Abstract
The circadian clock controls many circadian outputs. Although a large number of transcripts are affected by the circadian oscillator, very little is known about their regulation and function. We show here that the Drosophila takeout gene, one of the output genes of the circadian oscillator, is regulated similarly to the circadian clock genes Clock (Clk) and cry. takeout RNA levels are at constant high levels in ClkJRK mutants. The circadian transcription factor PAR domain protein 1 (Pdp1ε) is a transcription factor that had previously been postulated to control clock output genes, particularly genes regulated similarly to Clk. In agreement with this, we show here that Pdp1ε is a regulator of takeout. Takeout levels are low in flies with reduced Pdp1ε and high in flies with increased amounts of Pdp1ε. Furthermore, flies with reduced or elevated Pdp1ε levels in the fat body display courtship defects, identifying Pdp1ε as an important transcriptional regulator in that tissue.
Keywords: fat body, courtship, clock, Drosophila
Genetic and molecular analyses have yielded significant insight into the genes that constitute the core components of the Drosophila circadian clock (reviewed in refs. 1 –3). It is regulated by two interlocked transcription/translation–based feedback loops, the period/timeless (per/tim) and Clock (Clk) loops (4). In addition, the regulation of the nuclear entry of proteins, their degradation rate, and phosphorylation state are crucial regulatory steps that are controlled by and contribute to the circadian clock. The per/tim cycle starts with the binding of CLK/CYC heterodimers to E-box promoter elements of the per and tim genes and their subsequent transcriptional activation. Eventually, the newly formed PER and TIM proteins will enter the nucleus and inhibit CLK/CYC action, thereby inhibiting the transcription of their own genes (5 –7). Transcription of per and tim will resume once their protein levels have decreased sufficiently to release the inhibition of CLK/CYC. Rhythmic expression of Clock mRNA is regulated in the second loop, the Clock loop. CLK/CYC activate vrille (vri) and Pdp1ε, a transcriptional repressor and activator respectively, that have been shown to bind the Clock promoter competitively (8, 9). Although it was thought that this competition accounts for the oscillatory regulation of Clk mRNA, the fact that Clk mRNA levels are still high in ClkJRK mutants (4) and that the core oscillator is only minimally impacted in flies with increased or decreased PAR domain protein 1 (Pdp1ε) levels (depending on the allele/transgene and tissue examined) (10 –12), indicate that an as yet unknown activator is required for Clk transcription. It has recently been demonstrated that CLK protein levels are constant in cells and that it is the phosphorylation state of the protein that determines its binding to DNA and its transcriptional activator function (7).
Although locomotor activity is the best-characterized circadian output, the circadian clock regulates numerous other outputs such as sleep (13 –16), neuronal activity in olfactory neurons (17), and metabolism (18), and a number of tissues have been found to harbor autonomous clocks (19). This poses the question of the nature of the output genes that mediate these processes. Not surprisingly, because many of the main regulators are transcription factors, molecular screens for transcripts that are under circadian control, and change in mutants that affect the core clock, have revealed a large number of such transcripts (20 –27). However, very little is known about the function of these potential output genes and the processes they regulate. The takeout (to) gene has been consistently identified in these screens. Its RNA and protein have been shown to cycle with a circadian rhythm, with a peak at late night/early morning and a trough in the late morning, closely resembling the cycling pattern described for Clk mRNA (24, 25). In contrast to what has been described for Clk mRNA, however, takeout levels were found to be down-regulated in ClkJRK, cyco1, and tim01 circadian mutants (25). The simplest way to explain takeout down-regulation in Clk and cyc mutants was to assume that its transcription is regulated by CLK. This would make takeout a CLK target with an unusual circadian rhythm because the CLK targets per and tim cycle in almost perfect antiphase to takeout. This suggested a more complicated and unusual way of takeout transcriptional regulation.
In this paper we show that in wild-type strains, there are two types of takeout expressers: High level takeout expressers and low level takeout expressers. When transcript levels were examined in outcrossed strains that contain only the high-expressing variant, we found circadian regulation of takeout transcription similar to that of Clk. Consistent with this, we show that Pdp1ε, a circadian activator that had previously been postulated to control clock output genes, controls takeout levels. Flies with disrupted Pdp1ε levels in the fat body display courtship defects, identifying Pdp1ε as an important transcriptional regulator in that tissue.
Results
Circadian takeout RNA Expression Is Regulated Similarly to Clk RNA.
In the process of studying the transcriptional regulation of the takeout gene, we noticed that some laboratory strains expressed takeout at the high levels reported earlier (28), whereas others showed much lower levels of expression. None of the low expressing lines carried the previously described takeout1 (to1) mutation (in which no takeout RNA can be detected (24, 28). Two wild-type strains, Canton-S (CS) and Crimea showed high levels of expression, whereas the other lines showed much lower levels of takeout RNA. The effect is due to a cis-effect (Fig. S1). The finding that there are two distinct kinds of takeout expressing alleles has implications for the analysis of takeout expression in mutant flies. If, for example, the mutant strain carries a low expressing copy, whereas the strain it is compared to is a high expresser, it is impossible to determine whether takeout levels in the mutant are low because of the mutation or because there are inherent differences in the takeout alleles between the two strains. It is therefore necessary to have equally expressing takeout alleles present in all strains to be compared.
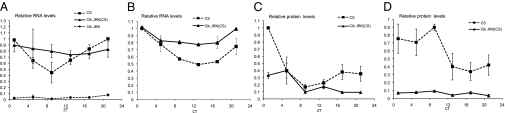
It has previously been demonstrated that takeout RNA levels cycle in a circadian manner with a peak between circadian time (CT) 21 and 1 and a trough around CT9, a rhythm similar to that of Clk and cry RNAs. But, whereas Clk and cry RNA levels are constant high in ClkJRK mutants, takeout RNA levels were reported to be nondetectable in ClkJRK and cyc01 mutants (24, 25), a regulation more like that found for per and tim, which both cycle with a different circadian phase than takeout. Thus, the regulation of takeout was unusual given the circadian profile of takeout RNA and protein. Given our findings of high- and low-expressing takeout alleles, we wondered whether the presence of differing takeout alleles in the strains compared might have influenced the results. To facilitate further examination of the circadian control of takeout in a Clk mutant background, we recombined the “high expressing” takeout allele from the Canton-S (CS) wild-type strain onto the ClkJRK mutant chromosome. We refer to this line as ClkJRK(CS). We verified the presence of the ClkJRK mutation by sequencing and monitoring circadian behavior and the presence of takeout(CS) by expression (see below). We then analyzed takeout RNA levels in these lines by quantitative real-time PCR (qPCR) and compared them to the levels in the original ClkJRK mutants (Fig. 1 A and B). We found that takeout RNA levels are constant at near-peak levels under light-dark (LD) and dark-dark (DD) conditions in ClkJRK(CS) flies in comparison with wild-type flies. Therefore, transcriptional regulation of takeout may be similar to that of Clk, consistent with their similar circadian profiles. The ClkJRK line was found to be a takeout low-expresser line.
Fig. 1.
(A and B) Circadian takeout RNA expression in ClkJRK(CS) males is constant at high levels. takeout qPCR analysis of male head RNA from flies collected at the indicated times under (A) LD (ZT) and (B) DD conditions (CT). Relative to mRNA levels were quantified as described in Materials and Methods. RNA levels were normalized to the amount in control CS flies at ZT1. Data are from three independent repeats. Original ClkJRK mutants, mutants carrying a high-expressing takeout allele ClkJRK(CS), and CS control flies were examined. (C and D) Takeout (TO) protein levels are under posttranscriptional control. Quantification of TO levels from Western blots. TO levels were quantified as described in Materials and Methods and normalized to wild-type levels. Data are from three independent repeats. Proteins from heads of wild-type CS and ClkJRK(CS) males were examined. (C) Flies were collected at the indicated times under LD conditions (ZT). (D) Flies were entrained for 3 days and collected at the indicated times on the first day of DD.
Complex Posttranscriptional Regulation of Takeout Protein Levels.
We next examined protein levels using a Takeout antibody we had previously generated (29). Flies were entrained and collected at different time points under 12-h light:12-h dark (LD) conditions, or under constant dark (DD) conditions following 3 days of LD entrainment. We found that under LD conditions, Takeout protein levels in wild-type closely follow RNA cycling with a peak around Zeitgeber Time (ZT) 1 (Fig. 1C). In the absence of light (DD conditions), the Takeout protein peak in wild-type is broader, suggesting a role for light in the degradation of the protein (Fig. 1D). Unexpectedly, in the ClkJRK(CS) mutants where RNA levels are at near-peak constant levels, Takeout protein amounts are very low, indicating posttranscriptional control of the protein. Under LD conditions, a small amount of protein is induced in the mutant in the beginning of the day, suggesting that protein levels can be directly controlled by light (Fig. 1C). In DD, Takeout protein levels are constant below trough levels in ClkJRK(CS) mutants (Fig. 1D).
Takeout Is a Target of PDP1ε Regulation.
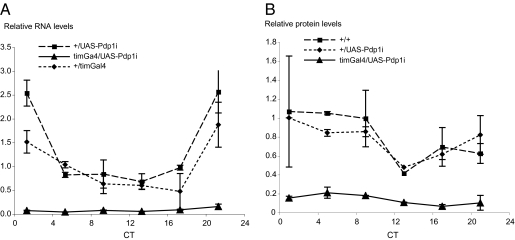
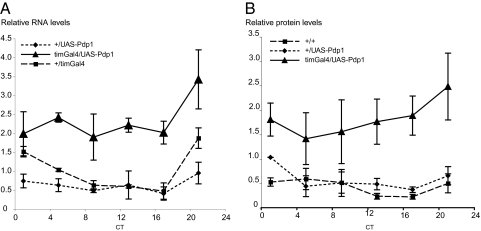
The results presented above indicate that the circadian regulation of takeout might be similar to that of Clk: both transcripts peak in the beginning of the day and are constant high in ClkJRK mutant flies. It has previously been suggested that in ClkJRK mutants an activator of Clk is constantly high, thus creating the high levels of Clk RNA. Because ClkJRK mutant RNA cannot give rise to a functional protein due to a nonsense mutation, the feedback loop that would normally reduce the levels of the activator is interrupted. It was previously shown that the CLOCK-CYCLE heterodimer activates the two transcription factors vri and Pdp1ε. Because both VRI and PDP1 bind the same regulatory element in Clk, the VRI/PDP1 ratio was thought to control the level of Clk transcription. This would predict that in flies with constitutively high levels of PDP1, Clk RNA levels should be very high. Flies with low levels of PDP1 should have low levels of Clk RNA. However, it was shown recently that Clk RNA and protein are not absent in flies expressing high or low levels of PDP1ε. Depending on which Pdp1 allele/transgene was used and which tissue was tested, Clk levels and the core circadian oscillator are either not affected (10, 11) or moderately impacted (12). However, interestingly, flies expressing high or low levels of PDP1ε were consistently arrhythmic in a locomotor activity assay, indicating that PDP1ε levels regulate oscillator output and thus control circadian output genes (10 –12). takeout has been identified in several screens as an output gene of the circadian oscillator. Given its similarity in circadian regulation to Clk, we hypothesized that it might be one of the genes controlled by Pdp1ε. We therefore examined takeout RNA and protein levels in flies with altered Pdp1ε levels. We made use of previously described transgenes that allow either overexpression of Pdp1ε (UAS-Pdp1), or its reduction by RNAi (UAS-Pdp1i) using the Gal4/UAS system (10). We used the tim-Gal4 driver to express the transgenes. tim-Gal4 is expressed in all clock cells, including the fat body (30). This is significant because takeout is preferentially expressed in the fat body of adult males (28). takeout RNA levels in the mutants were measured by qPCR and protein levels were assessed by Western blots using our Takeout antibody. Flies were entrained in a 12-h LD cycle for 3 days and collected at different time points during the first day of DD. RNA and protein was prepared from the heads of males and analyzed. Figs. 2 and 3 show expression of takeout RNA and protein in flies with either reduced PDP1 levels (timGal4/UAS-Pdp1i) or in flies that overexpress PDP1 (timGal4/UAS-Pdp1). In timGal4/UAS-Pdp1i flies, we observed very low levels of takeout RNA in comparison with the levels in the respective control strains, suggesting that PDP1 is required for takeout expression at wild-type levels (Fig. 2A). In agreement with the observed RNA levels, we also observed reduced protein levels in the timGal4/UAS-Pdp1i mutants (Fig. 2B). In contrast, when PDP1 was overexpressed, takeout RNA levels were significantly higher than in the control flies and so were protein levels (Fig. 3 A and B). Taken together these data indicate that takeout is a target of Pdp1 regulation. Because takeout is preferentially expressed in head associated fat body, and tim-Gal4 is expressed in these cells, we conclude that PDP1 is likely to be an important transcriptional regulator in the fat body.
Fig. 2.
takeout RNA and protein levels are low in flies with reduced Pdp1ε levels. takeout RNA (A) and protein (B) levels are strongly reduced under constant low levels of PDP1ε. PDP1 levels were reduced by expression of UAS-Pdp1i using the timGal4 driver. RNA and protein from heads of timGal4/UAS-Pdp1i males was compared to that of the corresponding control genotypes (+/UAS-Pdp1i and +/timGal4). Flies were entrained for 3 days and collected at the indicated times on the first day of DD. Data are from three independent repeats.
Fig. 3.
takeout RNA and protein levels are elevated in Pdp1ε over-expressing flies. takeout RNA (A) and protein (B) levels are increased in flies that overexpress PDP1ε. PDP1 was over-expressed by expression of UAS-Pdp1 by the timGal4 driver. RNA and protein from heads of timGal4/UAS-Pdp1 males was compared to that of the corresponding control genotypes (+/UAS-Pdp1 and +/timGal4). Flies were entrained for 3 days and collected at the indicated times on the first day of DD. Data are from three independent repeats.
PDP1 Mutants Show Reduced Male Courtship Behavior.
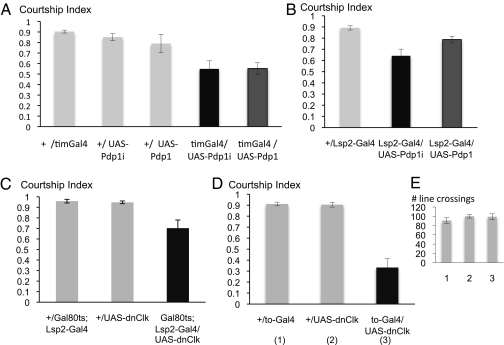
We have previously shown that the disruption of takeout leads to reduced male courtship (28). Given the low levels of takeout in the timGal4/UAS-Pdp1i mutants, we were wondering whether these males might have courtship defects when paired with target control females. As shown in Fig. 4A, males with low Pdp1 levels (timGal4/UAS-Pdp1i) show a significantly reduced courtship index. The courtship index is a measure of the fraction a male spends courting a female during the observation period. Courtship is also reduced in timGal4/UAS-PdP1 males that overexpress Pdp1 (Fig. 4A), suggesting that PDP1 is involved in maintaining controlled levels of proteins that are important for courtship. The observed reduction in courtship is more pronounced than that found in takeout mutant males alone (28), indicating that Pdp1 levels affect additional “courtship” genes. We have previously shown a similar courtship reduction to the one seen in the Pdp1 experiments in males with feminized fat body (29). Because timGal4 is expressed in fat body, we wondered whether the observed effect was due to Pdp1ε regulation of additional fat body genes besides takeout. To test this hypothesis, we used a fat body specific Gal4 driver that we had previously generated, Lsp2-Gal4 (29), to express UAS-Pdp1 and UAS-Pdp1i specifically in fat body (Figs. S2 and S3). We found similarly reduced courtship scores in these males (Fig. 4B), indicating that Pdp1ε has a role in regulating genes in the fat body that control courtship. The reductions are somewhat less pronounced than with the timGal4 driver. This may reflect differences in the temporal expression of the two drivers or the fact that Lsp2-Gal4 is a slightly weaker fat body driver than tim-Gal4 (Fig. S2). Alternatively, it may be due to the contribution of cells outside of the fat body.
Fig. 4.
Males with decreased or increased Pdp1ε levels or disrupted fat body clock show reduced courtship. Courtship indices (± SEM) of males toward wild-type virgin females. (A) timGal4 driven Pdp1 overexpression (timGal4/UAS-Pdp1) or reduction (timGal4/UAS-Pdp1i) results in reduced male courtship indices. Mutant males are compared to the corresponding control males. (B) Males with decreased or increased levels of PDP in the fat body show reduced courtship. The fat body specific Lsp2-Gal4 driver was used to express UAS-Pdp1i or UAS-Pdp1. n = 10 for all genotypes. (C and D) The courtship indices of males expressing a dominant negative form of Clk (dnClk) in the fat body is shown. (C) The fat body specific Lsp2-Gal4 driver was used to express dnClk. The presence of Gal80ts allows induction of dnClk only in adult males. Expression was induced by exposing mature males to 32 °C overnight. Experimental and control males were treated equally. Testing occurred 2–4 h later. (D) The to-Gal4 driver was used to express dnClk predominantly in fat body. (E) Activity assay of the genotypes in D. The number of line crossings was counted.
To examine whether the effect on courtship we observed in flies with altered Pdp1 levels was due to the disruption of the circadian clock in the fat body of these flies, we examined PER protein expression. We observed circadian oscillation of PER in all PER expressing tissues, including the fat body, indicating that the circadian clock was not abolished (Fig. S4). To test whether disruption of Pdp1 levels by a mutation in the circadian clock would be able to disrupt courtship, we next tested courtship behavior of flies with a disrupted circadian clock in the fat body. We made use of a previously described transgene that allows expression of a dominant negative form of Clock (UAS-dnClk) to disrupt the circadian clock (31). We first used the fat body specific Lsp2-Gal4 driver to express UAS-dnClk. However, fat body expression of dnClk using this driver proved lethal, possibly because of dominant effects of the transgene when expressed during development. Even when the Gal80ts system (32) was used to selectively induce dnClk only in adults, few adults were recovered, probably due to some inherent leakiness of the Gal80ts transgene during development. In the Gal80ts/UAS-dnClk; Lsp2-Gal4 males that could be recovered, we induced expression of dnCLK in adult males by overnight exposure to 32 °C, and tested the flies the next day. Males treated in this way showed reduced courtship, as shown in Fig. 4C, although the reduction was slightly less than that observed in Pdp1ε mutant flies. This might be due to limited disruption of the circadian clock in the surviving flies. It has recently been demonstrated that flies that express dnCLK in the fat body driven by a takeout-Gal4 (to-Gal4) driver (28), which is adult-specific, survive and show a disrupted circadian clock in the fat body (18). We therefore repeated the courtship experiments using to-Gal4 to express UAS-dnClk. The results are shown in Fig. 4D. We observed a strong reduction in courtship, similar to that found in Pdp1ε mutants. Taken together these results indicate that the observed phenotype in flies with altered Pdp1ε levels is at least in part due to the circadian function of Pdp1ε. Although it has previously been shown that these flies have defects in their metabolism, they were normal in a short term activity assay (Fig. 4E), indicating that the courtship defect is not caused by general sickness of the flies but rather due to the disruption of Pdp1ε and/or circadian regulation.
Discussion
Regulation of takeout RNA.
We have found that there are takeout high expressers and takeout low expressers. Interestingly, the observation of high- and low-expressing alleles may not be limited to the takeout gene but may extend to other fat body genes. Fuji et al. (33) have described strain differences in expression levels of other sex specific fat body genes. It is unknown what the biological significance of this dimorphism is. Strains collected from the wild from across Africa that differ in their hydrocarbon pheromone profiles (34) were found to be takeout low-expressers (Fig. S1), indicating selection against high levels of Takeout. The different expression levels clearly have implications for the analysis of expression of these genes in mutants. We have found that the original ClkJRK strain contains a low expressing copy of takeout. When a high expressing copy was recombined onto the ClkJRK chromosome, it became apparent that takeout RNA in Clk Jrk(CS) flies is constant at near-peak levels under LD conditions. Because takeout is mainly expressed in fat body, our results further confirm the presence of a functional circadian clock in that tissue, as has recently also been shown by Xu et al. (18). We have found that in wild-type, Takeout protein cycling closely follows RNA cycling under LD conditions, as has previously been described (25). In the absence of light, the Takeout protein peak in wild-type is broader, suggesting a role for light in the degradation of the protein. In ClkJRK(CS) mutants, despite near-wild-type constant levels of RNA, Takeout protein levels are constant below trough levels in DD, indicating additional posttranscriptional regulation. Under LD conditions, a small amount of protein is induced in the mutant at the beginning of the day, again suggesting that protein levels can be directly controlled by light. Control by light in addition to the circadian clock has also been observed for fat body regulated feeding rhythms (18).
Pdp1ε Regulates takeout RNA Levels.
In addition to regulation by Clk, our data show that takeout RNA expression is also regulated by PDP1 levels. takeout RNA levels are elevated approximately fivefold in flies that overexpress PDP1. That this is unlikely to reflect unspecific activation is demonstrated by the fact that in the opposite situation, when PDP1 levels are low due to reduction by RNAi, takeout levels are affected in the opposite way. In fact, this mode of regulation by Pdp1ε had previously been suggested for the Clk gene. However, recent experiments indicate that PDP1 is not a major activator of Clk transcription (10), but both increasing and decreasing the levels of Pdp1ε in clock cells disturbed circadian locomotor activity, indicating that Pdp1ε is required to regulate clock output genes. The molecular nature of these targets remains unknown. Genes with circadian rhythms that resemble those of Clk and takeout (peaks near dawn or early in the morning) are candidates for being Pdp1ε regulated output genes. Some of these are likely to be genes that are involved in locomotor activity, but there are probably others, like takeout, that have different roles and may be involved in other rhythmic outputs. A circadian function for very few of these genes has been identified to date. takeout was initially found in these screens and is among the best characterized clock output genes so far. We show here that takeout is regulated by Pdp1ε and that Pdp1ε is a transcriptional regulator in the fat body, a metabolic tissue. Disturbance of the circadian clock in fat body does not affect activity rhythms (18), indicating that fat body specific outputs are not involved in locomotor control. A role for the fat body clock in the control of feeding rhythms and circadian starvation resistance has recently been demonstrated, and at least one cyclically expressed metabolic gene identified (18). Interestingly, takeout has previously been implicated in the control of larval feeding behavior (24). It remains to be seen how the two observations are linked.
Although our data indicate similarities in the regulation of takeout and Clk or cry transcripts, there are important differences in the role of PDP1 as a regulator of to versus Clk that may be related to the more prominent role of Pdp1 in the control of output genes. It is unclear whether previous observations regarding the effect of per01 and tim01 mutations on to transcript levels need to be revised based on the status of the genetic backgrounds in these lines as high or low to expressors. Future experiments addressing this issue will shed further light on the extent of coregulation of to and Clk or cry.
We have shown here that the RNA levels of takeout are regulated by Pdp1ε; they are high when Pdp1ε is overexpressed and low when Pdp1ε levels are reduced. It has previously been shown that Pdp1ε levels are low in Clk mutants when measured in whole heads (8, 35). This would predict that takeout levels in Clk mutants should be low due to lowered PDP1 levels. However, we have observed fairly high constant takeout levels in ClkJRK(CS) mutants. This suggests that the low levels of Pdp1ε in the mutants are sufficient to activate takeout, or that there is a separate activator present. That there is an appreciable amount of PDP1 in ClkJRK flies is evident because these flies live, whereas null mutants for Pdp1ε die during development (8, 35). In contrast to what we have observed in ClkJRK(CS) mutants, takeout RNA and protein levels are directly correlated in Pdp1ε over- and underexpressing flies, suggesting a disruption of the circadian translational control of takeout in the mutants. The role of Pdp1ε in the regulation of takeout transcription is likely to be indirect because we have not been able to find PDP1 binding sequences or PDP1 binding to the takeout promoter. Furthermore, the regulatory elements in takeout that mediate circadian expression have not been identified yet.
Pdp1ε Regulated Fat Body Genes Are Involved in Male Courtship.
Takeout is predominantly expressed in male fat body and takeout mutant males have reduced courtship (28). That male specific factors from the fat body play an important role in male courtship has been demonstrated by the fact that specific feminization of just this tissue significantly reduces courtship to a degree that is beyond the reduction observed in takeout mutants (29). This indicates that male factors other than takeout also play a role. We speculate that some of these factors are also regulated by PDP1 because flies with disturbed PDP1 levels in the fat body show courtship defects similar to those observed in flies with feminized fat body.
Circadian control of mating (but not courtship) has been described. However, in both of these studies it was noted that male courtship did not show a rhythm but that the rhythm was set by the female. Tauber et al. (36) have found preferential mating around dusk and overall higher levels during the subjective night than in the subjective morning. These rhythms were dependent on the clock gene per. Sakai and Ishida (37) observed mating rhythms in wild-type females, which were abolished in tim and per mutants. The only description of a male circadian courtship activity rhythm to date is by Fuji et al. (38). These authors observed a distinct shift in activity pattern when a male and a female fly were housed together. Male–female couples show high levels of “close-proximity” (courtship) activity throughout the night and early morning. The rhythm is dependent on the clock genes in the brain and antennae and is dependent on the male’s circadian rhythm. It remains to be seen whether this circadian output behavior is regulated by Pdp1ε-regulated genes in the brain and/or the fat body.
Materials and Methods
RNA Northern blots and hybridizations were performed as described in (28).
RNA Quantification.
takeout mRNA levels were assayed by qPCR. Flies were entrained in a 12-h LD cycle for at least 3 days, collected every 4 h, and immediately stored at −80 °C. For DD collections, flies were entrained for 3 days in a 12-h LD cycle and collected every 4 h on the first day of DD. Total RNA was isolated from male fly heads using TRIZOL (Invitrogen). To eliminate genomic DNA contamination, each sample was treated with DNaseI (Promega). First-strand cDNA was synthesized from 1 μg of RNA using oligodT primers and SuperScript II (Invitrogen). For qPCR, TaqMan assays were performed using the following to primers and probes: forward primer, 5′-GCCTTTTGGTCTCGGTGGAT-3′; reverse primer, 5′-GCCATCACCATACTTACAAGGTTTT-3′; probe 6FAM-TCCCCGAAGATC -MGBNFQ. Ribosomal protein 49 mRNA (rp49) was used as the internal loading control. The primers and TaqMan probe for rp49 were as follows: forward primer, 5′-CTGCCCACCGGATTCAAG-3′; reverse primer, 5′-CGATCTCGCCGCAGTAAAC-3′; probe VICCCTCCAGCTCGCGCACGTTG-MGBNFQ. Reactions were run on an Applied Biosystems Prism 7000. The relative levels of to and rp49 RNAs were calculated based on standard curves for to and rp49 that were run in each assay. to levels were normalized to rp49 at each time point.
Takeout Western blots were performed as described in ref. 29. Flies were entrained in a 12-h LD cycle for at least 3 days, collected every 4 h, and immediately stored at −80 °C. For DD collections, flies were entrained for 3 days in a 12-h LD cycle and collected every 4 h on the first day of DD. Protein was extracted from male heads. Quantitation of Western blots: The relative levels of TAKEOUT (TO) were quantified as the ratio of the TO band intensity to that of a nonspecific background band using Quantity One1-D Analysis software (Bio-Rad). These relative TO levels were normalized to TO levels in the wild-type control at ZT1 or its highest level.
Fly Stocks.
The UAS-PDP1i and UAS-PDP1 transgenic strains were as described in ref. 10. The timGal4 driver (39), the fat body driver 3.1 kb Lsp2-Gal4 (29) and the to-Gal4 driver (28, 29) have been described before. ClkJRK flies were as described (40). Generation of ClkJRK(CS):ClkJRK, ry flies were crossed to CS flies. Individual recombinant progeny of ClkJRK(CS), ry/CS females were then screened for the absence of ry and the presence of ClkJRK by arrhythmicity in a locomotor assay (10). (Location of genes: Clk 66A, ry 87D, to 96C). The DNA region around the C to T amino acid replacement that changes Q776 into a stop codon in ClkJRK was amplified by PCR and sequenced to verify the presence of the mutation. At the same time, the number of glutamines in the longest polyglutamine repeat were confirmed to be 25, as had previously been described for ClkJRK (40). UAS-dnClk transgenic lines were as described (31). The Gal80ts system was used to conditionally express dnCLK (32). P{tubP-GAL80ts}20/UAS-dnClk; Lsp2-Gal4 flies were grown and kept after eclosion at 18 °C. To inactivate Gal80ts, 7- to 8-day-old males were transferred to the restrictive temperature of 32 °C overnight for at least 18 h and tested after an additional 2–3 h at room temperature. Control flies were treated in the same way. All fly strains were reared on medium containing corn meal, yeast, agar, and Tegosept at 25 °C, except for the Gal80ts experiments, for which flies were reared at 18 °C.
Courtship assays were performed as described in ref. 28.
Supplementary Material
Acknowledgments
We thank William Mattox (University of Texas M.D. Anderson Cancer Center) for help with the identification of the polymorphisms in the wild-type strains and for discussions. We thank Gregg Roman (University of Houston) for providing bench space during parts of this project, and Fanny Ng and Pete Taylor for helpful discussions. We further thank Michael Rosbash (Brandeis University) for anti-PER antibodies. This work was supported by National Science Foundation Grants IOS-0416476 and IOS-0919697 to B.D.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906422107/DCSupplemental.
References
- 1.Benito J, Zheng H, Ng FS, Hardin PE. Transcriptional feedback loop regulation, function, and ontogeny in Drosophila. Cold Spring Harb Symp Quant Biol. 2007;72:437–444. doi: 10.1101/sqb.2007.72.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall JC. Genetics and Molecular Biology of Rhythms in Drosophila and Other Insects. Adv Genet. 2003;48:1–280. doi: 10.1016/s0065-2660(03)48000-0. [DOI] [PubMed] [Google Scholar]
- 3.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 5.Darlington TK, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 6.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cyran SA, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 9.Glossop NR, et al. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 10.Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim C, Lee EK, Choe J. Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster. Biochem Biophys Res Commun. 2008;364:294–300. doi: 10.1016/j.bbrc.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, et al. An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator. J Neurosci. 2009;29:10920–10927. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19:386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sehgal A, et al. Molecular analysis of sleep: Wake cycles in Drosophila. Cold Spring Harb Symp Quant Biol. 2007;72:557–564. doi: 10.1101/sqb.2007.72.018. [DOI] [PubMed] [Google Scholar]
- 16.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 18.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 20.Ceriani MF, et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claridge-Chang A, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 24.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 25.So WV, et al. takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol Cell Biol. 2000;20:6935–6944. doi: 10.1128/mcb.20.18.6935-6944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda HR, et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 27.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazareva AA, Roman G, Mattox W, Hardin PE, Dauwalder B. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3:e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 32.McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: A synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Fujii S, Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–5363. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sureau G, Ferveur JF. Co-adaptation of pheromone production and behavioural responses in Drosophila melanogaster males. Genet Res. 1999;74:129–137. doi: 10.1017/s0016672399003936. [DOI] [PubMed] [Google Scholar]
- 35.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 36.Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr Biol. 2003;13:140–145. doi: 10.1016/s0960-9822(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 37.Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci USA. 2001;98:9221–9225. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 40.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.