Abstract
Using electron microscopy and image processing, we have observed myosin 5a modified with lever arms of different lengths (four, six, and eight calmodulin-binding IQ domains) and orientations walking along actin filaments. Step lengths were dependent on lever length: 8IQ > 6IQ > 4IQ, which is consistent with myosin 5a having evolved to walk straight along actin. Lead heads were mostly in the prepowerstroke state, tethered there by the trail head. However, improved image processing showed that in 5–10% of molecules the lead motor was in the postpowerstroke state. This is a unique attached state of myosin, where the motor domain has completed its powerstroke at the expense of severe lever distortion, but with little cargo movement. Postpowerstroke lead heads were seen in both wild-type and modified lever molecules, mostly where there was least strain. These data allow the strain dependence of the equilibrium between pre- and postpowerstroke conformations to be measured. Slow rates of ADP dissociation observed from lead heads of these molecules can be explained by the unfavorable equilibrium between the pre- and postpowerstroke conformations preceding ADP loss.
Keywords: actin, powerstroke, electron microscopy, kinetics, molecular motor
The myosin 5a molecular motor occurs widely and is especially abundant in neurons, forming ≈0.3% of brain protein (1). The two heads in a molecule walk hand-over-hand along actin filaments, pulling diverse vesicle cargoes (2). Heads attach to actin through their motor domains, which also contain ATPase activity. Extending from the motor domains are levers that produce force and movement by swinging through ≈80°, brought about by movement of the converter region of the motor domain (3). The ends of the levers join at the tail, the other end of which binds cargo. The myosin 5a lever consists of six IQ motifs that bind calmodulin family light chains, which is among the longest in the 30 or so myosin classes now known (4, 5). This extended length allows the two heads to span the actin filament helical pseudorepeat of ≈36 nm, which results in approximately straight walking along filaments.
Myosin 5a walks processively, taking many steps for each encounter with an actin filament (6). By contrast, muscle myosin 2 attaches only briefly and takes single steps that are smaller, 5–10 nm (7, 8). Processivity is thought to result from phosphate dissociation from the lead head being much more rapid than ADP dissociation from the trail head, a higher affinity of the M.ADP.Pi state for actin than in myosin 2, and a slower rate of ADP release from the lead head as a result of backward mechanical strain (9 –14). This enables the lead head to form the strongly bound A.M.ADP state before ADP dissociation from the trail head, which allows the trail head to bind ATP and dissociate from actin. Rate-limiting dissociation of ADP from the trail head results in myosin 5a being strongly bound to actin for more than 95% of its working cycle—a high so-called duty cycle. The high duty cycle also facilitates study of the walking mechanism (15); as a consequence, myosin 5a walking is now the best understood of any myosin class.
Much progress in understanding the structure and dynamics of myosin 5a walking has come from mechanical experiments and electron microscopy on individual molecules. Single-molecule mechanics showed that the head working stroke occurs in substeps of 20 and 5 nm that correlate with Pi and ADP release (3, 16, 17). The high duty cycle allowed imaging of a myosin walking during ATPase. This was observed in actin filaments sparsely decorated with myosin 5a by negative stain electron microscopy, to a resolution of ≈2 nm. Single-particle image averaging of these data revealed that the heads mostly bound 13 actin subunits apart (36 nm), which would result in approximately straight walking, although some molecules were attached 11 and 15 subunits apart (30 nm and 41 nm) (18).
Sparse decoration electron microscopy also showed that in the doubly attached molecule the levers project from the back and front of the lead and trail motor domains, respectively. This revealed the different positions of the converter subdomain that cause the lever to swing. This in turn showed that lead and trail motors were at the start and end of their powerstrokes, (19). Subsequently, 3D cryo-electron microscopy reconstructions were obtained of actin saturated with single myosin 5a heads with AMPPNP or ADP.AlF4 bound, which are thought to show the prepower state; these models were at 4.2 and 5.4 nm resolution, respectively (20). The myosin 5a data remain the only images of a myosin attached to actin in prepowerstroke conformation.
Movement of the head–tail junction produced by the lever of a lead head swinging to become a trail head is ≈25 nm, but the step distance is 36 nm (16). It is thought that the extra distance the new lead head reaches through to find its binding site results from a diffusive search for an actin subunit at a similar orientation to the one bound by the trail head. This search is driven by Brownian motion and means that the doubly attached molecule is distorted, which is important for strain-dependent steps of the ATPase linking the enzymatic and walking cycles. Lead and trail head shapes compared with head crystal structures docked onto actin gave some indication of the extent of the distortion. This seemed to be mainly in the lead head lever, either bending throughout its length, or at the “pliant region” where it joins the motor domain, which is due to the IQ α-helix melting locally (19).
Myosin 5a walking has also been explored in single-molecule experiments with optical tweezers and fluorescence imaging with one nanometer accuracy (FIONA) using mutant molecules in which lever length was varied by adding or deleting IQ motifs and hence the number of light chains bound. Mutant molecules were constructed with levers composed of four or eight light chains, or six light chains with two alanines inserted between IQs 3 and 4 to give ≈180° rotation between the two halves of the lever. The altered molecules were found to move processively along actin filaments, although not as well as the wild type (12, 21). There was also greater variability in step size, especially in 8IQ molecules (22).
Here we used electron microscopy and stopped-flow fluorescence to study binding to actin of the same mutant myosin 5a types as those used above. Step lengths were dependent on lever arm length: 8IQ > 6IQ > 6IQ+2Ala > 4IQ, which is consistent with myosin 5a having evolved with 6IQ domains to bind actin subunits at similar azimuths to walk straight along actin. The rates of dissociation of the fluorescent hydrolysis product deac-aminoADP from the lead head were slowest from the wild-type molecule and increased in the order 6IQ < 8IQ ≈ 6IQ+2Ala < 4IQ. Improved image processing of sparsely decorated filaments allowed whole-molecule image averages to be produced, resulting in greater clarity. This revealed a unique conformation of attached lead heads, which shows severe curvature of the lead head lever resulting from the motor domain having completed its powerstroke. Cargo movement usually directly accompanies the powerstroke, and this unique attached head shape demonstrates decoupling of these processes.
Results
Microscopy of Mutant Myosin 5a.
Using negative staining electron microscopy, mutant 4IQ, 6IQ+2Ala, and 8IQ myosin 5a molecules were first examined not attached to actin and compared with wild-type molecules. All of the preparations showed two-headed molecules similar to the images of myosin 5a we reported previously (19) and with apo head contour lengths increasing in proportion to the number of IQ motifs (Table 1). A linear fit to the data (r 2 = 0.99) indicates that the long axis of the motor domain is 8.8 nm and that each IQ motif with calmodulin bound adds 3.8 nm.
Table 1.
Summary of WT and mutant myosin 5a properties
| Construct | Head length (nm ± SD) | Trail head k (s-1)* | Lead head k (s-1)* | Ifast/Itotal observed* | Ifast/Itotal calculated† | Kc (n)‡ | Kc (n − 2) |
| S1 | — | 0.49§ | — | — | — | — | — |
| 4IQ | 23.5 ± 1.9 | 0.55 | 0.17 | — | — | — | — |
| 6IQ+2Ala | 32.0 ± 3.3 | 0.44 | 0.041 | 0.73 | 0.70 | ND | 0.63 (11) |
| 8IQ | 39.2 ± 4.0 | 0.50 | 0.04 | 0.72 | 0.63 | ND | 0.85 (13) |
| 6IQ | 31.8 ± 2.8 | 0.49 | 0.015 | 0.56 | 0.57 | 0.02 (13) | 0.81 (11) |
*Data from Fig. S1.
†Fraction of the ADP molecules expected to dissociate rapidly = 0.5 + (n j-2)/2, where n j-2 is the mole fraction bound with lowest strain.
‡Equilibrium constant (Kc) between pre- and postconformations at the most common head spacing (n) and at two subunits less separation on the actin genetic helix (n − 2).
§See ref. 25.
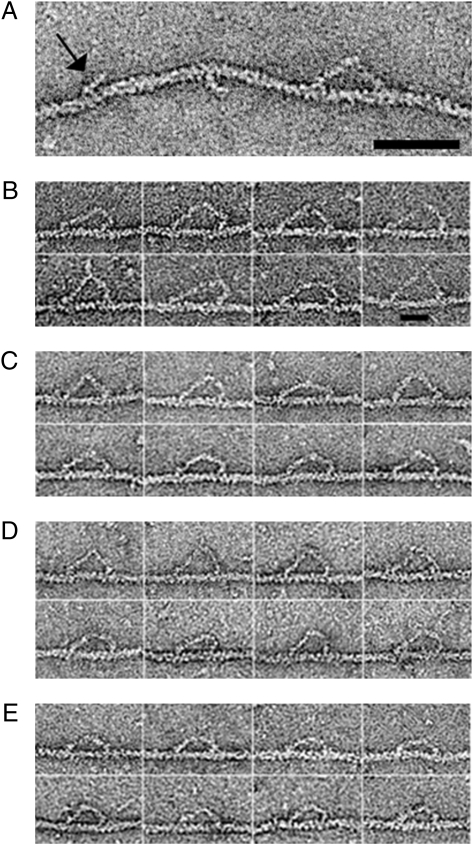
Fig. 1A shows an actin filament with a wild-type myosin 5a molecule attached. Also bound are two separate N-ethyl-maleimide (NEM)-treated myosin 2 heads (S1). NEM treatment makes the S1 bind strongly at the rigor angle of ≈45°, pointing toward the “barbed” end of the filament, and is not dissociated by ATP. This reveals directly the filament polarity, which in turn shows the walking direction of myosin 5a and thus which are the lead and trail heads in the molecule.
Fig. 1.
(A) Wild-type myosin 5a walking in ATP. Also attached are two single NEM-treated myosin 2 heads, one of which is indicated (arrow). These demonstrate the polarity of the actin filament. (Scale bar, 50 nm.) (B–E) Montages of wild-type and mutant myosin 5a HMM molecules bound to actin in 0.5 μM ATP, all walking to the right. (B) 8IQ, (C) wild type, (D) 2Ala-6IQ, (E) 4IQ. (Scale bar, 25 nm.)
Fig. 1 B–E were prepared using the polarity information and show montages of wild-type and mutant molecules, all walking to the right. As we previously reported, the appearance of attached wild-type molecules is asymmetric and quite variable. Asymmetry arises because the molecule appears skewed in the walking direction. This is because the lead head generally appears bent, but this can either be a sharp bend where the lever joins the motor domain, or a more gradual bend throughout the lever. In the latter cases the lever is bowed forward and away from the actin filament. Trail heads are generally straighter, but in some cases the lever is bowed toward the actin filament. A similar appearance of skewing in the walking direction and mixtures of straight and bent levers was seen in all of the mutant molecules but was most obvious in the 8IQ construct.
Heads of doubly attached molecules were found on the same side of actin filaments and spaced well apart in all cases. Attachment of the heads to adjacent actin subunits, as occurs with myosin 2 (23), was not seen; nor were molecules straddling both sides of an actin filament. Approximately 40% of both wild-type and mutant molecules attached by a single head, with the free head at a variety of angles suggestive of hinging at the head–tail junction. There were also some unattached molecules in the background. However, when ATP was omitted, or after sufficient time for it all to be hydrolyzed, there were very few unattached molecules, which demonstrates that almost all of the myosin was competent to bind actin in an ATP-dependent manner. ATP-free mixtures left for ≈1 min before preparing a grid showed bundling of the filaments, indicating cross-linking by the myosin.
Determination of Step Length.
Windowed images of doubly attached molecules were brought into alignment and classified by single-particle image processing. Images of walking to the left were mirrored and combined with those walking to the right. Classification aims to achieve homogeneous image groups that can then be averaged. The number of classes chosen depends on the variability in appearance, with more variability requiring more images to achieve homogeneity, as was the case here. Class averages containing more than approximately 30 images of similar structure generally result in greatly improved detail and signal-to-noise over raw images.
Averaging reveals the actin filament subunits, which become clearly visible angled toward the barbed end of the filament (Fig. 2). Subunits between the heads of attached myosin 5a can then be counted directly to produce histograms of head–head spacing for the different constructs (Fig. 2). The wild-type histogram had three peaks, with the major central peak corresponding to heads binding 13 actin subunits apart (77% of molecules). However, there was some binding 11 and 15 subunits apart (13% and 10% of molecules, respectively), that is, to the adjacent subunits on the same long-pitch actin helix. These data are similar to what we reported previously and demonstrate that myosin 5a heads span approximately the pseudorepeat of the actin filament helix; they are therefore consistent with the molecule walking approximately straight along actin filaments (18).
Fig. 2.
Details of stepping. Histograms show motor domain separation in raw images (gray bars) and image averages (black bars). Raw image distances were measured between motors using SPIDER. Distances between heads in image averages were measured between the attached actins. (Insets) Class averages obtained by segregating images according to motor separation. Classification used reference-free methods after windowing and aligning myosins attached by both heads. (Scale bar, 50 nm.)
Image averaging and classification produced only two step lengths for each mutant molecule type (Fig. 2, black bars and Insets). Thus, 8IQ heads were mainly (74%) 15 subunits apart, with a minority (26%) spaced by 13 subunits. 4IQ heads were spaced 11 (87%) and 13 (13%) subunits apart. 6IQ+2Ala molecules attached 11 (40%) and 13 (60%) subunits apart. However, there was evidence in measurements from the raw images of wider step distributions (Fig. 2, gray bars) that were more similar to those determined by optical tweezers and FIONA (12, 21). The absence of class averages with heads spaced other than 11, 13, and 15 subunits was probably due to the small numbers of molecules at extreme separations or variable appearance.
Details of Attached Head Shape.
Image averaging also revealed detail in the myosin attachment. In our previous work, lead and trail heads were windowed and averaged separately, but we have now been able to average entire molecules (Fig. 2, Insets). Lead and trail motor domains both appear similar, with each binding slightly toward the barbed end of its actin subunit, which is consistent with an unchanging stereospecific attachment. The motor domains of both wild-type and mutant molecules mostly have a roughly triangular profile, which results because the SH3 domain is a distinctive protrusion at the front in this view (19). The levers are visible in the averages but are less distinct owing to their variable shapes.
The asymmetric appearance of attachment results because the forward-pointing lever of the trail head emerges nearly overlapping the SH3 domain, which gives the whole trail head a relatively straight appearance. Because of the different position of its converter, in the prepowerstroke position, the lead head lever emerges from the rear of its motor domain and is also angled backward. The lead head SH3 domain therefore protrudes alone at the front, giving the lead head the appearance of being bent at the motor domain/lever junction. The motor domains of the mutant molecules all showed these features.
Postpowerstroke Lead Motors.
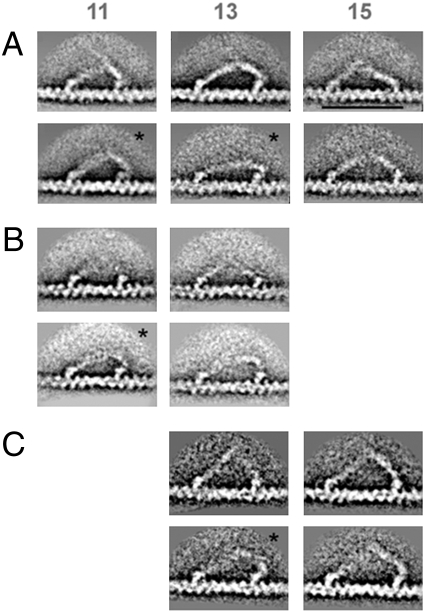
We previously showed that in the majority of cases the lead head lever projected from the rear of its motor domain, demonstrating that the lead converter was in the prepowerstroke state (18, 19). This conclusion remains unchanged and is important because there are still very few structural data about myosin heads attached to actin at the start of the powerstroke. However, a more detailed analysis revealed a different attached head state in a small proportion of lead heads. Fig. 3A shows segregation of the wild-type molecules into subclasses and reveals that in a fraction of cases (asterisks) the lead head lever emerges from the front and then bends strongly backward over the motor domain. This indicates that in these molecules the converter was in the postpowerstroke position, as in trail heads.
Fig. 3.
Subclasses of (A) wild-type, (B) 6IQ+2Ala, and (C) 8IQ molecules bound 11, 13, and 15 actin subunits apart. (Upper) Straight lead levers in prepowerstroke conformation. (Lower) Curved lead levers, some attached to a motor in postpowerstroke conformation (asterisks). (Scale bar, 50 nm.)
This behavior was seen in approximately 7% of wild-type lead heads and mainly occurred in short, 11-actin-subunit steps. We considered the possibility that the postpowerstroke conformation was produced by calmodulin dissociation, but it was also observed in the presence of excess (2 μM) calmodulin. Similar behavior was also observed in a small proportion of the mutant image averages, in the short step classes of the 8IQ and 6IQ+2Ala constructs (Fig. 3 B and C). These image classes therefore demonstrate a unique type of myosin cross-bridge behavior. The extra flexibility of the long myosin 5a lever, compared with other myosins, allows the lead head converter transition without detachment of the trail head. This occurs at the expense of severe distortion of the lead head lever near the motor. It was also accompanied by a small forward movement of the head–tail junction, estimated in the range 2–6 nm.
Kinetics of ADP Dissociation.
ADP dissociation from lead and trail heads was investigated using the ATP analog deac-aminoATP and double-mixing stopped-flow fluorescence. Construct was mixed with deac-aminoATP and, after a delay for substrate binding and hydrolysis to be completed, was then mixed with actin containing 2 mM ADP to measure a single turnover of product dissociation. The large (≈20-fold) fluorescence reduction of deac-aminoADP when released allows accurate measurement of rates (24, 25). Product dissociation of wild-type myosin 5a-deac-aminoADP.Pi mixed with actin is biphasic, with the fast phase from the trail head, 0.5 s−1, the same as from unstrained single heads. The slow phase from the lead head is ≈30-fold slower, 0.015 s−1 (Table 1 and Fig. S1), consistent with our earlier report (25).
Rates of product dissociation from the mutant trail heads were all similar, but those from lead heads varied in the order 6IQ < 8IQ ≈ 6IQ+2Ala < 4IQ (Table 1 and Fig. S1). For each myosin it would be expected that the rate of deac-aminoADP dissociation might vary with strain differences in the slightly different step lengths shown in Fig. 2, and that there would therefore be more than the two rates from single populations of trail and lead heads. This may be reflected in the deviations from the double exponential fits shown by the nonrandom residuals in Fig. S1. However, any differences were insufficient to allow determination of the rates of the individual processes, which were dominated by the largest amplitudes.
Discussion
Myosin 5a has one the longest levers in the 30 or so myosin classes now known. This allows its heads to span the actin filament 36-nm helical pseudorepeat and thereby walk approximately straight. What functional advantage straight walking confers on myosin 5a is still not clear, but it may avoid collisions of vesicle cargos, for example with membranes where actin filaments run close to these. It may also enable transport along bundles of actin filaments where following a helical track is impossible.
The 36-nm steps result from a strong preference of the lead head to attach to an actin subunit at an azimuthal angle similar to the one bound by the trail head. These steps span 13 actin subunits, and the main deviations from this are to adjacent subunits along the same long-pitch actin helical strand, producing 30- or 41-nm steps (18). The less frequent attachments are to subunits rotated only ±28°, assuming 13/6 filament symmetry. The data therefore suggest that the working stroke of myosin 5a lies along the actin filament axis, placing the detached head at an appropriate azimuth for straight walking. The mutant molecule data thus provide further evidence for the conclusion that myosin 5a evolved to walk straight.
Considerations of myosin 5a walking have usually assumed a 13/6 actin filament helix; however, the observed distance between the points where the long-pitch helices cross varies by ≈20%, indicating substantial departures from exact helicity (26). This disorder was modeled and proposed to result from rotational freedom between successive actin subunits (27). The distribution of myosin 5a binding sites may therefore be affected by conformational flexibility in the actin filament. Binding of the second head may thus apply a torque to the filament, so that attachments to 11 or 15 subunits apart are rotated less than the 28° predicted for a 13/6 helix. Consistent with this type of interpretation, the step distributions of the mutants in Fig. 2 are in good agreement with those predicted from a mechanochemical model of stepping that included up to 5° departures in the subunit arrangement from 13/6 symmetry (28).
Our step-size histograms from native myosin 5a are similar to those reported from optical tweezer and FIONA data (12, 22) but are somewhat narrower than those obtained from mutant molecules with these methods. Some of these differences may have resulted from greater noise in the tweezer and FIONA measurements, but these histograms also peaked further from the predominant native molecule steps of 36 nm. 4IQ molecules were reported to have ≈26-nm steps (±10 nm), which indicated heads spaced at nine or seven actin subunits that might reach around the actin filament by ≈60° or even 90° (12). FIONA data from the 8IQ mutant showed variable steps from 11 to 20 actin subunits (21, 22). The narrower stepping range seen by microscopy may have been because molecules taking extreme steps were so distorted as to be unrecognizable, perhaps because of increased flexibility in the mutant molecules.
Two Alanine Mutant.
The mutant with two alanine residues inserted after the third IQ (6IQ+2Ala) still bound mostly 11 or 13 actin subunits apart, similar to wild-type molecules, although the distribution was skewed toward shorter steps. This was unexpected because if the heavy chain α-helix continued through the added residues, the two halves of the lever will have been mutually rotated by approximately half a turn. The free head would then be expected to be turned away from the actin filament, impeding its rebinding as a lead head. Construction of the 4IQ and 8IQ mutants should also have inserted a 120° turn between the lever ends. That all these alterations had little effect on processivity may thus provide additional evidence that heads can rotate about their long axis at the head–tail junction to facilitate attachment to actin. Electron microscopy consistent with such behavior has previously been reported for myosin 2 (29, 30), and free movement of the detached head over a wide range during stepping has been reported for myosin 5a (17).
Alternatively, it may be that the insertion of the two alanines disrupted the IQ α-helix, resulting in flexibility. Sharp kinks in the middle of the lever suggestive of flexibility were not seen in either the actin-bound or free molecules of this mutant. However, the 6IQ+2Ala mutant studied by FIONA walked with a wider distribution of steps, suggestive of increased flexibility (22); it also gave a working stroke approximately half that of the wild-type molecule, again suggesting lever flexibility under load (21). Overall, the data suggest the lever is unbroken but weakened by this insertion.
Step Length Distribution.
The step lengths distributions were used to calculate the relative free energies of attachment, ∆∆Go, of the different molecules. Curves through the data were fit to the equation for a Hookean spring, ∆∆Go = 0.5 × k(i − B)2 + C, where i is the number of subunits between the bound heads (equal to 2.75i nm), k is the spring constant, B is the distance of minimum energy for each mutant, and C is a constant (Fig. S2). The data were fit by allowing the values of k and B to vary for each construct. Within experimental error, values for k (0.28 ± 0.07 kcal/mole)/(actin subunit)2 and C (0.26 kcal/mole) were the same in all cases. The distances fit to B are 10.7 (4IQ), 12.3 (6IQ+2Ala), 12.8 (6IQ), and 14.3 (8IQ) actin subunits. The spring constant translates to an apparent stiffness of 0.26 pN/nm per molecule, which can be compared with the rather wide range of values reported for the stiffness of skeletal acto-myosin cross-bridges (0.6–3.0 pN/nm) (31 –34) and for single myosin 5a heads, 0.2 pN/nm (10). Note that the apparent stiffness we obtained is a measurement of the flexibility of the lead head binding irreversibly to actin and contains both linear and twist terms, which is not the same as the stiffness of a single bound head.
Postpowerstroke Lead Motors.
The postpowerstroke lead head is an alternative attached state of myosin. In it the converter completes its movement, but the lead head lever severely distorts to accommodate this, with only small forward movement of the head–tail junction. Thus the lead head is in a very different conformation from the postpowerstroke trail head, and an attached cargo would be largely unmoved by the converter transition. Incidence of this behavior was strongly dependent on step length (Table 1). It was seen in 7% of all wild-type molecules but not at the maximum 15-actin-subunit spacing. It was most common in heads spaced by 11 subunits (45%) and was much less frequent (2%) at the 13-subunit spacing. The latter figure corresponds to Kc ≈0.02 between the pre- and postpowerstroke converter positions when the heads are at the most common 13-subunit separation.
It is reasonable to postulate that ADP is released only from postpowerstroke lead head motors and that release from the prepowerstroke state can be neglected. We have assumed that the equilibrium between the pre- and postpowerstroke conformations is rapid and small, but our data would also be consistent with a model in which a slow conversion between the pre- and postpowerstroke conformations limits ADP dissociation. An equilibrium constant of 0.02 would be expected to reduce the rate of deac-aminoADP dissociation from the lead head from ≈0.5 s−1 to ≈0.01 s−1, which is in good agreement with the measured value of 0.015 s−1 for wild-type myosin 5a. The equilibrium constant of 0.005 expected from previous measurements of ADP dissociation from lead heads (25) is only 4-fold smaller and is still in reasonably good agreement. Kc increases to ≈0.8 in 11-subunit steps, which corresponds to a more favorable free energy of −2.4 kcal/mole for the pre- to postpowerstroke transition. It also indicates that the deac-aminoADP dissociation rate will only be reduced to 0.22 s−1, making lead heads at this spacing kinetically indistinguishable from the fast, trail head population. The dependence of Kc on step length leads to an estimate that 5.6 pN is required to drive lead heads from postpowerstroke to prepowerstroke. This is comparable to the force (3–5 pN) required to make myosin 5 take backward processive runs (35).
Postpowerstroke lead motors were also observed in subclasses of the 6IQ+2Ala (17%) and 8IQ (10%) mutant molecules, mainly at the 11- and 13-subunit separations, respectively (Fig. 3 B and C). The fact that they were mainly found in shorter steps suggests that their occurrence depends critically on the internal strain with both heads attached to actin. It is therefore not surprising that the powerstroke is most readily completed where the force to overcome is lowest.
It is possible that slowing dissociation of ADP from the lead heads assists both processive motion and the thermodynamic efficiency of the motor. Fig. 4 shows key intermediates of the chemical and mechanical pathways. Processive movement will be maintained as long as at least one head has an empty active site or ADP bound (states 1→5). State 5 is partitioned between states 1 and 6 in the ratio of the rate constants of phosphate and ADP dissociation from state 5 (k5→1/k5→6). This favors state 1 by >20-fold in the native molecule. Flux through states 11 and 12 is reduced to a low level in the native molecule and in the mutant molecules by the slow rates of ADP dissociation from the lead head (1↔9→11 and 2↔10→12).
Fig. 4.
Principal intermediates of the hydrolysis mechanism. Solid arrows joining states 1–5 indicate the principal processive pathway. Intermediates in the box show the prepowerstroke (1 and 2) to postpowerstroke (9 and 10) change. Dashed arrows (1→9→11→5) are a “futile pathway” that hydrolyzes ATP without stepping. Dotted arrows indicate side paths (5→8, 2→8, and 11→8) leading to dissociation and termination of processivity. Rapidly binding and dissociating lead heads are indicated by  . T, ATP; D, ADP; P, phosphate.
. T, ATP; D, ADP; P, phosphate.
Image averages showing both pre- and postpowerstroke lead motors suggest an equilibrium between these states, which is consistent with the idea that myosin 5a is distorted and under tension when attached by both heads. This also suggests that the unfavorable equilibria (K2–10 and K1–9 << 1 in Fig. 4) between the pre- and postpowerstroke conformations produced by strain from the attached trail head is what reduces the rate of ADP dissociation from the lead head. Such a model suggests that ADP dissociation from postpowerstroke lead motors occurs at a rate similar to the unstrained ADP dissociation from the trail head, or from single myosin 5a heads attached to actin. The model also predicts that in less strained steps (i.e., 11 subunits apart for wild type and 6IQ-2Ala and 13 for 8IQ) where the equilibrium constant between pre- and postpowerstroke conformations is near 1, ADP dissociates at rates similar to those of single-headed myosin 5a. As a result, the observed fraction of ADP rapidly dissociating from 8IQ and 6IQ+2Ala is expected to increase relative to wild type. Comparison of the two right-hand columns in Table 1 shows that the observed fractions of rapidly dissociating ADP are in good agreement with those calculated from the fraction of trail heads plus lead heads in less strained conformations.
A similar equilibrium between pre- and postpowerstroke lead motors was recently proposed, based on “traveling wave tracking” of a bead attached to the myosin 5a tail, using optical tweezers (36). Reversible 5-nm forward movement was observed as the first part of the walking step, which is comparable to the 2–6-nm movement of the head–tail junction produced by the transition seen here. The main movement observed would then occur after detachment of the trail head after it binds ATP (steps 3→4→5→1 in Fig. 4). Also consistent with the unique postpowerstroke lead head conformation in Fig. 3 are recent brief reports of high-speed atomic force microscopy (AFM) images of myosin 5a walking (37, 38). These articles reported strong and reversible bending of the lead head lever just before trail head detachment and the main forward movement, although the resolution of the AFM was not sufficient to show whether the bending resulted from converter movement. There is thus now substantial evidence that in doubly attached myosin 5a the lead head undergoes a strain-dependent conformational change between pre- and postpowerstroke positions of the converter, accompanied by only small movements of cargo.
Methods
Myosin 5a heavy meromyosin (HMM) was constructed by truncating a full-length cDNA clone of mouse myosin 5a (residues 1–1,091). This fragment, linked to a C-terminal FLAG epitope tag, DYKDDDDK, and calmodulin were expressed in Sf9 cells by coinfection with baculovirus vectors and purified as previously described (15). Myosin was stored in 0.5 M KCl, 0.1 mM EGTA, 1 mM DTT, and 10 mM Mops, pH 7.0. The mutant lever constructs were 4IQ, 6IQ+2ala, and 8IQ (21). Concentrated G-actin (100 μM), in 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP, and 2 mM Tris-HCl, pH 8.0, at 0 °C was purified from rabbit skeletal muscle. G-actin was either lyophilized with 10 mg sucrose/mg protein and dialyzed against the required buffer before use in stopped-flow experiments, or stored drop-frozen in liquid nitrogen for electron microscopy. F-actin solutions used in electron microscopy experiments were treated with 0.5 U/mL apyrase (Sigma A6535) to convert any ADP or ATP present to AMP. F-actin in stopped-flow experiments contained a 2-mM ADP chase and was treated for 1 h with 1 mM glucose and 0.1 U/mL hexokinase to remove any contaminating ATP.
Proteins used in electron microscopy were diluted into 25 mM KCl, 10 mM Mops, 1.0 mM EGTA, 1.0 mM MgCl2, and 2 mM K-phosphate, pH 7.0, at room temperature. Myosin 2 heads (S1) used to reveal the actin filament polarity were treated with NEM as previously described (39). Microscopy specimens were made by mixing 0.4 μM NEM-S1 in 2 μM ATP with an equal volume of 0.2 μM myosin 5a-HMM; this was then mixed with an equal volume of F-actin (1–2 μM) to give a final concentration of 0.5 μM ATP, 0.1 μM NEM-S1, 0.1 μM HMM, and 0.5–1 μM actin. This solution was applied to carbon-coated grids and stained as previously described (40) using 1% uranyl acetate within 10–15 s. Images were recorded at a nominal magnification of 40,000×, and whole micrographs were digitized as 16-bit images using an Imacon Flextight 848 (Imacon A/S) scanner with a pixel size corresponding to 0.53 nm at the specimen. Image averages were produced by alignment and K-means clustering classification using SPIDER software (41). Double-mixing stopped-flow fluorescence was as previously described (24, 25).
Supplementary Material
Acknowledgments
We thank M. Webb for deac-aminoATP, J. Sleep for discussion, M. Walker for help with EM and B. Belknap and S. Cartwright for technical assistance. This work was funded by National Institutes of Health Grant EB00209.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906907107/DCSupplemental.
References
- 1.Cheney RE, et al. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 2.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 3.Forkey JN, Quinlan ME, Shaw MA, Corrie JET, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 4.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 7.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 8.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DCS. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 9.Rief M, et al. Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 12.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci USA. 2002;99:14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 14.Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, et al. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J Biol Chem. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- 16.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 17.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 18.Walker ML, et al. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 19.Burgess S, et al. The prepower stroke conformation of myosin V. J Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volkmann N, et al. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto T, et al. Neck length and processivity of myosin V. J Biol Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto T, Yildez A, Selvin PR, Sellers JR. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, et al. Electron microscopy of thin filaments decorated with a Ca2+-regulated myosin. J Mol Biol. 1980;140:35–55. doi: 10.1016/0022-2836(80)90355-1. [DOI] [PubMed] [Google Scholar]
- 24.Forgacs E, et al. Kinetic mechanism of myosinV-S1 using a new fluorescent ATP analogue. Biochemistry. 2006;45:13035–13045. doi: 10.1021/bi060712n. [DOI] [PubMed] [Google Scholar]
- 25.Forgacs E, et al. Kinetics of ADP dissociation from the trail and lead heads of actomyosin V following the power stroke. J Biol Chem. 2008;283:766–773. doi: 10.1074/jbc.M704313200. [DOI] [PubMed] [Google Scholar]
- 26.Hanson J. Axial period of actin filaments: Electron microscope studies. Nature. 1967;213:353–356. [Google Scholar]
- 27.Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- 28.Vilfan A. Influence of fluctuations in actin structure on myosin V step size. J Chem Inf Model. 2005;45:1672–1675. doi: 10.1021/ci050182m. [DOI] [PubMed] [Google Scholar]
- 29.Knight P, Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J Mol Biol. 1984;177:461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- 30.Winkelmann DA, Lowey S, Press JL. Monoclonal antibodies localize changes on myosin heavy chain isozymes during avian myogenesis. Cell. 1983;34:295–306. doi: 10.1016/0092-8674(83)90160-5. [DOI] [PubMed] [Google Scholar]
- 31.Piazzesi G, et al. Skeletal muscle performance determined by modulation of number of myosin motors rather than motor force or stroke size. Cell. 2007;131:784–795. doi: 10.1016/j.cell.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 32.Mehta AD, Finer JT, Spudich JA. Detection of single-molecule interactions using correlated thermal diffusion. Proc Natl Acad Sci USA. 1997;94:7927–7931. doi: 10.1073/pnas.94.15.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veigel C, Bartoo ML, White DCS, Sparrow JC, Molloy JE. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys J. 1998;75:1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewalle A, Steffen W, Stevenson O, Ouyang ZQ, Sleep J. Single-molecule measurement of the stiffness of the rigor myosin head. Biophys J. 2008;94:2160–2169. doi: 10.1529/biophysj.107.119396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhardt JC, Clemen AE, Jaud J, Rief M. Myosin-V is a mechanical ratchet. Proc Natl Acad Sci USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cappello G, et al. Myosin V stepping mechanism. Proc Natl Acad Sci USA. 2007;104:15328–15333. doi: 10.1073/pnas.0706653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ando T, et al. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 38.Ando T, Uchihashi T, Fukuma T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog Surf Sci. 2008;83:337–437. [Google Scholar]
- 39.Cande WZ. Preparation of N-ethylmaleimide-modified heavy meromyosin and its use as a functional probe of actomyosin-based motility. Methods Enzymol. 1986;134:473–477. doi: 10.1016/0076-6879(86)34113-2. [DOI] [PubMed] [Google Scholar]
- 40.Walker M, Knight P, Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 41.Frank J, Verschoor A, Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.