Abstract
Prevailing theories suggest that luminal cells are the origin of prostate cancer because it is histologically defined by basal cell loss and malignant luminal cell expansion. We introduced a series of genetic alterations into prospectively identified populations of murine basal/stem and luminal cells in an in vivo prostate regeneration assay. Stromal induction of FGF signaling, increased expression of the ETS family transcription factor ERG1, and constitutive activation of PI3K signaling were evaluated. Combination of activated PI3K signaling and heightened androgen receptor signaling, which is associated with disease progression to androgen independence, was also performed. Even though luminal cells fail to respond, basal/stem cells demonstrate efficient capacity for cancer initiation and can produce luminal-like disease characteristic of human prostate cancer in multiple models. This finding provides evidence in support of basal epithelial stem cells as one target cell for prostate cancer initiation and demonstrates the propensity of primitive cells for tumorigenesis.
Keywords: AKT, AR, ETS, ERG, CD49f
Although prostate cancer is the most common malignancy diagnosed in males in the Western world, the cell of origin of the disease remains unclear (1). Several approaches have been used to identify the cell of origin of different cancers. One approach involves the creation of mouse models in which oncogenic lesions are introduced in alternative cell types of a tissue using cell-type specific promoters. Clevers and colleagues (2) recently used this technique to demonstrate that deletion of adenomatous polyposis coli in crypt stem cells results in rapid formation of intestinal adenomas, while deletion in transit-amplifying cells results in transient disease that does not progress. An alternative approach uses in vivo transplantation models to investigate the tumorigenic potential of prospectively identified populations of cells following ex vivo genetic modification. Huntly et al. (3) used this approach to demonstrate that BCR-ABL can induce myeloid disease in mice when introduced into HSC but not more committed progenitors, while other fusion oncogenes can initiate disease from both stem cells and progenitors (4, 5).
The prostate is comprised of several types of epithelial cells that may serve as targets for cancer initiation, including basal, luminal, and neuroendocrine cells. Prostate stem cells are thought to reside in the basal cell layer of the ducts, and in the mouse they predominate in the region of the gland that is proximal to the urethra (6, 7). Clinical observations that the majority of human prostate cancer cells express luminal cell markers have led many to propose that these cells are the source of prostate cancer initiation (8). The neoplasms that develop in the Rb−/−p53−/− knockout mouse model express the stem cell marker Sca-1 and arise in the proximal region of the gland (9). In a PTEN null mouse model there is a preferential expansion of basal cells compared to luminal cells, suggesting disease in these mice is propagated by basal cells (10). Several recent reports have also shown that progenitor cells with luminal characteristics can initiate prostate cancer following PTEN deletion. Korsten et al. (11) demonstrated that PSA-driven PTEN deletion specifically in luminal cells results in prostatic hyperplasia, and suggest luminal-specific progenitors as the candidate cell of origin in this model. Shen and colleagues (12) found that a bipotent, self-renewing population of castration-resistant NKX3.1-expressing cells (CARNs) can produce high-grade PIN/carcinoma lesions following inducible deletion of PTEN. It is not clear in these studies whether these populations represent the only cells capable of cancer initiation.
We have developed an approach analogous to those used for the study of the hematologic malignancies to study prostate cancer initiation. The tumorigenic potential of different prostate cell subpopulations was assessed using an in vivo transplantation assay that utilizes fetal urogenital sinus mesenchymal (UGSM) cells to induce the growth of dissociated adult prostate cells (13). Using FACS to prospectively identify prostate cell subpopulations with the distinct properties of basal/stem cells, luminal cells, and stromal cells, we find that basal/stem cells are more efficient targets for transformation than luminal cells following the introduction of multiple alternative oncogenic stimuli.
Results
Prospective Identification of Prostate Basal/Stem, Luminal, and Stromal Cells by Sca-1 and CD49f Expression.
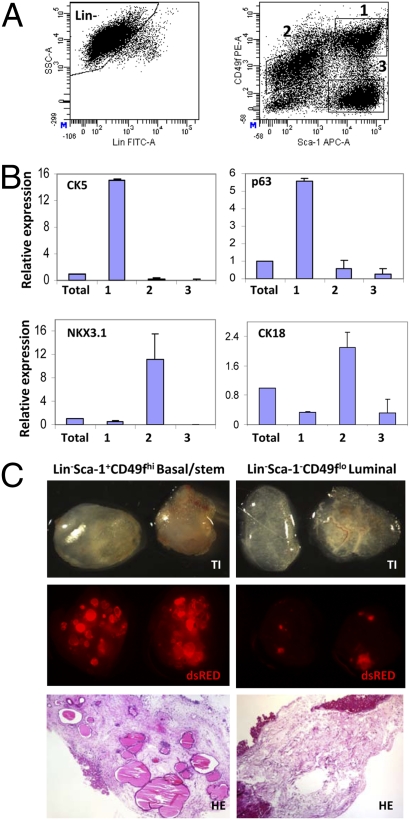
Enrichment for murine prostate epithelial stem cells can be achieved using FACS to sort cells with a Lin(CD45/CD31/Ter119)−Sca-1+CD49f+ cell-surface profile (14). Further scrutiny of Sca-1 and CD49f expression on prostate cells revealed three discrete populations (Fig. 1A). Quantitative PCR (qPCR) analysis of the lineage status of each population demonstrates that Lin−Sca-1+CD49fhi cells express low levels of the luminal cell markers cytokeratin (CK) 8, CK18, and NKX3.1 (Fig. 1B and Fig S1A). These cells express high levels of CK5, CK14, and p63, and possess a basal-like phenotype (15). Lin−Sca-1−CD49flo cells express high levels of the luminal cell markers and low levels of the basal cell markers. Lin−Sca-1+CD49f− cells express high levels of the stromal cell marker Vimentin (VIM), but not the epithelial markers. Immunocytochemical (ICC) analysis demonstrates that the protein expression of these lineage markers corresponds with mRNA expression (Fig. S2A).
Fig. 1.
Prospective identification of basal/stem, luminal, and stromal cells by Sca-1 and CD49f expression. (A) FACS analysis for lineage markers (CD45, CD31, Ter119), and Sca-1 vs. CD49f after gating on Lin− cells. (Lin-Sca-1+CD49fhi (1) = 19.4 ± 3.6%; Lin-Sca-1−CD49flo (2) = 21.9 ± 4.6%; Lin−Sca-1+CD49f- (3) = 19.6 ± 2.5%) (B) qPCR analysis for expression of prostate lineage markers in each population. Expression is relative to total unsorted prostate cells. (C) Each population was sorted from β-actin dsRED animals, and equal numbers (50,000) were mixed with UGSM and implanted in SCID mice for 8 weeks. Transillumination (TI, Top) and fluorescent (dsRED, Middle) images of each graft are shown. (Bottom) H&E staining of representative tissue sections from each graft. (Original magnification, 10×.)
The properties of each population were investigated using previously defined functional assays (13, 14). Figs. S1 C and D show that Lin−Sca-1+CD49fhi cells form large colonies of primitive cells that express both CK5 and CK8. Lin-Sca-1−CD49flo cells form small colonies of cells that exclusively express CK8, suggesting these cells have more limited proliferative and differentiation potential. Lin-Sca-1+CD49f− cells form sheets of spindle-shaped cells resembling stromal cells that express the stromal cell-marker smooth-muscle actin. Only the Lin−Sca-1+CD49fhi cells are capable of forming spheres in three-dimensional culture as previously demonstrated (Fig. S1C Bottom) (14).
The regenerative potential of these populations was evaluated using the in vivo prostate regeneration assay. Fig. 1C shows that ductal structures were only observed in grafts generated from Lin−Sca-1+CD49fhi cells. Analysis of grafts harvested after short incubation periods (1–3 weeks), however, revealed that transplanted cells could also be identified in the other grafts by flow cytometry, suggesting these cells remain viable in vivo (Fig. S3 A–C). Immunohistochemical (IHC) analysis showed that grafts regenerated from Lin−Sca-1−CD49flo cells contain scattered clusters of CK5-CK8+ luminal cells, and grafts from Lin−Sca-1+CD49f− cells contain SMA+ stromal cells (Fig. S3D). This protocol can be used to enrich for cells with the phenotypic and functional properties of basal-like stem cells (Lin−Sca-1+CD49fhi), luminal cells (Lin−Sca-1−CD49flo), and stromal cells (Lin−Sca-1+CD49f−). The viability of each population in the in vivo regeneration assay also validates the use of this assay for comparison of their oncogenic potential following genetic manipulation.
Androgen receptor (AR) signaling plays an important role in both prostate development and tumorigenesis. Most studies in the human report AR expression is restricted to the luminal cell layer; several studies in the mouse report widespread expression in both basal and luminal cells (16 –18). qPCR and Western blot analyses demonstrate that both mRNA and protein for AR can be identified at high levels in basal/stem, luminal, and stromal cell fractions (Fig. S1 A and B). ICC analysis shows clear nuclear expression of AR in cytospins of basal/stem and luminal cells (Fig. S2B). AR is expressed in the nucleus and cytoplasm of stromal cells. We questioned whether AR-mediated transcription was active in each cell type by examining expression of known androgen regulated genes. Cells from each population were sorted from ARR2 Pb-Lux transgenic mice expressing luciferase under regulation of the androgen-regulated probasin promoter (19). Despite the roughly equal expression of AR in basal/stem and luminal cell fractions by qPCR and Western blot, luciferase activity was approximately 7-fold lower in basal/stem cells compared to luminal cells (Fig. S1B). qPCR analysis for several known androgen regulated genes also showed these genes were expressed at higher levels in luminal cells (Fig. S1B). This suggests that even though AR is expressed in both epithelial cell types, other factors may regulate AR-mediated transcription in basal/stem cells.
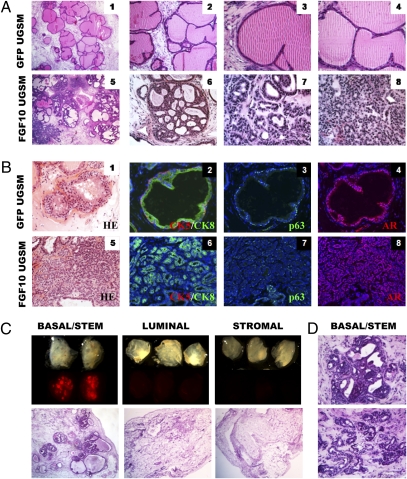
Basal/Stem Cells Produce Multifocal Glandular Carcinoma in Response to Paracrine Stimulation of FGF Signaling.
Incubation of naïve adult prostate cells with FGF10-expressing UGSM cells results in the regeneration of a multifocal disease that histologically is similar to human prostate cancer (20). Fig. 2A shows that grafts are characterized by the extensive proliferation of small, single-layered compact glands ranging in pathological appearance and Gleason score. IHC analysis shows that the majority of small cancerous glands are comprised of CK8+ luminal-type cells and lack CK5+ basal cells (Fig. 2B). This is striking because human prostate cancer is defined by expansion of malignant luminal cells and loss of basal cells. The tumor cells in the grafts are positive for AR, although p63 expression is abundant, which is not frequently observed in the human disease.
Fig. 2.
Paracrine stimulation of basal/stem cells with FGF10 results in multifocal adenocarcinoma. (A) Prostate cells were combined with control GFP-UGSM or FGF10-UGSM and implanted in vivo. Panels 1 and 5 show low power images of H&E stains of tissue sections from representative grafts. (Original magnification, 40×.) High-power images show examples of the different pathological phenotypes observed. In Panel 7, the cancerous glands are well formed with open lumens, characteristic of Gleason 3 human prostate cancer. Panels 6 and 8 are characteristic of Gleason graded 4 prostate cancer. (Original magnification panels 2 and 6, 100×, panels 3, 4, 7, 8, 200×.) (B) IHC analysis for lineage marker expression in GFP and FGF10 grafts (original magnification, 200×). (C) Each fraction was sorted from β-actin dsRED mice, combined with FGF10-UGSM and implanted in vivo. (Upper) TI and fluorescent images of each graft. (Lower) Low power images of H&E stains of tissue sections from representative grafts. (Original magnification, 40×.) (D) Representative images of pathological phenotypes observed in tissue sections from basal/stem cells stained with H&E. (Original magnification, 200×.)
No clear mechanism for the development of the small glandular structures observed in human prostate cancer is known. Previous studies have shown that FGFs induce branching in many epithelial organs (21). FGF10 knockout mice fail to produce several branched organs, including the prostate (22). We tested if the phenotype observed in the FGF10 model is a manifestation of excess branching of the epithelium. Prostate cells from β-actin dsRED and β-actin GFP transgenic mice were sorted by FACS using single-cell gating. Red and green cells were mixed together in combination with control or FGF10-UGSM and implanted in the regeneration assay. Fig. S4A shows that regenerated grafts contain many GFP+ and dsRED+ ducts, indicating that the multifocal disease induced by FGF10 is polyclonal like human prostate cancer. Low-power analysis using a dissecting microscope shows that ducts in FGF10 grafts exhibit dramatic branching architecture and contain an abundance of small acini compared to control grafts (Fig. S4A Right). As suspected, these acini appear in histological sections as numerous small glandular structures (Fig. S4B). Fluorescence microscopy analysis of tissue sections shows that regions of different pathological grade in FGF10 grafts represent the outgrowth of different clones, as they tend to segregate with color marker expression (Fig. S4B).
To determine whether FGF10-induced disease can originate from basal/stem cells, luminal cells, or both, equal numbers of dsRED cells from each population were mixed with control or FGF10-UGSM and implanted in vivo. qPCR analysis shows that both basal/stem and luminal cells express the receptors for FGF10 (Fig. S4C). Fig. 2C shows that dsRED signal was observed in grafts generated from basal/stem but not luminal or stromal cells. Cancerous glands regenerated from basal/stem cells possess a similar range of pathological phenotypes as observed from unfractionated prostate cells (Fig. 2D). Oncogenic stimulation of basal/stem cells produces a luminal-dominant disease.
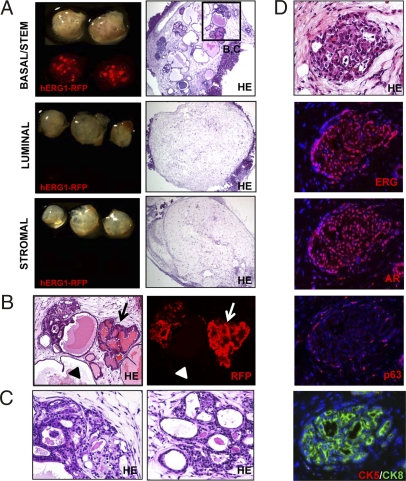
Overexpression of ERG1 in Basal/Stem Cells Results in Ductal Dysplasia and PIN Lesions.
Chromosomal translocations involving ETS family transcription factors are present in up to 70% of human prostate cancers and 20% of PIN lesions, making them the most common class of genetic lesions present in the disease (23). Probasin-driven overexpression of ETV1 or ERG results in the development of low-grade PIN lesions in transgenic mice, and lentiviral-mediated overexpression in naïve epithelial cells results in PIN lesions in the prostate regeneration assay (24 –26). We increased ERG expression in basal/stem, luminal, and stromal cell fractions following ex vivo lentiviral-mediated gene transfer. To test whether each population was competent for lentiviral infection, cells were sorted from dsRED mice, transduced with lentivirus carrying a GFP construct via spinduction, and implanted in the prostate regeneration assay. FACS analysis of grafts harvested 1 week later shows the presence of GFP+dsRED+ cells in each graft (Fig. S5).
Sorted prostate basal/stem, luminal, and stromal cells were transduced with lentivirus carrying hERG1 and RFP. Cells were also infected with virus carrying only RFP as control. Equal numbers of transduced cells from each population were implanted in the regeneration assay and harvested 8 weeks later. Fig. 3A shows that RFP signal was only observed in grafts from basal/stem cells. Low magnification images of tissue sections from each graft show the presence of ductal structures in basal/stem cell grafts; no growth of transformed cells was observed in luminal or stromal cell grafts (Fig. 3A).
Fig. 3.
Increased expression of hERG1 in basal/stem cells results in focal dysplasia and PIN lesions. (A) Basal/stem, luminal, and stromal cells were transduced with hERG1-RFP or RFP control lentivirus. (Left) TI and fluorescent images of each regenerated graft. (Right) Low-power images of H&E stains of tissue sections from representative grafts. (Original magnification, 40×.) (B) High-power images show hERG1-RFP infected (arrows) and uninfected (arrowheads) ducts in basal/stem cell grafts. (Original magnification, 100×.) (C) Histological phenotype of H&E stains of hERG1-RFP infected ducts. (Original magnification, 200×.) (D) IHC analysis for prostate lineage marker expression in hERG1-RFP infected ducts. (Original magnification, 200×.)
Histological analysis of basal/stem cell grafts demonstrates that ERG+RFP+ ducts exhibit focal dysplasia and PIN lesions (Fig. 3B Right, arrows). As reported in transgenic models, these ducts display epithelial cell stratification and loss of cell polarity, with nuclear pleomorphism and atypia (Fig. 3C) (24, 25). The epithelial cells also exhibit a signet ring cell like appearance, similar to the pathology observed in human prostate cancer samples harboring the TMPRSS2-ERG gene fusion (27). Adjacent ERG-RFP- ducts in these grafts (Fig. 3B Right, arrowheads) display normal ductal architecture and cytologic features.
IHC analysis shows that ERG+RFP+ ducts express high levels of hERG1 and AR, and p63+ cells can be identified along the basement membrane (Fig. 3D). As reported in transgenic models, there is an expansion of CK8+ luminal cells and concomitant loss of CK5 expression in these glands.
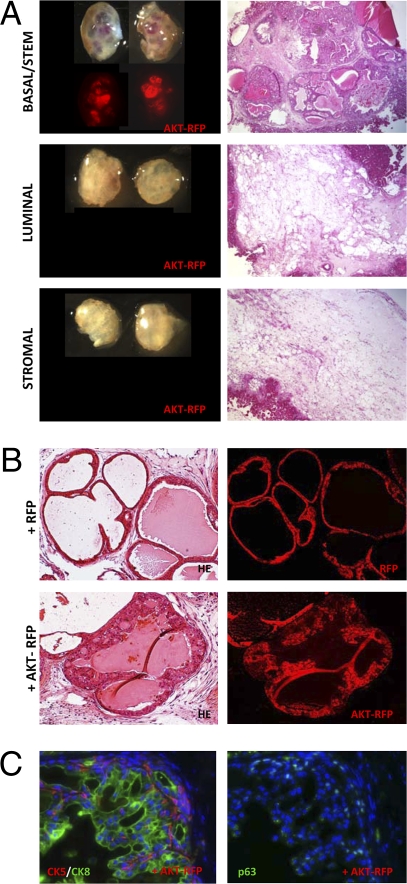
Basal/Stem Cells Regenerate Epithelial Hyperplasia and PIN Lesions in Response to Activated AKT Signaling.
Mutations and deletions in PTEN are present in up to 30% of primary and 63% of metastatic prostate tumors, making them one of the most common classes of genetic alterations observed in prostate cancer (28, 29). Mice with prostate-specific expression of an activated form of the downstream intermediate AKT1 develop PIN lesions (30), and lentiviral-mediated introduction of activated AKT1 into naïve prostate epithelial cells results in PIN lesions in the prostate regeneration assay (31). Equal numbers of basal/stem, luminal, and stromal cell fractions were transduced with lentivirus carrying a construct containing myristoylated AKT1 and RFP or RFP only for control. Fig. 4A shows that only grafts regenerated from basal/stem cells transduced with AKT-RFP display RFP signal. Low-power images of tissue sections from each graft show basal/stem cells regenerated prostatic tubules containing PIN lesions (Fig. 4A), while luminal and stromal grafts lacked growth of transformed cells (Fig. 4A).
Fig. 4.
Constitutive activation of AKT in basal/stem cells results in epithelial hyperplasia and PIN lesions. (A) Basal/stem, luminal, and stromal cells were transduced with AKT1-RFP or control RFP lentivirus. (Left) TI and fluorescent images of regenerated graft. (Right) Low power images of H&E stains of tissue sections from representative grafts. (Original magnification, 40×.) (B) High-power images show histological phenotype of RFP infected (Upper) and AKT1-RFP infected (Lower) ducts in basal/stem cell grafts. (Original magnification, 200×.) (C) IHC analysis for prostate lineage marker expression in AKT-RFP infected ducts. (Original magnification, 200×.)
PIN lesions in AKT+RFP+ ducts are characterized by epithelial cell stratification and nuclear atypia (Fig. 4B). IHC analysis shows that, in contrast to FGF10 and ERG1 models, expansion of both CK5+ and CK8+ cells is present (Fig. 4C). p63+ cells can also be found around the periphery of the ducts (Fig. 4C). These data demonstrate that basal/stem cells represent a more efficient target for AKT-mediated transformation than luminal cells. This is also consistent with our findings using the prostate specific PTEN null model that only Lin−Sca-1+CD49fhi basal/stem cells from these mice form PIN lesions when implanted in the regeneration assay (32).
AKT and AR Synergize to Promote Progression to Poorly Differentiated Prostate Carcinoma from Basal/Stem Cells.
Many mechanisms for ligand-independent activation of AR signaling have been suggested (33), including phosphorylation by AKT (34). Analysis of the AKT and AR genes in the prostate regeneration assay has shown that activated AKT synergizes with increased AR expression to promote prostate cancer progression and androgen-independent disease (35).
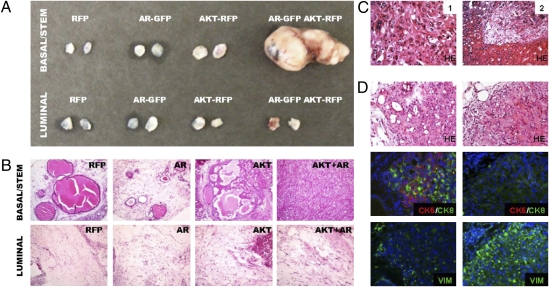
To investigate the response of basal/stem and luminal cell fractions to combined activation of AKT and AR signaling, each population was transduced with RFP, AKT1, AR, or both AKT1 and AR lentiviruses, and implanted in the regeneration assay. Fig. 5A shows that large tumors were generated from basal/stem cells tranduced with AKT1 and AR. Interestingly, even potent stimulation with these two oncogenes did not result in growth or transformation of luminal cells (Fig. 5A).
Fig. 5.
AKT and AR synergize to promote cancer progression from basal/stem cells. (A) Basal/stem and luminal cells were transduced with RFP, AR-GFP, AKT-RFP, or both AR-GFP and AKT-RFP lentiviruses. Overview image shows relative size of regenerated grafts. The two basal/stem cell grafts transduced with both viruses merged together. (B) H&E stains show histological appearance of regenerated tissue from representative grafts. (Original magnification, 100×.) (C) High-power images show representative fields of carcinoma cells (1) and invasion into the kidney parenchyma (2) in basal/stem cell grafts transduced with both the AR-GFP and AKT-RFP viruses. (Original magnification, 200×.) (D) IHC staining shows regions of cells expressing CK5 and CK8 (Left) and regions expressing VIM (Right). (Original magnification, 200×.)
Histological analysis of grafts from basal/stem cells infected with AKT1 virus displayed PIN lesions as expected (Fig. 5B). Basal/stem cells infected with AR virus regenerated small ductal structures that occasionally contained low grade PIN lesions. AKT+AR basal/stem cell grafts displayed a high grade, poorly differentiated carcinoma with focal spindle-cell morphology. Areas typical for carcinoma contained polygonal-shaped tumor cells that form sheets with tight cell junctions (Fig. 5C -1). In some grafts, invasion into the mouse renal parenchyma can also be observed (Fig. 5C -2).
IHC analysis shows variable expression of epithelial and stromal markers. Some regions of cells express CK5 and CK8 (Fig. 5D Middle); others express VIM and not the epithelial markers (Fig. 5D Lower). This finding suggests AKT and AR activation may induce basal/stem cells to progress to a mesenchymal phenotype. These data demonstrate that basal/stem cells can produce advanced disease in response to increased AKT and AR activation.
Discussion
The finding that basal/stem cells respond more efficiently to tumorigenic signals has clinical relevance. Metastatic prostate cancer is often treated with hormonal therapy that targets AR signaling. This treatment initially achieves dramatic therapeutic responses because mature prostate cells are dependent on androgen for viability, but it eventually fails in nearly all patients. If prostate cancer initiates from basal/stem cells in which AR signaling is inactive, hormonal therapy may only interfere with a much later step in tumor progression. The more primitive tumor-initiating cells survive hormonal therapy and eventually give rise to progeny that have developed a mechanism to escape hormonal therapy. More effective therapies may be those that target the more primitive tumor-initiating cells directly, such as the transformed basal/stem cells.
Although the populations identified in this study have clear properties of basal/stem, luminal, and stromal cells, there most likely still exist significant heterogeneity within each fraction. There is evidence for the existence of intermediate transit-amplifying cells, as well as several types of luminal-like progenitor cells in the prostate (36, 37). Luminal-like progenitors, such as CARNs, have been shown to be capable of producing cancerous lesions in response to PTEN deletion during prostate regeneration. It is unclear what the function of these cells is in the normal prostate in steady-state conditions, or where these cells reside in the fractions identified in this study. It would be very interesting in future studies to identify markers to prospectively identify and sort CARNs so their tumorigenic efficiency in response to multiple oncogenic stimuli can be directly compared to the populations identified in this study.
It was not clear in these experiments whether basal/stem cells undergo normal development and branching morphogenesis before the onset of the PIN or carcinoma. Many luminal cells die following transplantation, and those that survive do not appear to divide or form ductal structures in this assay. It is important to note that this is may compromise their capacity to respond to oncogenic insults. Although we investigated the consequences of several common oncogenic signals, there may be some combination of oncogenic events that can induce disease in a mouse luminal cell that we did not identify in this study.
Materials and Methods
Animal Strains and Tissue Collection.
The β-actin GFP [C57BL/6-TgN(ACTbEGFP)1Osb], β-actin dsRED [Tg(ACTB-DsRed.MST)1Nagy/J], and CB.17SCID/SCID mouse strains were purchased from The Jackson Laboratory. ARR2 Pb-Lux transgenic mice were a generous gift from Hong Wu (University of California Los Angeles) and Charles Sawyers (Memorial Sloan Kettering). Eight- to 16-week-old C57BL/6 or B6.SJL-Ptprc inbred mice originally purchased from The Jackson Laboratory were used for all experiments.
All animal experiments and surgical procedures were performed under Division of Laboratory Animal Medicine regulations of the University of California, Los Angeles. Prostate cells were dissociated by mincing and collagenase digestion as previously described (38). UGSM was harvested from embryonic day 16 fetuses as previously described (13).
Lentiviral Vectors, Virus Preparation, and Transduction.
The FU-CGW, FU-CRW, FU-mAKT1-CGW, FU-AR-CGW, andFU-ERG1-CRW lentiviral vectors, and the MSCV-FGF10 retroviral vector have been described previously (13, 26, 35). Lentivirus preparation, titering, and infection of dissociated prostate cells were performed as previously reported (20).
Quantitative Reverse Transcription-PCR.
Total RNA, reverse transcription, and quantitative reverse transcription-PCR were performed as previously described using an ABI 7700 instrument. Primer sequences used are listed in Table S1. Primers for AR, calreticulin, FKHBP51, TMEFF2, and aquaporin 5 were purchased from Superarray. Relative quantification of gene expression was calculated according the Pfaffl method. Standard curves and melting curves using cDNA from unfractionated murine prostate were performed to validate the use of each primer pair. Target gene expression in each sorted prostate cell subpopulations was normalized to the amount of actin they contained. For qPCR analysis of prostate cell fractions, prostates from 6 to 10 mice were digested as described and sorted by FACS using single-cell gating. The data reported represents the averaged results from three independent sorting experiments.
Histology, IHC, and ICC.
Histological and IHC analyses were performed as described previously (13). For preparation of cytospins, prostates from six mice were digested and sorted by FACS into basal/stem, luminal, and stromal cell fractions. Cytopspins were fixed with cold acetone (4 °C) for 2 min and washed with 1 × PBS.
Sections and cytospins were stained with H&E or the antibodies listed in Table S2. For fluorescence visualization of cytokeratin 5 and 8, sections were stained with directly labeled secondary antibodies. Visualization of p63 and AR was performed using biotinylated secondary antibodies and fluorescently labeled streptavidin.
FACS and Flow Cytometry.
Prostate cells were suspended in DMEM 10% FBS and stained with antibody for 20 min at 4 °C. Antibodies and dilutions that were used are listed in Table S2. FACS analysis was performed using the BD FACS Canto and FACSDiva software. Cell sorting was done using the BD FACS Aria (BD Biosciences).
In Vitro Assays.
Colony and sphere assays were performed using previously published protocols. Colony sizing was done using Image Pro-6.2 software to draw regions of interest around the perimeter of colonies for calculation of the surface area of each colony.
In Vivo Prostate Regeneration Assay.
The prostate regeneration assay was performed as previously described (13). For oncogene experiments, prostates from 6 to 10 mice were digested and sorted by FACS into each fraction. Equal numbers of cells from each fraction ranging from 25,000 to 100,000 cells were mixed with 1 to 5 × 105 UGSM cells, and resuspended in rat tail collagen (Becton Dickinson). Approximately two to five replicates of each sample condition were performed in each experiment. Each type of experiment was repeated at least three times, and the data shown represent grafts harvested from one representative experiment.
Luciferase Enzyme Assay.
Cell lysates were prepared from each fraction using a cell lysis buffer (Promega). In vitro luciferase activity (relative light units) was assessed following the addition of the D-luciferin substrate using an LMax II384 luminometer (Molecular Devices).
Supplementary Material
Acknowledgments
We thank Andrew Goldstein, Rita Lukacs, Houjian Cai, Stephanie Shelly, Mireille Riedinger, and Donghui Cheng for technical assistance and suggestions. This work is supported in part by funds from the Prostate Cancer Foundation (D.A.L.); National Institutes of Health 5 K12 HD001400, the Ovarian Cancer Research Fund, the Stewart and Lynda Resnik Prostate Cancer Foundation Grant at University of California Los Angeles, and the Stein/Oppenheimer Clinical Translational Seed Grant (to S.M.); National Institutes of Health Grant 1K99/R00 CA125937 Pathway to Independence (to L.X.); and Grant RSG-07-092-01-TBE from the American Cancer Society, Grant PC061456 from the Department of Defense Prostate Cancer Research Program, a Prostate Cancer Foundation Challenge grant, and a University of California Los Angeles Specialized Program of Research Excellence Developmental Research grant (to J.H.). Y.Z. is an associate of the Howard Hughes Medical Institute. O.N.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913873107/DCSupplemental.
References
- 1.Carson CC., 3rd Carcinoma of the prostate: overview of the most common malignancy in men. N C Med J. 2006;67:122–127. [PubMed] [Google Scholar]
- 2.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 3.Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 5.Cozzio A, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English HF, Santen RJ, Isaacs JT. Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate. 1987;11:229–242. doi: 10.1002/pros.2990110304. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimura A, et al. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J Cell Biol. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada H, et al. Keratin profiles in normal/hyperplastic prostates and prostate carcinoma. Virchows Arch. 1992;421:157–161. doi: 10.1007/BF01607049. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z, Flesken-Nikitin A, Nikitin AY. Prostate cancer associated with p53 and Rb deficiency arises from the stem/progenitor cell-enriched proximal region of prostatic ducts. Cancer Res. 2007;67:5683–5690. doi: 10.1158/0008-5472.CAN-07-0768. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, et al. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korsten H, Ziel-van der Made A, Ma X, van der Kwast T, Trapman J. Accumulating progenitor cells in the luminal epithelial cell layer are candidate tumor initiating cells in a Pten knockout mouse prostate cancer model. PLoS One. 2009;4:e5662. doi: 10.1371/journal.pone.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology. 1988;122:552–562. doi: 10.1210/endo-122-2-552. [DOI] [PubMed] [Google Scholar]
- 16.Mirosevich J, et al. Androgen receptor expression of proliferating basal and luminal cells in adult murine ventral prostate. J Endocrinol. 1999;162:341–350. doi: 10.1677/joe.0.1620341. [DOI] [PubMed] [Google Scholar]
- 17.Bonkhoff H, Remberger K. Widespread distribution of nuclear androgen receptors in the basal cell layer of the normal and hyperplastic human prostate. Virchows Arch. 1993;422:35–38. doi: 10.1007/BF01605130. [DOI] [PubMed] [Google Scholar]
- 18.Masai M, et al. Immunohistochemical study of androgen receptor in benign hyperplastic and cancerous human prostates. Prostate. 1990;17:293–300. doi: 10.1002/pros.2990170405. [DOI] [PubMed] [Google Scholar]
- 19.Ellwood-Yen K, Wongvipat J, Sawyers C. Transgenic mouse model for rapid pharmacodynamic evaluation of antiandrogens. Cancer Res. 2006;66:10513–10516. doi: 10.1158/0008-5472.CAN-06-1397. [DOI] [PubMed] [Google Scholar]
- 20.Memarzadeh S, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies JA. Do different branching epithelia use a conserved developmental mechanism? Bioessays. 2002;24:937–948. doi: 10.1002/bies.10161. [DOI] [PubMed] [Google Scholar]
- 22.Donjacour AA, Thomson AA, Cunha GR. FGF-10 plays an essential role in the growth of the fetal prostate. Dev Biol. 2003;261:39–54. doi: 10.1016/s0012-1606(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 23.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Klezovitch O, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlins SA, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zong Y, et al. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosquera JM, et al. Morphological features of TMPRSS2-ERG gene fusion prostate cancer. J Pathol. 2007;212:91–101. doi: 10.1002/path.2154. [DOI] [PubMed] [Google Scholar]
- 28.Dahia PL. PTEN, a unique tumor suppressor gene. Endocr Relat Cancer. 2000;7:115–129. doi: 10.1677/erc.0.0070115. [DOI] [PubMed] [Google Scholar]
- 29. Sellers WR, Sawyers CL. (2002) Somatic Genetics of Prostate Cancer: Oncogenes and Tumor Suppressors (Loppincott Wiliams & Wilkins, Philadelphia)
- 30.Majumder PK, et al. Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proc Natl Acad Sci USA. 2003;100:7841–7846. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulholland DJ, et al. Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555–8562. doi: 10.1158/0008-5472.CAN-08-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 34.Wen Y, et al. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000;60:6841–6845. [PubMed] [Google Scholar]
- 35.Xin L, et al. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc Natl Acad Sci USA. 2006;103:7789–7794. doi: 10.1073/pnas.0602567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue Y, Smedts F, Debruyne FM, de la Rosette JJ, Schalken JA. Identification of intermediate cell types by keratin expression in the developing human prostate. Prostate. 1998;34:292–301. doi: 10.1002/(sici)1097-0045(19980301)34:4<292::aid-pros7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 37.van Leenders G, Dijkman H, Hulsbergen-van de Kaa C, Ruiter D, Schalken J. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab Invest. 2000;80:1251–1258. doi: 10.1038/labinvest.3780133. [DOI] [PubMed] [Google Scholar]
- 38.Shen MM, Wang X, Economides KD, Walker D, Abate-Shen C. Progenitor cells for the prostate epithelium: roles in development, regeneration, and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:529–538. doi: 10.1101/sqb.2008.73.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.