Abstract
Metastasis leads to the death of most cancer patients, and basal breast cancer is the most aggressive breast tumor type. Metastasis involves a complex cell migration process dependent on cytoskeletal remodeling such that targeting such remodeling in tumor cells could be clinically beneficial. Here we show that Hormonally Up-regulated Neu-associated Kinase (HUNK) is dramatically down-regulated in tumor samples and cell lines derived from basal breast cancers. Reconstitution of HUNK expression in basal breast cancer cell lines blocked actin polymerization and reduced cell motility, resulting in decreased metastases in two in vivo murine cancer models. Mechanistically, HUNK overexpression sustained the constitutive phosphorylation and inactivation of cofilin-1 (CFL-1), thereby blocking the incorporation of new actin monomers into actin filaments. HUNK reconstitution in basal breast cancer cell lines prevented protein phosphatase 2-A (PP2A), a phosphatase putatively acting on CFL-1, from binding to CFL-1. Our investigation of HUNK suggests that the interaction between PP2A and CFL-1 may be a target for antimetastasis therapy, particularly for basal breast cancers.
Metastasis is a hallmark of cancer and remains the major cause of cancer-related mortality (90%) (1). Currently, there is no FDA-approved drug that specifically blocks metastasis. Metastasis is a multistep process that requires a cancer cell to leave a primary tumor, intravasate, survive in the blood, extravasate, migrate, invade through basement membranes and connective tissues, and establish a viable tumor in a distant site (2). Cytoskeletal reorganization and cell movement underlie all these events (3), and disruption of these processes could therefore constitute an effective anticancer approach.
The major subtypes of breast cancer (luminal A/B/C, HER-2, and basal) can be distinguished by their gene expression profiles (4). The basal subtype is the most aggressive, has the worst prognosis, and shows the greatest extent of metastasis (4, 5). To identify molecules whose expression varies by breast cancer subtype and might be linked to metastasis, we screened the online data of Sorlie and colleagues (4) for candidate promoters and suppressors of metastasis. Our hypothesis was that the mRNA expression of kinases involved in cell migration and invasion should be altered in basal breast cancers relative to the mRNA profiles of other breast cancer subtypes. One molecule emerging from this screen was Hormonally Up-regulated Neu-associated Kinase (HUNK), an 80-kDa protein (6). HUNK was down-regulated almost threefold in basal cancer samples compared to the other subtypes. HUNK contains a 260-aa domain predicted to have serine/threonine kinase activity; however, to date, no clear kinase activity, substrates, interacting proteins, or indeed physiological role for HUNK have been identified (6, 7). In normal murine mammary cells, HUNK levels vary with the hormonal cycle (6). With respect to transformed cells, Wertheim et al. recently reported that HUNK is highly expressed in cells derived from HER-2 and luminal breast cancers and suggested that HUNK kinase function is essential for the ability of these cells to establish metastases in a MMTV-MYC mouse model (8). We have examined HUNK function in basal breast cancer cells and show that, in contrast to cells from HER-2 and luminal tumors, it is the loss of HUNK expression that is necessary for basal cancer cell metastasis. Our results indicate that HUNK is a metastasis suppressor for the basal breast cancer subtype.
Results
HUNK Expression Is Down-Regulated in Basal-Like Breast Cancer Cell Lines.
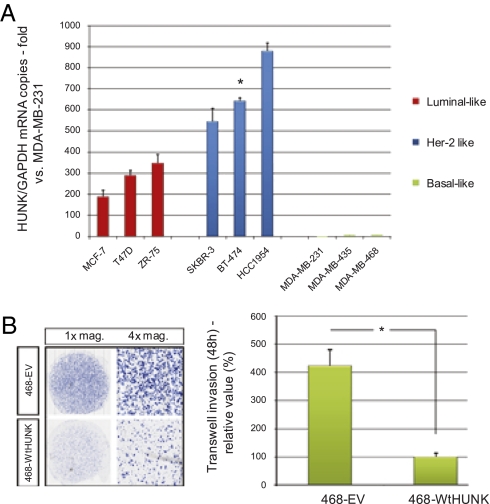
We compared HUNK expression by qRT-PCR (normalized to GAPDH expression) in a panel of human breast cancer cell lines representing the basic subtypes (4). HUNK was almost undetectable (Ct > 33) among the three basal-like cell lines examined but elevated by >100-fold in luminal and HER-2 cell lines (Fig. 1A).
Fig. 1.
HUNK expression in basal-like breast cancer cell lines and effects on migration/invasion. (A) HUNK/GAPDH ratios in the indicated human breast cancer cell lines were normalized to the ratio of the cell line with the lowest value (MDA-MB-231). (B) (Left) ScanScope XT imaging of the bottom membranes of 468-WtHUNK and 468-EV cells in the transwell migration assay. (Right) Quantitation of the total surface of the bottom membrane covered by cells, normalized to the mean for 468-WtHUNK and expressed as a percentage. For all figures, * indicates P < 0.05.
HUNK Reconstitution Decreases Cell Migration and Invasion.
To determine the effect of HUNK on cell migration, we cloned the wild type (Wt) HUNK cDNA with a triple FLAG tag (3× FLAG) into pcDNA3.1 and reconstituted stable HUNK expression in the basal breast cancer cell line MDA-MB-468. Examination of three 468-EV and 468-WtHUNK clones showed that transfection did not alter either the replication rate or the relative plating efficiency (Fig. S1). We next measured migration and invasion capacity using a Boyden chamber assay and found that cells expressing WtHUNK were less able to migrate and invade than controls (Fig. 1B). When transient siRNA-mediated knockdown of HUNK was performed in 468-WtHUNK cells, they underwent phenotypic reversal and showed restored migration and invasion capabilities (Fig. S2).
To exclude cell type-specific or clone-specific effects, we generated stable transfectants using pBIG2(WtHUNK or empty) to achieve an inducible “tet-on” system responsive to either tetracycline or doxycycline (Dox). The cells transfected were the basal line MDA-MB-231D3H2LN-luc (231Luc) established after two in vivo passages in NIH-III mice. The 231Luc cell line expresses luciferase, has a high xeno-engraftment success rate, shows rapid in vivo growth, and is highly metastatic to lymph nodes (9). In response to Dox, 231LucBig-WtHUNK (BIG-HUNK) cells showed an increase in HUNK expression from undetectable to levels similar to those in HER-2 cell lines (Fig. S3A). Furthermore, inducible HUNK expression did not affect 231Luc cell replication (Fig. S3 B and C). When we compared the invasion capacities of BIG-HUNK and BIG-EV cells, we found that induction of HUNK reduced the ability of tumor cells to migrate ( Fig. S4). These data implied that reconstitution of HUNK expression might interfere with metastasis in basal breast cancers.
HUNK Acts Downstream of RHO and RAC and Regulates Cofilin-1 Phosphorylation.
The small GTPases RHO and RAC are important for cell migration because they control cytoskeletal rearrangement and motility through effects on stress fibers, focal adhesion areas, and membrane ruffles (10, 11). Mechanistically, RHO and RAC isoforms integrate stimuli from a cell’s microenvironment and transduce signals from receptor tyrosine kinases, G proteins, or integrins. The combined influence of all these proteins and kinases is reflected in the final activation status of the RHO or RAC molecule: GDP bound (inactive) or GTP bound (active) (10, 11). To determine if HUNK affects RHO/RAC signaling nodes governing motility, we explored the degree of RHO/RAC activation in HUNK-expressing cells. We treated three 468-EV and three 468-WtHUNK clones with 40 nM epidermal growth factor (EGF) and assessed levels of total and activated RHO and RAC. No significant differences were detected between 468-EV and 468-WtHUNK clones (Fig. S5), suggesting that HUNK functions downstream of RHO and RAC.
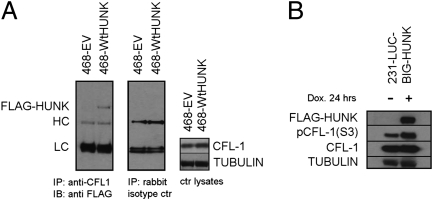
To identify proteins interacting with HUNK, we performed mass spectrometric analysis of proteins acquired by incubating anti-3× FLAG antibody-coated beads with extracts of 468-WtHUNK and 468-EV cells. This analysis revealed cofilin-1 (CFL-1) among the proteins pulled down with FLAG-HUNK. CFL-1 and its regulatory proteins form a system that is the downstream effector of the signaling pathway triggering cytoskeletal rearrangement in response to stimuli in the tumor microenvironment. Alterations to the CFL-1 system have been noted in invasive tumors (12). We subjected 468-EV and 468-WtHUNK cells to CFL-1 coimmunoprecipitation (co-IP) and probed for FLAG. The results demonstrated the association of CFL-1 with HUNK (Fig. 2A). Co-IP did not occur when isotype-control antibody was used, and there were no differences in total CFL-1 originally present. These data argue that HUNK interacts with CFL-1; whether this interaction is direct remains to be determined.
Fig. 2.
HUNK sustains constitutive phosphorylation of CFL-1. (A) Co-IP of HUNK and CFL-1 in 468-WtHUNK and 468-EV cells treated with anti-CFL-1 (Left) or rabbit isotype control (Center). (Right) Control lysate. LC/HC, light/heavy Ig chain. (B) Inducible (231Luc) HUNK reconstitution results in increased phosphorylation of CFL-1 Ser3.
CFL-1 is regulated by phosphorylation on serine-3 [pCFL-1(S3)]. Unphosphorylated CFL-1 is active and allows actin turnover and cytoskeletal rearrangement, whereas pCFL-1(S3) is inactive and halts actin polymerization (12). To investigate whether HUNK affects CFL-1 phosphorylation, we evaluated levels of pCFL-1(S3) in 468-EV and 468-WtHUNK cells. Only when HUNK was expressed was Ser-3 of CFL-1 highly phosphorylated (total CFL-1 was unchanged) (Fig. S6). In addition, immunoblotting of cells showing inducible HUNK expression (BIG-HUNK) revealed an increase in pCFL-1(S3) levels upon Dox treatment (Fig. 2B). These data indicate that HUNK sustains CFL-1 phosphorylation.
HUNK Reconstitution Halts Cytoskeletal Remodeling and Actin Polymerization.
To visualize the effects of HUNK on cytoskeletal remodeling, we subjected 468-WtHUNK and 468-EV clones to overnight serum starvation followed by EGF stimulation. Cytoskeletal rearrangement in response to EGF was detected via rhodamine–phalloidin staining and confocal imaging. In the absence of EGF, cells of both genotypes showed similar cytoskeletal structures (Fig. S7A Left). However, in response to EGF, 468-EV cells reorganized their actin and generated new axial stress fibers, establishing an axis for polarization and cell movement. In contrast, EGF-stimulated 468-WtHUNK cells showed no change to their cytoskeletons (Fig. S7A Right). Furthermore, the blot in Fig. S8 shows that the absence of HUNK does not cause an increase of actin in the cells. We obtained similar results when we assessed the cytoskeletal rearrangement, upon inducing HUNK expression, in BIG-HUNK cells during the transition from cell suspension to cell adhesion (Fig. S7 B and C). Thus, HUNK reconstitution halts cytoskeletal rearrangement.
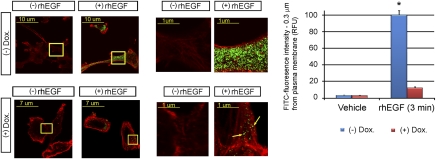
The cytoskeletal remodeling implicated in metastasis requires active actin polymerization (1, 2, 12, 13). Existing actin filaments are severed to generate barbed ends that allow the integration of new actin monomers. The polymerization of these monomers into existing filaments leads to changes in the shape and direction of the cell and generates the protrusion forces that drive cell movement. When CFL-1 is inactivated (phosphorylated), actin is not severed, new monomers cannot integrate into filaments, and the cytoskeleton cannot reorganize. We used confocal microscopy to examine the effect of HUNK on the incorporation of new FITC-actin monomers into the cytoskeleton. In the absence of Dox (no HUNK), EGF-stimulated BIG-HUNK cells showed intense incorporation of new actin monomers that did not occur in the presence of Dox (HUNK expressed), indicating a lack of actin-severing activity (Fig. 3). This failure to polymerize actin in the presence of HUNK is consistent with the lack of motility of these cells observed in the invasion assays.
Fig. 3.
HUNK reconstitution blocks actin polymerization. (Left) Confocal images of the incorporation of FITC-actin monomers (green) into preexisting rhodamine-labeled actin (red). EGF-stimulated 231LucBig-WtHUNK cells induced to express HUNK by Dox addition fail to incorporate new actin monomers. (Center) Magnification of the areas framed by the yellow squares on the Left. Arrows, areas of de novo incorporation of actin monomers. (Right) Quantitation of green fluorescence.
HUNK Suppresses Metastasis and Prolongs Survival in Mouse Engraftment Models.
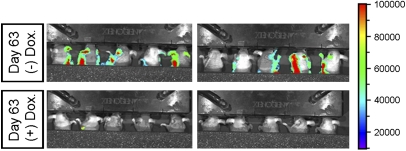
To explore the effect of HUNK on metastasis in vivo, we employed two well-established murine engraftment cancer models. First, we compared the incidence of metastasis of BIG-HUNK-expressing cells in the presence or absence of Dox (HUNK vs. no HUNK expression) in an orthotopic xenograft model. Because 231Luc cells metastasize to the regional lymph nodes, the incidence of homolateral lymph node invasion is a useful endpoint. We injected ∼1 × 106 BIG-HUNK cells into the right mammary fat pads of NIH-III mice (n = 20) and primary tumors were allowed to form in the absence of Dox for 10 days. When these animals were intraperitoneally injected with luciferin followed by luminescence imaging, each mouse showed a well established primary tumor and no secondary metastatic foci (Fig. S9 A and B). From day 11 on, one group (n = 10 mice) was fed regular water, whereas the other group (n = 10) was fed Dox “ad libitum” (1 mg/mL in water) to induce HUNK expression in the tumor cells. At 9 weeks postxenografting, the experiment had to be terminated due to the growth of the primary tumors in both groups to >1.5 cm. Before euthanasia, dorsal imaging (in which the strong signal from the primary tumor was shielded) was conducted and revealed lymph node metastases in 9/10 (−) Dox mice (no HUNK; Fig. 4 Upper) but metastases in only 1/10 (+) Dox mice (HUNK expressed; Fig. 4 Lower) (Fisher’s P < 0.001). Mice that received 231LucBig-EV cells and underwent Dox treatment showed a rate of lymph node metastasis (Fig. S10) that was equivalent to (−) Dox mice injected with the same cells, indicating that it was not the action of Dox alone that was responsible for the decreased rate of metastasis.
Fig. 4.
HUNK suppresses metastasis in vivo. Tumors were allowed to form for 10 days. From day 11 on, 10 mice received oral Dox to induce HUNK expression (Lower), resulting in a blockade of metastatic spread to the lymph nodes. The remaining 10 mice received regular water (no HUNK expression, Upper) and showed extensive metastasis to secondary sites.
Our second model was based on the homografting of B16-F10 murine melanoma cells, a highly metastastic cell line that naturally does not express HUNK (qRT-PCR Ct > 35). Upon injection into the tail vein of a C57BL/6 mouse, B16-F10 cells arrest in the lung capillaries and form primary tumors that eventually spread systemically to secondary sites. The derivative B16-F10Luc strain expresses luciferase. We generated stable transfectants of B16-F10Luc cells expressing WtHUNK or EV and systemically injected ∼1 × 105 cells into the tail vein of C57BL/6 mice. Animals (n = 10/group) injected with B16-F10Luc-WtHUNK cells showed decreased metastasis and increased survival compared with mice injected with B16-F10Luc-EV cells (Fig. S11). These results suggest that HUNK is a bona fide suppressor of metastasis rather than a classical tumor suppressor because primary tumor growth is not affected by HUNK expression and only the development of secondary tumors is compromised.
HUNK Maintains CFL-1 Phosphorylation by Disrupting the Ability of Protein Phosphatase 2-A (PP2A) to Bind CFL-1.
To elucidate the mechanism by which HUNK suppresses metastasis, we first focused on the role of CFL-1 in cell migration. We transiently transfected 468-WtHUNK or 468-EV cells with either control siRNA or siRNA against CFL-1 and examined cell migration in the transwell assay. Knockdown of CFL-1 dramatically suppressed the ability of 468-EV cells to invade but had only a minimal effect on the migration of HUNK-expressing cells (Fig. S12A). Next, we took advantage of a dominant negative mutant of CFL-1 (CFL-1S3D) that mimics constitutive phosphorylation of CFL-1 at Ser3; CFL-1S3D is unable to bind and sever actin (14). We engineered 231Luc cells to express CFL-1S3D and showed that migration in vitro is suppressed when CFL-1 is constitutively phosphorylated (Fig. S12B). Similarly, mice orthopically xenografted with 231Luc-CFL-1S3D cells were unable to generate locoregional metastases at 9 weeks, as opposed to mice engrafted with 231Luc-EV cells (Fig. S12C). Thus, in our system at least, CFL-1 is necessary for cell migration, and constitutive phosphorylation of Ser3 is sufficient to stall cell invasion in vitro and in vivo. These data are consistent with our HUNK reconstitution experiments, in which CFL-1 was constitutively inactive. Thus, we believe that HUNK’s suppressive effect on the metastasis of basal breast cancer cells is due largely to HUNK’s ability to alter CFL-1 phosphorylation.
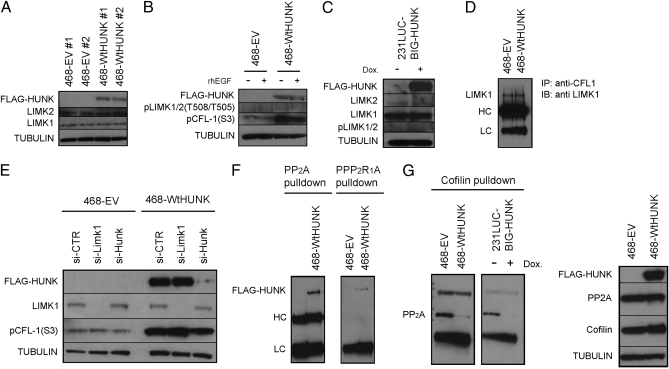
We sought to explain how HUNK maintains constitutive CFL-1 phosphorylation. RHO and RAC recruit and activate the kinases ROCK and PAK, both of which can phosphorylate and activate LIMK1/2, which are putative CFL-1 kinases (15). Dephosphorylation and activation of CFL-1 is mediated by phosphatases such as cronophin (CRNPH), slingshot (SSH), and protein phosphatases I and IIA (PP1 and PP2A) (12). Our data indicated that HUNK interacted with CFL-1 downstream of RHO/RAC activation, so that HUNK might increase pCFL-1 through activation of PAK/ROCK/LIMK1/2, direct CFL-1 phosphorylation, and/or inactivation of CRNPH, SSH, PP1 or PP2A. Neither mass spectrometry nor co-IP revealed any interaction between HUNK and ROCK or PAK. Furthermore, we have been unable to demonstrate unequivocally that the kinase domain of HUNK can in fact phosphorylate a non-HUNK substrate. Baseline levels of LIMK1/2 in 468-WtHUNK and 468-EV cells were similar (Fig. 5A), and no differences in LIMK1/2 phosphorylation were observed in response to EGF stimulation (Fig. 5B). The same result was observed when HUNK expression was induced in BIG-HUNK cells (Fig. 5C).
Fig. 5.
HUNK’s effect on CFL-1 phosphorylation is mediated through PP2A. (A) Western blot showing equal levels of LIMK1/2 in 468-WtHUNK and 468-EV clones. (B and C) No changes in LIMK1/2 expression or phosphorylation in response to 5-min EGF stimulation of (B) 468-WtHUNK or 468-EV cells or (C) BIG-HUNK cells minus or plus Dox. In both cases, EGF stimulation leads to CFL-1 dephosphorylation (activation). (D) Western blot showing equal levels of LIMK1 bound to immunoprecipitated CFL-1 in 468-WtHUNK and 468-EV cells. (E) Western blot showing that transient siRNA silencing of LIMK1/2 in 468-WtHUNK cells (Wt) has no effect on CFL-1 Ser3 phosphorylation, unlike transient siRNA silencing of HUNK. (F) Western blot showing that the C subunit of PP2A (PP2A), as well as its A subunit (PP2R1alpha), co-IP with HUNK in 468-WtHUNK cells. (G) (Left) Western blot showing in 468-based and 231Luc-based transfectants that anti-CFL-1 pulls down the PP2A C subunit only in the absence of HUNK. (Right) HUNK has no effect on levels of the PP2A C subunit.
To further rule out that LIMK1/2 are implicated in the link between HUNK and CFL-1, we investigated whether HUNK physically interacted with LIMK1/2. Immunoprecipitation of LIMK1 from 468-WtHUNK cells pulled down HUNK but the reverse was not true (Fig. S13A). However, transient knockdown of CFL-1 in 468-WtHUNK cells revealed that the interaction between LIMK1 and HUNK occurred only in the presence of CFL-1 (Fig. S13B). This suggests that LIMK1, CFL-1, and HUNK are members of a multiprotein complex. Furthermore, anti-CFL-1 co-IP in 468-WtHUNK and 468-EV cells showed that HUNK reconstitution did not change the total levels of LIMK1 bound to CFL-1 (Fig. 5D). Finally, transient siRNA-mediated silencing of LIMK in 468-WtHUNK and 468-EV cells did not induce significant changes in CFL-1 phosphorylation regardless of HUNK, in contrast to the decrease in pCFL-1(S3) observed upon transient silencing of HUNK (Fig. 5E).
We could not detect endogenous CRNPH or SSH in any of our 231Luc-based or 468-based transfectants. Endogenous PP1 was detectable in our transfectants but did not show any interaction with HUNK or variation in levels in the presence or absence of HUNK (Fig. S14). In contrast, HUNK appeared to have a profound effect on the ability of PP2A to bind to CFL-1. PP2A is a phosphatase in which the catalytic C subunit is bound to several different B regulatory subunits via a scaffolding A subunit (15). The B subunits organize in heteromultimeric combinations and determine the substrate specificity of the multimeric PP2A complex. More than 70 A-B-C combinations are possible and many substrates have been described, but the regulation and determination of substrate specificity of PP2A are not understood (16). Our mass spectrometric data predicted that HUNK would interact with the PP2A A and C subunits, a finding confirmed by co-IP in 468-WtHUNK cells (Fig. 5F). Moreover, we showed using both our 231Luc-based and 468-based transfectants that, even though total levels of the PP2A C subunit were not affected by HUNK (Fig. 5G Right), this subunit was no longer able to bind CFL-1 in the presence of HUNK (Fig. 5G Left). Other substrates of PP2A, including ERK and AKT, showed no differences in phosphorylation in 468-WtHUNK vs. 468-EV cells either under resting conditions or following EGF stimulation (Fig. S15). Thus, the effects of HUNK on PP2A appear to be restricted to PP2A’s binding to CFL-1. These data suggest that, in basal breast cancers, the microenvironmental stimuli that drive the binding of PP2A to CFL-1 do so by down- regulating HUNK. This PP2A-CFL-1 binding results in vigorous dephosphorylation of CFL-1 and thus a reduction in the amount of phosphorylated CFL-1 available to support metastasis.
Discussion
Despite the advent of the PARP inhibitor BSI-201 as a treatment for basal breast cancer, the median overall survival of patients with this metastatic disease is <9 months (17). Novel approaches are therefore needed to improve the course of this disease. Metastasis depends on cell motility, which in turn depends on a multiprotein system that regulates the polymerization/depolymerization of new actin microfilaments (1, 2, 11–14, 18). We hypothesize that disruption of this system in a tumor-specific manner could form the basis of a new therapy.
Our data confirm that the HUNK gene encodes a protein that is highly expressed in HER-2 and luminal breast cancers but also demonstrate that HUNK is suppressed in basal breast cancers, in line with the gene expression study of Sorlie et al. (4). We have not found measurable HUNK in normal breast epithelial cell lines (HMEC, MCF-10A) and have identified no changes in HUNK when breast cells (benign or transformed, ER+ or −) are cultured with 17-β-estradiol. We also have not detected any kinase activity attributable to HUNK in our basal-like cell lines. Our results stand in contrast to those of the Chodosh group, who reported that levels of HUNK in normal murine mammary gland varied with the estrus cycle (6). This group has also described up-regulation of HUNK in several (unspecified) human breast cancer cell lines and in a mammary-specific MYC-transgenic mouse model (8). In that study, it was suggested that HUNK kinase activity is important for the metastasis of HER-2 and luminal breast cancer cells. These discrepancies remain to be resolved.
We found that the suppression of HUNK in basal-like breast cancer cell lines was the key to their capacity to metastasize. In vitro motility and invasion were substantially impaired, although not totally suppressed, upon HUNK reconstitution in two basal-like cell lines, whether the reconstitution was stable or inducible, and in multiple clones. Moreover, transient siRNA silencing of HUNK in stable HUNK transfectants partially recapitulated the aggressive basal phenotype. These effects on migration were independent of cell replication. In vivo, HUNK reconstitution resulted in a near-complete suppression of metastasis. We hypothesize that cytoskeletal rearrangement and actin polymerization, the two events most impaired by HUNK reconstitution, play a role in other processes necessary for metastasis, such as cell detachment or reattachment, shape deformation for intravasation and extravasation, or the epithelial–mesenchymal transition. We postulate that a HUNK-induced decrease in actin polymerization may have pleiotropic effects on events of the metastatic cascade. Such effects could cause tumor cells to migrate at rates below the threshold necessary to produce clinically evident metastasis. In addition to our mouse studies, we have examined the effects of transient or stable HUNK knockdown in benign human fibroblasts and in transformed human breast cancer cell lines with naturally high HUNK levels. These results confirm an increase in the migration/invasion capacity of human tumor cells upon HUNK depletion. It would thus be interesting to obtain annotation of the cell lines examined by the Chodosh group (8) and compare our findings.
The Chodosh group has reported that deletion of HUNK on an MMTV-MYC background diminishes metastases, as measured by direct observation of necropsied lungs (8). However, the mechanism underlying this reduction was not explored. These data conflict not only with our observations but also with those of Korobko et al., who found that high HUNK expression was not associated with metastasis in a small series of human breast tumors (19). We have attempted to tease out a full biochemical chain of events that, at least for basal breast cancers, can explain how HUNK could interfere with a tumor cell’s ability to metastasize. We show that HUNK’s effects on the cytoskeletal remodeling machinery are downstream of RHO and RAC, that there is interaction between HUNK and CFL-1, and that increased HUNK is consistently linked to increased p-CFL-1(S3). Although the data in Fig. 2B suggest that CFL-1 is moderately phosphorylated even when 231Luc-BIG-HUNK clones are not exposed to Dox, we believe that this result is due to vector leakiness because, in the presence of Dox, HUNK levels are increased by 10-fold compared to Luc EV clones. However, other mechanisms (20) are possible.
Luminal and HER-2 breast cancer cell lines must differ from basal cell lines in their regulation of the cytoskeletal machinery, because the former obviously move and remodel their cytoskeletons in the presence of HUNK. Only in basal cell lines does it appear that loss of HUNK is necessary for the acquisition of a metastatic phenotype. In addition, we show that EGF induces actin polymerization and organizes axial stress fibers in basal-like tumor cells and that these events do not occur if HUNK is reconstituted in these cells. Finally, cytoskeleton reorganization in basal cells is preceded by CFL-1 dephosphorylation, an event we show is disrupted by HUNK because HUNK inhibits the main CFL-1 phosphatase, PP2A. Our additional demonstrations that HUNK does not affect LIMK1/2 or directly phosphorylate CFL-1 further support our contention that the effects of HUNK on CFL-1 are restricted to the PP2A pathway.
Our study shows that HUNK fulfills all of the criteria for a tumor metastasis suppressor (21), at least in basal breast cancer: HUNK affects cell motility but not replication in vitro; HUNK’s effects on motility are conserved in an orthotopic xenograft model; and HUNK expression does not disrupt the growth of the primary tumor but diminishes metastases. Although we have yet to explore our hypothesis in vivo in a HUNK knockout mouse, it not clear that doing so would be helpful. In humans, the distinction between HER-2 and luminal and basal breast cancer subtypes is profound, well established, and associated with specific prognoses and clinical decisions. In mice, the various tumor subtypes that develop spontaneously in the mammary gland cannot be completely superimposed on the human subtypes and indeed overlap in characteristics such as keratin expression pattern (22). This mismatch extends to the MMTV-MYC transgenic mouse used by Chodosh and colleagues to cross to their HUNK knockout mutant. Although the MMTV-MYC transgenic mouse was the first mouse model developed for breast cancer (23), the tumors that develop in these animals do not parallel any of the known human subtypes. Information more pertinent to the human situation would be gained from crossing the HUNK knockout to a luminal model such as the P18-deficient mouse (24), a HER-2 model such as the MMTV-NEU mouse (25), or a basal model such as a BRCA-deficient mouse (26).
Our work implies that perturbation of the interaction between PP2A and CFL-1 may be useful for the therapy of human basal breast cancers. It remains to define the role of HUNK in normal cells and in transformation, if any; to clarify HUNK’s kinase activity and substrate, if any; and to investigate how HUNK disrupts the ability of PP2A to bind to CFL-1 while sparing its capacity to bind to other substrates such as ERK or AKT. Resolution of these issues may lead to a new avenue of treatment for the most malignant of breast cancers.
Materials and Methods
Additional details on the above and methods for mass spectrometry, immunoblotting, antibodies, siRNA, and quantitative RT-PCR appear in SI Materials and Methods.
Plasmids.
Human HUNK cDNA was cloned into pCDNA3.1(G418) or pBIG2. CFL-1 was cloned into pCDNA3.1 Myc/His (G418).
Cell Lines and Transfections.
Cell lines were from ATCC except 231Luc and B16-F10Luc (Caliper Life Sciences). Tetracycline-free or estrogen-free sera used for HUNK induction studies were from Clontech. For stable transfections, MDA-MB-468 or 231Luc or B16-F10Luc cells (107) were electroporated with 20 μg pCDNA3/PBIG2 (EV), pCDNA3/PBIG2 containing Wt HUNK, or pCDNA3Myc/His containing Wt CFL-1 or CFL-1 S3D and cultured under selection until colonies formed. Stable MDA-MB-468 and B16-F10Luc transfectants, or 231LucBig transfectants, were maintained in the presence of 1 mg/mL G418 or 200 μg/mL hygromycin, respectively. In the pBIG system, the expression of the Tet-on transactivator is controlled by the TK* promoter and activates the CMV* promoter in the presence of 2 μg/mL Dox (27).
Actin Polymerization.
Triplicate samples of BIG-HUNK cells (5 × 104) were allowed to attach overnight to coverslips placed in 24-well plates containing medium with or without 2 μg/mL Dox. Cells were serum starved for 24 h before permeabilization, using a saponin-based buffer (28) that maintained viability. Permeabilized cells were stimulated for 5 min with 40 nM EGF (Sigma) or vehicle in the presence of saponin and FITC-labeled actin monomers (Cytoskeleton) as described (28). Cells were fixed and prepared for confocal microscopy, using standard procedures. Preexisting filamentous actin was labeled with rhodamine–phalloidin (red). The intensity of the green signal represents the amount of new actin monomers incorporated after stimulation and is related to the degree of activation of CFL-1 and its capacity to generate new barbed ends of actin filaments. The amount of FITC-fluorescence at 0.3 μm from the plasma membrane was quantitated in relative fluorescence units and expressed as a percentage (100% = value of EGF-stimulated, non-Dox-treated BIG-HUNK cells).
Confocal Imaging.
Fixed, permeabilized, and blocked (PBS/5% goat serum) cells were stained for 1 h with rhodamine–phalloidin. Images were acquired with a Carl Zeiss LSM700 microscope and Fluoview software.
Animals.
Female NIH-III (Charles River) and male/female C57BL/6 (Jackson) 6- to 8-week-old mice were maintained as approved by the University Health Network Review Board (protocol AUP 1338.2). Imaging of animals containing luciferase-expressing engrafted cells was performed using an IVIS instrument (Caliper Life Sciences).
Statistical Analyses.
Group measurements were compared using a two tailed Student’s t distribution. A Fisher’s exact test was used for percentage comparisons. Error bars represent the standard error of the mean. Each number/result shows data representative of at least three independent experiments. Each data point is the mean of (at least) triplicates. Survival analysis was performed with the Kaplan–Meier method, and significance was calculated using the log-rank test. All statistical tests were calculated with the SPSS v.13 program.
Supplementary Material
Acknowledgments
We thank Mary Saunders, Ph.D., for critical review and scientific editing of this manuscript. Tak W. Mak is the recipient of Ontario Cancer Research Network 2006 Grant 05NOV00203. Miguel Quintela-Fandino is the recipient of Spanish Ministry of Education Grant EX-2006-0728 for PostDoctorate Trainees, “Ministerio de Educacion y Ciencia/Fullbright.”
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914492107/DCSupplemental.
References
- 1.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Boyer B, Tucker GC, Vallés AM, Franke WW, Thiery JP. Rearrangements of desmosomal and cytoskeletal proteins during the transition from epithelial to fibroblastoid organization in cultured rat bladder carcinoma cells. J Cell Biol. 1989;109:1495–1509. doi: 10.1083/jcb.109.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner HP, et al. Developmental role of the SNF1-related kinase Hunk in pregnancy-induced changes in the mammary gland. Development. 2000;127:4493–4509. doi: 10.1242/dev.127.20.4493. [DOI] [PubMed] [Google Scholar]
- 7.Gardner HP, et al. Cloning and characterization of Hunk, a novel mammalian SNF1-related protein kinase. Genomics. 2000;63:46–59. doi: 10.1006/geno.1999.6078. [DOI] [PubMed] [Google Scholar]
- 8.Wertheim GB, et al. The Snf1-related kinase, Hunk, is essential for mammary tumor metastasis. Proc Natl Acad Sci USA. 2009;106:15855–15860. doi: 10.1073/pnas.0906993106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005;7:R444–R454. doi: 10.1186/bcr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boivin D, Bilodeau D, Béliveau R. Regulation of cytoskeletal functions by Rho small GTP-binding proteins in normal and cancer cells. Can J Physiol Pharmacol. 1996;74:801–810. [PubMed] [Google Scholar]
- 11.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meberg PJ, Ono S, Minamide LS, Takahashi M, Bamburg JR. Actin depolymerizing factor and cofilin phosphorylation dynamics: Response to signals that regulate neurite extension. Cell Motil Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg Y. Protein phosphatase 2A: Who shall regulate the regulator? Biochem Pharmacol. 1999;57:321–328. doi: 10.1016/s0006-2952(98)00245-7. [DOI] [PubMed] [Google Scholar]
- 16.Janssens V, Goris J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Shaughnessy JOC, et al. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): Results of a randomized phase II trial. J Clin Oncol. 2009;27(18S):3. [Google Scholar]
- 18.Yang N, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 19.Korobko IV, Zavalishina LE, Kiselev SL, Raĭkhlin NT, Frank GA. [Proteinkinase MAK-V/Hunk as a possible dianostic and prognostic marker of human breast carcinoma] Arkh Patol. 2004;66:6–9. [PubMed] [Google Scholar]
- 20.Kim YB, et al. Cell adhesion-dependent cofilin serine 3 phosphorylation by the integrin-linked kinase.c-Src complex. J Biol Chem. 2008;283:10089–10096. doi: 10.1074/jbc.M708300200. [DOI] [PubMed] [Google Scholar]
- 21.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9:253–264. doi: 10.1038/nrc2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikaelian I, et al. Proteotypic classification of spontaneous and transgenic mammary neoplasms. Breast Cancer Res. 2004;6:R668–R679. doi: 10.1186/bcr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 24.Pei XH, et al. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell. 2009;15:389–401. doi: 10.1016/j.ccr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy A, et al. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- 27.Strathdee CA, McLeod MR, Hall JR. Efficient control of tetracycline-responsive gene expression from an autoregulated bi-directional expression vector. Gene. 1999;229:21–29. doi: 10.1016/s0378-1119(99)00045-1. [DOI] [PubMed] [Google Scholar]
- 28.Chan AY, et al. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci. 1998;111:199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.