure 5.
Relative amount of the Obp1 mRNA in adult chemosensory tissues of An. stephensi. The y-axis is the relative level of AsteObp1 mRNA as determined by the 2−ΔΔCT method. Detailed data analyses are shown in Table 1 and Supplementary Material Tables S1-S3. The results are shown as the mean and a range specified by 2−(ΔΔCT+SD) and 2−(ΔΔCT−SD), where SD is the standard deviation, for three or more biological replicates each from independent collections. (A) Comparison of AsteObp1 expression levels in different female chemosensory tissues. (B) Comparison of AsteObp1 expression levels in the antenna of male, female and blood-fed females. (C) Comparison of AsteObp1 expression levels in the maxillary palp and proboscis of male, female and blood-fed females. (D) Comparison of AsteObp1 expression levels in the legs of male, female and blood-fed females. The relative amount of AsteObp1 is significantly different between different chemosensory tissues in the females (Panel A; P < 0.001, One-way ANOVA with Tukey's post test). The relative amount of AsteObp1 in antennae of non-blood fed females and blood-fed females, and the relative amount of AsteObp1 in legs of male and female mosquitoes are not significantly different (Panels B-D) (P > 0.05, One-way ANOVA with Tukey's post test). In all cases, the calibrator sample for normalization show an average value of 1.
Table 1.
Relative mRNA levels of Anopheles stephensi odorant-binding protein 1 gene (AsteObp1) in female antennae, maxillary palp and proboscis, and legs
| Tissue | Obp1CT | Rps4CT | ΔCT(Obp1CT − Rps4CT) | ΔΔCT(ΔCT − ΔCTcalibrator) | Normalized Obp1 amount relative to the calibrator, 2 −ΔΔCT |
|
|---|---|---|---|---|---|---|
| Antenna | replicate1 | 14.322 | 18.968 | −4.647 | ||
| replicate2 | 14.226 | 19.225 | −5 | |||
| replicate3 | 14.7 | 19.681 | −4.982 | |||
| Average | 14.416 +/− 0.145 | 19.292 +/− 0.208 | −4.876 +/− 0.254 | −9.838 +/− 0.254 | 915 (767.5 − 1090) | |
| Maxillary palp and Proboscis * |
replicate1 | 23.73 | 18.08 | 5.651 | ||
| replicate2 | 22.889 | 18.565 | 4.323 | |||
| replicate3 | 23.183 | 18.272 | 4.912 | |||
| Average | 23.267 +/− 0.247 | 18.306 +/− 0.141 | 4.962 +/− 0.284 | 0 +/− 0.284 | 1 (0.82 − 1.217) | |
| Legs | replicate1 | 28.209 | 18.913 | 9.295 | ||
| replicate2 | 26.354 | 18.042 | 8.312 | |||
| replicate3 | 27.125 | 18.35 | 8.776 | |||
| Average | 27.229 +/− 0.538 | 18.435 +/− 0.255 | 8.794 +/− 0.595 | 3.833 +/− 0.595 | 0.07 (0.046 − 0.106) |
represents the calibrator.
5-day-old female mosquitoes were used for each tissue collection.
Data presentation is according to Livak & Schmittgen (2001), with average and standard deviation (SD). Data in the final column are presented as the mean and a range specified by 2−(ΔΔCT+SD) and 2−(ΔΔCT−SD). All replicates are biological replicates.
We also determined expression levels of AsteObp1 in different tissues between males, females and blood-fed females. We observed a significantly higher expression of AsteObp1 in the female antennae (~7-fold) and maxillary palp and proboscis (~80-fold) than those in males (Fig. 5B-D). The AsteObp1 expression in legs was not significantly different between male and females. However, the expression in legs was increased by > 2-fold in blood-fed females. Given the low level of transcripts in the legs, it is unclear whether the difference before and after blood-feeding is biologically relevant. Bloodmeal did not affect the mRNA level of AsteObp1 in the antennae of female mosquitoes. However, we observed a ~20-fold reduction of AsteObp1 gene expression in maxillary palp and proboscis of blood-fed females compared to that of females before a bloodmeal.
Conservation of OBP1 protein structure among divergent mosquito species
The deduced amino acid sequences of OBP1 from An. gambiae, An. stephensi, Ae. aegypti and C. p. quinquefasciatus were aligned in Fig. 6. Mosquito OBP1 proteins show 76% to 86% amino acid identity, as well as six cysteines whose positions are absolutely conserved. The predicted secondary structure of AsteOBP1 includes six α-helices (Fig. 6; solid bars above the peptide sequence), consistent with the predicted structures from AgamOBP1 and other insect OBPs (Leal et al., 1999; Sandler et al., 2000; Lartigue et al., 2004; Kruse et al., 2003; Mohanty et al., 2004; Wogulis et al., 2006). The N-terminal signal peptide sequence (Fig. 6, underlined) of mosquito OBP1 is less conserved than the rest of the protein.
Figure 6.
Alignment of Anopheles stephensi odorant-binding protein 1 gene (AsteObp1) with its possible mosquito orthologues. All identical amino acid residues are shaded. The six conserved cysteines are shown in bold and indicated with asterisks. Predicted signal peptides are underlined in the sequences. The α-helices of AsteOBP1 are indicated by solid bars above the amino acid sequence. Species abbreviations are Aste, Anopheles stephensi; Agam, Anopheles gambiae; Aaeg, Aedes aegypti; Cqui, Culex pipiens quinquefasciatus.
Selection pressure acting on mosquito Obp1 genes
Pairwise comparisons of mosquito Obp1 sequences were used to determine the rates of synonymous (dS) and nonsynonymous (dN) codon substitutions and the ratio of dN/dS was calculated to provide an estimate of the selection pressure on Obp1 genes in An. gambiae and An. stephensi. OBP1 orthologues from divergent mosquito species such as Ae. aegypti and C. p. quinquefasciatus were not included in the dN/dS ratio calculations, because the dS values were saturated and were not reliable. The coding regions of Obp1 were included in the analysis except the 5′ exon regions, which encode the signal peptides that are highly divergent among OBPs. The dN value between AsteObp1 and AgamObp1 is 0.022 and the dS is 0.596. The dN/dS ratio is far less than 1, suggesting that Obp1 is evolving under strong purifying selection for a possibly important function in mosquitoes.
Discussion
In this study, we report identification and characterization of Obp1 gene from the Asian malaria mosquito An. stephensi. We named it Obp1 gene in An. stephensi (AsteObp1) because it shows sequence identity and conservation of gene structures with the An. gambiae Obp1 (AgamObp1). Qualitative and quantitative RT-PCR analysis of AsteObp1 reported here showed predominant expression in the antennae and a strong female-bias. This is consistent with strong expression of AgamObp1 in female head (Biessmann et al., 2005) and supports that Obp1 may play an important role in adult female antennae, the primary olfactory tissue. Female-biased expression of AsteObp1 in olfactory organs, such as antennae and maxillary palp and proboscis, may result from a larger number of olfactory sensilla in females than in males (McIver, 1982).
Our analysis of AsteObp1 expression in larval and pupal stages and quantitative RT-PCR analysis of AsteObp1 in dissected adult olfactory organs offer several new insights. First, we detected relatively strong expression of AsteObp1 in late pupae, which either suggests a possible chemosensory role of this gene in pupae or a simple correlation with the onset of development of chemosensory tissues during pupation. Further study is needed to differentiate the above two possibilities. Second, AsteObp1 transcripts were also detected, albeit at low levels, in gustatory organs, such as legs and wings. It is possible that AsteObp1 may have a gustatory role. Alternatively, these gustatory tissues might have olfactory capabilities that are yet to be understood in mosquitoes.
Third, although we did not observe significant difference in either the antennae or the leg between non-bloodfed females and females 24 h after blood-feeding, we detected ~ 20-fold reduction of AsteObp1 transcript level in female maxillary palp and proboscis. This is very interesting as it indicates a possible involvement of AsteObp1 in blood-feeding behavior. The ORNs of maxillary palpal sensilla in mosquitoes are important in olfaction including detection of CO2 and 1-octen-3-ol for host localization (Gillies, 1980; Grant et al., 1995; Grant & O'Connell, 1996; Dekker et al., 2001; Syed & Leal, 2007; Lu et al., 2007). The reduction of AsteObp1 (this paper) and AsteObp7 transcripts (Sengul & Tu, 2008) in pooled maxillary palp and proboscis after blood-feeding may reflect the decreased need for host-seeking or blood-feeding. It will be interesting to see if such reduction is also observed in Obp1 homologues of other mosquito species.
Comparative analysis suggests that Obp1 orthologues underwent little structural changes in mosquitoes. In fact, we determined a strong purifying selection (dN/dS< 1) among anopheline Obp1 genes, which is consistent with its proposed functional importance. A better understanding of the functional role of OBPs in olfaction may come from the structural studies of different OBPs in various insect species. In mosquitoes, the crystal structure of OBP1 in An. gambiae has been determined (Wogulis et al., 2006), representing the only known OBP structure from mosquitoes. This protein is crystallized as a dimer with a unique binding pocket, consisting of a single tunnel running through both subunits. It has been found that AgamOBP1 undergoes a pH dependent conformational change, which was initially proposed for the ligand (i.e. bombykol) binding and releasing mechanism for PBP in B. mori (BmorPBP) (Horst et al., 2001). The alignment of AsteOBP1 with AgamOBP1 and other mosquito orthologues showed a high level of conservation in the overall protein structure (Fig. 6). The availability of the primary and secondary structure of AsteOBP1 will help future modeling studies that compare and contrast the two anopheline OBP1 proteins.
Understanding the regulatory elements that play a role in Obp1 expression may provide insights into the function of this gene in mosquito olfaction. For this purpose, a comparative approach was used to uncover possible regulatory elements of Obp1 gene in An. gambiae and An. stephensi by determining the conservation in the non-coding regions. Previously, by using a similar approach, we have determined conserved motifs in the 5′ upstream sequences of Obp7 gene in the three anopheline mosquitoes, including An. gambiae, An. stephensi and An. quadriannulatus (Sengul & Tu, 2008). Although we were unable to experimentally verify the transcription start site of AsteObp1, predictions based on the AgamObp1 gene model points to a conserved transcription start site and upstream TATA box (Fig. 3A). In the 5′ upstream regions of Obp1 genes, we observed conserved binding sites for Broad-complex isoforms and RXR-VDR transcription factors both in An. gambiae and An. stephensi. However, much of the conservation in other regions did not give any hits for a known transcription factor. It is possible that these are novel binding sites that may likely play a role in Obp gene regulation in mosquitoes, which need to be experimentally tested. We have also identified a possible odorant receptor gene orthologue of the An. gambiae GPROr66 gene, approximately 10 kb apart from the AsteObp1 gene (Fig. 2A). It would be interesting to determine whether the close chromosomal localization of Obp1 and Or66 genes in both species has any effect on the regulation of these genes.
Experimental procedures
Mosquitoes and dissections
Anopheles stephensi Indian strain were reared at 27°C with 75% relative humidity. Conditions for dissection and tissue collection were previously described (Sengul & Tu, 2008).
Generation of digoxigenin-labelled, single-stranded DNA (ssDNA) probes and Bacterial Artificial Chromosome (BAC) library screening
An amino acid alignment of AgamOS-E (EAA01090; Vogt, 2002), AgamOS-F (EAA09615; Vogt, 2002), AgamOBP1 (AF4377884; Biessmann et al., 2002), D. melanogaster OS-F (U02542; McKenna et al., 1994), and OS-E (U02543; McKenna et al., 1994) has been performed using Clustal X (1.8) (Thompson et al., 1997). This alignment was used to design the following degenerate primers; OBP1F1: 5′-TGYTAYATGAAYTGYYTNTTYCA corresponding to the motif CYMVCLFH and OBP1R1: 5′-CARCAYTTRTGNARCCARAANGC corresponding to the motif AFWLHKCW. The genomic DNAs were isolated from An. stephensi mosquitoes using DNAzol Genomic DNA Isolation Reagent (Molecular Research Center, Inc., Cincinnati, OH, USA). Degenerate primers (mentioned above) were used to amplify a 186 bp sequence in the second exon region of the AsteObp1 gene. The PCR product was cloned using pGEM-T-Easy vector systems (Promega, Madison, WI, USA) and confirmed by sequencing (Virginia Bioinformatics Institute, Blacksburg, VA, USA). The plasmid that contains Obp1 gene sequence was used as a template in an asymmetric PCR using the primer, OBP1probe: 5′-GTTTGTTTCACGAGGCCAAG, and DIG-dUTP labelling mixture (Roche Diagnostics, Indianapolis, IN, USA) to generate ssDNA probe. The DIG-labelled ssDNA probe was used to screen the BAC library of An. stephensi under moderate conditions. The average insert size of the An. stephensi BAC library is 125 kb and the library represents ~10x coverage of the genome. Hybridization was carried out at 55°C overnight. Two set of washes were performed at 55°C with 2X and 0.5X SSC (Saline Sodium Citrate). Details about the construction and screening of the An. stephensi BAC library were previously described (Sengul & Tu, 2008).
BAC DNA sequencing
The positions of the positive clones were determined using the 384-well plate overlay. Single colony preps were made by dipping a sterile inoculating loop into stab (BACs are supplied as stabs in agar) and drawing parallel lines on the LB/chloramphenicol (12.5μg/ml) plates followed by incubation overnight at 37°C. One positive BAC clone per each screening was selected for BAC DNA isolation that was used to check for the coverage of clones by PCR before sequencing. Sequencing was done at TIGR (The Institute for Genomic Research, Rockville, MD) using Sanger chemistry and the shotgun approach.
Identification of AsteObp1 and its possible mosquito orthologues
Contigs obtained from An. stephensi BAC sequencing were compared to AgamObp1 using BLAST (Altschul et al., 1990). Sequences that had significant match to AgamObp1 and nearby coding sequences were further analyzed using BLAST and VISTA alignment (http://genome.lbl.gov/vista/index.shtml). The orthologues of OBP1 were obtained from the NCBI database and they include AaegOBP1 from Ae. aegypti (AY189223; Ishida et al., 2004), and CquiOBP1 from C. p. quinquesfasciatus (AF468212; Ishida et al., 2002).
Fluorescent in situ hybridization (FISH) analysis of AsteObp1
MapOBP1L: 5′-GCTCGTGTTTGCTTCAGTTG and MapOBP1R: 5′-ACC CACACCGATTAAAGTGC primer pair was used to amplify a 1054 bp genomic sequence of the AsteObp1 gene, which was confirmed by sequencing (Virginia Bioinformatics Institute). Gel purified PCR product was used to make probe labeling with Cy5-AP3-dUTP (GE Healthcare, Little Chalfont, Buckinghamshire, UK) using Random Primers DNA Labeling System (Invitrogen, Carlsbad, CA, USA). Fluorescently labelled Obp1 gene probes was used for in situ hybridization on the polytene chromosomes of An. stephensi females prepared from ovarian nurse cells. Chromosome preparation and hybridization were performed as previously described (Sharakhova et al., 2006).
Expression analysis of AsteObp1 by RT-PCR
Total RNA was isolated from various An. stephensi developmental stages and manually dissected 1-day-old and 5-6 days old male and female adult tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Following DNase treatment (Ambion, Inc., Austin, TX, USA), samples were reverse transcribed using Superscript II reverse transcriptase (Invitrogen) at 42°C for 1.5 hr followed by heat inactivation at 70°C for 15 minutes. All cDNA synthesis reactions were carried out in the absence of reverse transcriptase (−RT) in parallel for each sample. RT-PCR amplification of cDNAs were carried out using gene specific primers, RTOBP1L: 5′-GTTTGTGTCGGGTTGATGTG and RTOBP1R: 5′-TTGTCGCAAAGATTTTCACC, that span the first intron sequence of AsteObp1 gene to control for any genomic DNA contamination. Accordingly, PCR products with an expected size of 356 bp from cDNA amplification can be differentiated from genomic DNA amplification of a 477 bp product. We used AsteRps4 (EU883624; Sengul & Tu, 2008) as an internal control in the RT-PCR reactions to determine the integrity of cDNA templates. RT-PCR primer pair Rps4L: 5′-CACGAGGATGGATGTTGGAC and Rps4R: 5′-ATCAGGCGGAAGTATTCACC amplified a cDNA product of about 262 bp in length. The optimal annealing temperature for the RT-PCR was 60°C for both AsteObp1 and AsteRps4 genes. Both reactions were run for a total of 35 cycles and are not quantitative. PCR products were analysed by 2% agarose-gel electrophoresis.
Quantitative real-time PCR
For quantitative real-time PCR, total RNA from selected tissues was isolated from adult 6-day-old male, 5-day-old female and 6-day-old blood-fed females to perform cDNA synthesis as mentioned above. Our analyses included at least three biological replicates for all samples, each from independent collections. The quantitative real-time PCR was performed using the TaqMan probe-based chemistry with an ABI Prism 7300 Sequence Detection System (SDS; Applied Biosystems, Foster City, CA, USA). For each 25 μl PCR reaction, 2 μl of cDNA was used with 9.25 μl nuclease-free water, 12.5 μl 2 X TaqMan Universal PCR Master Mix (Applied Biosystems), and 1.25 μl 20 X assay mix designed by ABI. The thermocycler programme consisted of 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. As the relative mRNA amount of AsteObp1 was being determined, parallel TaqMan assays were also carried out for the control gene, AsteRps4. The assay mixture for AsteObp1 and AsteRps4, consisted of 5′FAM (5′-carboxyfluorescein), nonfluorescent quencher (NFQ) labelled probes and the primer pairs as follows:
AsteObp1primerF: 5′-AACCGCTGCACGATATTTGC-3′
AsteObp1primerR: 5′-GGATCTCTTCGTCGCTGAACTTTT-3′
AsteObp1probe: 5′FAM-ATCGCTTCCTCAGTAACAC-NFQ
AsteRps4primerF: 5′-TTCGCACCGATCCGAACTAC-3′
AsteRps4primerR: 5′-GAAGTATTCACCGGTCTTGTGGAT-3′
AsteRps4probe: 5′FAM-TTGATCACATCCATGAAACC-NFQ
All TaqMan PCR data were analyzed using SDS Software based on the comparative method (ΔΔCT) (Livak & Schmittgen, 2001) as previously described (Sengul & Tu, 2008). We used ΔCTvalues for statistical analysis of real-time PCR data. One-way ANOVA with Tukey's post test was performed using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego, CA, USA). Significance in comparisons was assumed if P < 0.05 was obtained in the appropriate test.
Rapid Amplification of cDNA Ends (RACE)
3′ RACE were performed using the BD Smart RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA, USA) according to the manufacturer's instructions. After the initial cDNA synthesis, PCR was performed using the adapter primer and the gene specific primer (3′RACE: 5′-GGCAAACGATGCCTCTATCC). The PCR products were cloned into pGEM-T-Easy vector (Promega, Madison, WI, USA) and confirmed by sequencing (Virginia Bioinformatics Institute).
Sequence analyses of OBP1
The gene structure of AsteObp1 was confirmed by RT-PCR and RACE. The peptide alignment of OBP1 orthologues was generated by using CLUSTAL X (1.8) (Thompson et al., 1997). The following parameters were used for the alignment: pairwise gap penalty (open=35, extension=0.75), multiple gap penalty (open=15, extension=0.3). N-terminal signal peptide sequences of OBP1 were predicted by the SIGNALP V 3.0 programme (Bendtsen et al., 2004; http://www.cbs.dtu.dk/services/SignalP/). Putative α-helical regions were predicted using JPRED (Cuff & Barton, 1999; http://www.compbio.dundee.ac.uk/~www-jpred/). The results of Jpred were confirmed with Swiss-Model, a web based homology modeling program (http://swissmodel.expasy.org/).
Sequence comparison and transcription factor binding site (TFBS) predictions
The genome comparisons to identify conservation in coding and non-coding regions of OBP1 from An. stephensi and An. gambiae were established by using MLAGAN (Multi-LAGAN; Brudno et al., 2003; http://genome.lbl.gov/vista/lagan/submit.shtml). The cut-off for sequence conservation was 75% over 100 bp. The An. gambiae genome annotation (Holt et al., 2002) was used to determine the coding regions in the alignment. The alignment was visualized by VISTA (Mayor et al., 2000), which gave a VISTA plot output calculating the percent identity over the window at each base pair. The x-axis showed the relative position of the An. gambiae genomic DNA, and the y-axis shows the % identity between An. stephensi and An. gambiae sequences. ConSite program (Sandelin et al., 2004; http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite/) is used to predict potential transcription factor binding sites for the 5′ upstream sequences of Obp1, up to 1 kb region from the translation start site, of the aligned sequences between An. gambiae and An. stephensi. The cut-off for sequence conservation was 74%, and transcription factor score threshold was set to 80%.
dN and dS calculations
Synonymous and nonsynonymous mutation rates were analyzed by the method of Nei & Gojobori (1986). The SNAP program (Synonymous/Nonsynonymous analysis program; http://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html; Korber, 2000) was used to calculate dN, dS and to determine dN/dS ratios for mosquito OBP1 orthologues.
Supplementary Material
Acknowledgements
We thank Dr Maria V. Sharakhova for help with in situ hybridizations; Thomas R. Saunders for rearing An. stephensi mosquitoes. We also thank three anonymous reviewers for their constructive comments. This work was supported by National Institutes of Health Grant AI063252.
Nomenclature
- AsteObp1
Anopheles stephensi odorant-binding protein 1 gene
- AsteOBP1
Anopheles stephensi odorant-binding protein 1 protein
- AgamObp1
Anopheles gambiae odorant-binding protein 1 gene
- AgamOBP1
Anopheles gambiae odorant-binding protein 1 protein
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Walter MF, Dimitratos S, Woods D. Isolation of cDNA clones encoding putative odourant binding proteins from the antennae of the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol Biol. 2002;11:123–132. doi: 10.1046/j.1365-2583.2002.00316.x. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Nguyen QK, Le D, Walter MF. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol Biol. 2005;14:575–589. doi: 10.1111/j.1365-2583.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, NISC Comparative Sequencing Program. Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Dekker T, Takken W, Carde RT. Structure of host odour plumes influences catch of Anopheles gambiae s.s. and Aedes aegypti in a dual-choice olfactometer. Physiol Entomol. 2001;26:124–134. [Google Scholar]
- Dekker T, Steib B, Carde RT, Geier M. L-lactic acid: a human-signifying host cue for the anthropophilic mosquito Anopheles gambiae. Med Vet Entomol. 2002;16:91–98. doi: 10.1046/j.0269-283x.2002.00345.x. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies MT. The role of carbon dioxide in host-feeding my mosquitoes (Diptera: Culicidae) Bull Entomol Res. 1980;70:525–532. [Google Scholar]
- Grant AJ, Wigton BE, Aghajanian JG, O'Connell RJ. Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J Comp Physiol [A] 1995;177:389–396. doi: 10.1007/BF00187475. [DOI] [PubMed] [Google Scholar]
- Grant AJ, O'Connell RJ. Electrophysiological responses from receptor neurons in mosquito maxillary palp sensilla. Ciba Found Symp. 1996;200:233–248. doi: 10.1002/9780470514948.ch17. [DOI] [PubMed] [Google Scholar]
- Ha TS, Smith DP. A pheromone receptor mediates 11-cis-Vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26:8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Nicole Fox A, Zwiebel LJ, Carlson JR. Olfaction: mosquito receptor for human-sweat odorant. Nature. 2004;427:212–213. doi: 10.1038/427212a. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Horst R, Damberger F, Luginbuhl P, Guntert P, Peng G, Nikonova L, Leal WS, Wuthrich K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci USA. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Cornel AJ, Leal WS. Identification and cloning of a female antenna-specific odorant-binding protein in the mosquito Culex quinquefasciatus. J Chem Ecology. 2002;28:867–871. doi: 10.1023/a:1015253214273. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Leal WS. Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Mol Biol. 2002;32:1775–1780. doi: 10.1016/s0965-1748(02)00136-4. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Chen AM, Tsuruda JM, Cornel AJ, Debboun M, Leal WS. Intriguing olfactory proteins from the yellow fever mosquito, Aedes aegypti. Naturwissenschaften. 2004;91:426–431. doi: 10.1007/s00114-004-0551-7. [DOI] [PubMed] [Google Scholar]
- Justice RW, Dimitratos S, Walter MF, Woods DF, Biessmann H. Sexual dimorphic expression of putative antennal carrier protein genes in the malaria vector Anopheles gambiae. Insect Mol Biol. 2003;12:581–594. doi: 10.1046/j.1365-2583.2003.00443.x. [DOI] [PubMed] [Google Scholar]
- Korber B. Computational Analysis of HIV Molecular Sequences. In: Rodrigo Allen G., Learn Gerald H., editors. HIV Signature and Sequence Variation Analysis. Kluwer Academic Publishers; Dordrecht, Netherlands: 2000. pp. 55–72. Chapter 4. [Google Scholar]
- Kruse SW, Zhao R, Smith DP, Jones DNM. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat Struct Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue A, Gruez A, Briand L, Blon F, Bezirard V, Walsh M, Pernollet JC, Tegoni M, Cambillau C. Sulfure single-wavelength anomalous diffraction crystal of a pheromone-binding protein from the honeybee Apis mellifera L. J Biol Chem. 2004;279:4459–4464. doi: 10.1074/jbc.M311212200. [DOI] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464:85–90. doi: 10.1016/s0014-5793(99)01683-x. [DOI] [PubMed] [Google Scholar]
- Li ZX, Pickett JA, Field LM, Zhou JJ. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. Arch Insect Biochem Physiol. 2005;58:175–189. doi: 10.1002/arch.20047. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu T, Qiu YT, Wang G, Kwon JY, Rutzler M, Kwon HW, Pitts RJ, van Loon JJ, Takken W, Carlson JR, Zwiebel LJ. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- McIver SB. Sensilla of mosquitoes (Diptera: Culicidae) J Med Entomol. 1982;19:489–535. doi: 10.1093/jmedent/19.5.489. [DOI] [PubMed] [Google Scholar]
- McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J Biol Chem. 1994;269:16340–16347. [PubMed] [Google Scholar]
- Meijerink J, Braks MAH, van Loon JJA. Olfactory receptors on the antennae of the malaria mosquito Anopheles gambiae are sensitive to ammonia and other sweat-borne components. J Insect Physiol. 2001;47:455–464. doi: 10.1016/s0022-1910(00)00136-0. [DOI] [PubMed] [Google Scholar]
- Mohanty S, Zubkova S, Gronenborg AM. The solution NMR structure of Antheraea polyphemus PBP provides a new insight into pheromone recognition by PBP. J Mol Biol. 2004;337:443–451. doi: 10.1016/j.jmb.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai KS, Black IV WC. Mosquito genomes: structure, organization, and evolution. Adv Genet. 1999;41:1–33. doi: 10.1016/s0065-2660(08)60149-2. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:249–252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem Biol. 2000;7:143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- Sengul MS, Tu Z. Characterization and expression of the odorant-binding protein 7 gene in Anopheles stephensi and comparative analysis among five mosquito species. Insect Mol Biol. 2008;17:631–645. doi: 10.1111/j.1365-2583.2008.00837.x. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Syed Z, Leal WS. Maxillary palps are broad spectrum odorant detectors in Culex quinquefasciatus. Chem Senses. 2007;32:727–738. doi: 10.1093/chemse/bjm040. [DOI] [PubMed] [Google Scholar]
- Takken W, Knols BGJ. Odor-mediated behavior of afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- Takken W, van Loon JJA, Adam W. Inhibition of host-seeking response and olfactory responsiveness in Anopheles gambiae following blood feeding. J Insect Physiol. 2001;47:303–310. doi: 10.1016/s0022-1910(00)00107-4. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Pleawniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;22:4673–4680. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG. Odorant binding proteins of the malaria mosquito Anopheles gambiae: possible orthologues of the OS-E and OS-F OBPs of Drosophila melanogaster. J Chem Ecology. 2002;28:2371–2376. doi: 10.1023/a:1021009311977. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Callahan FE, Rogers ME, Dickens JC. Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera) Chem Senses. 1999;24:481–95. doi: 10.1093/chemse/24.5.481. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wogulis M, Morgan T, Ishida Y, Leal WS, Wilson DK. The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism. Biochem Biophysics Res Commun. 2006;339:157–164. doi: 10.1016/j.bbrc.2005.10.191. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003;12:549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- Xu PX, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol. 2008;17:147–163. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.