Abstract
Like animals, the mature plant body develops via successive sets of instructions that determine cell fate, patterning, and organogenesis. In the coordination of various developmental programs, several plant hormones play decisive roles, among which auxin is the best-documented hormonal signal. Despite the broad range of processes influenced by auxin, how such a single signaling molecule can be translated into a multitude of distinct responses remains unclear. In Arabidopsis thaliana, lateral root development is a classic example of a developmental process that is controlled by auxin at multiple stages. Therefore, we used lateral root formation as a model system to gain insight into the multifunctionality of auxin. We were able to demonstrate the complementary and sequential action of two discrete auxin response modules, the previously described SOLITARY ROOT/INDOLE-3-ACETIC ACID (IAA)14-AUXIN REPONSE FACTOR (ARF)7-ARF19–dependent lateral root initiation module and the successive BODENLOS/IAA12-MONOPTEROS/ARF5–dependent module, both of which are required for proper organogenesis. The genetic framework in which two successive auxin response modules control early steps of a developmental process adds an extra dimension to the complexity of auxin’s action.
Keywords: AUXIN/INDOLE-3-ACETIC ACID, AUXIN RESPONSE FACTOR, cell cycle, lateral root
Unlike animals, plants produce new organs primarily postembryonically. The formation of these new structures follows a precise pattern that guarantees an optimal spacing of plant organs and that contributes to their functionality. To investigate how a general signal, such as auxin, can be translated into various distinct responses (1), we used the postembryonic development of lateral roots in Arabidopsis thaliana as a model process. Lateral roots originate from a few asymmetrically dividing pericycle cells and develop according to a highly regular pattern (2 –4). Each step in this process is controlled predominantly by the phytohormone auxin, and a number of AUXIN RESPONSE FACTOR (ARF) (transcriptional regulators) and AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) (inhibitors of ARF) proteins are implicated in lateral root development (3 –5). Most prominently, the SOLITARY ROOT (SLR)/IAA14-ARF7-ARF19–mediated auxin response module is required for cell cycle activation and controls the initial, asymmetric pericycle cell divisions (4, 6, 7). Whereas genetic stimulation of the basic cell cycle machinery is capable of bypassing the SLR/IAA14-mediated control on cell division, it is insufficient for de novo lateral root organogenesis (7). Previous analyses of auxin response markers (8 –10) have suggested that additional AUX/IAA-ARF–mediated signaling might be required for lateral root development, however.
Here we demonstrate that the capacity of pericycle cells to form new lateral roots in response to auxin is enhanced considerably on genetic stimulation of the basic cell cycle machinery, arguing for the existence of an additional auxin response module during lateral root initiation. We show that in dividing pericycle cells, the BODENLOS (BDL)/IAA12-MONOPTEROS (MP)/ARF5–mediated auxin response guarantees organized lateral root patterning downstream of SLR/IAA14.
Results and Discussion
Cell Cycle Activation Sensitizes Pericycle Cells for Auxin-Induced Lateral Root Initiation.
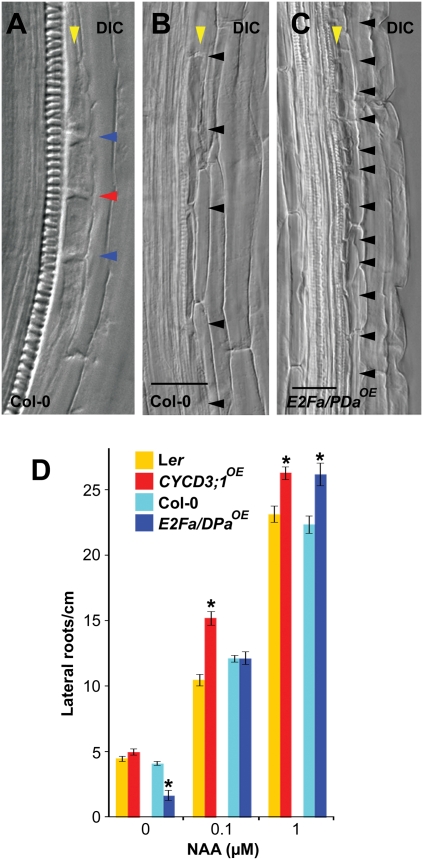
To explore whether during lateral root formation other auxin response modules are active after or besides the SLR/IAA14-dependent signaling, we released cell cycle regulation in the pericycle from its SLR/IAA14-dependent repression via overexpression of core cell cycle regulators. WT roots with enhanced cell cycle activity caused by 35S-driven overexpression (OE) of the heterodimeric G1-S E2Fa/DPa transcription factor (E2Fa/DPaOE) displayed stretches with small pericycle cells that clearly deviated from the larger WT pericycle cells or the typical lateral root initiation pattern (Fig. 1 A–C). Tight control of patterned pericycle cell division has been shown to be required for de novo lateral root formation (7, 11, 12). Consequently, the lateral root density of the E2Fa/DPaOE seedlings—in which the cell cycle is not inhibited in dividing pericycle cells—was significantly reduced (by >60%) and, interestingly, significantly higher in the presence of auxin compared with WT seedlings (Fig. 1D). A similar significant increase in lateral root density was observed in auxin-treated plants with 35S-driven overexpression of the G1-S–related D-type cyclin CYCD3;1 (CYCD3;1OE), which exhibited ectopic cell divisions in the shoot (13) but no initial decrease in lateral root density in the absence of auxin (Fig. 1D). The increase in the number of primordia on auxin treatment was greater for E2Fa/DPaOE than for CYCD3;1OE at both concentrations used, possibly reflecting the greater cell division competence obtained by overexpression of the heterodimeric E2F-DP transcription factors. We conclude that although genetic stimulation of the basic cell cycle machinery is not sufficient for de novo lateral root formation, it enhances the capacity of pericycle cells to divide and to form new lateral roots in response to auxin.
Fig. 1.
Auxin response module after cell cycle activation. (A) WT (Col-0) lateral root initiation. Arrowheads indicate adjacent (red) and short cell boundaries (blue). (B and C) Pericycle cell boundaries (black arrowheads) in a Col-0 (B) and E2Fa/DPaOE (C) root regions. The yellow arrowhead indicates single pericycle layer. DIC, differential interference contrast (Nomarski) imaging. (Scale bars: 50 μM.) (D) Number of lateral roots/cm in WT (Col-0 and Ler), CYCD3,1OE, and E2Fa/DPaOE (mean ± SEM) at different NAA concentrations. Seedlings 5 days after germination were transferred to NAA for 5 days. *Statistically significant differences for values compared with WT as determined by Student's t test (P < 0.05).
Other Auxin Response Modules After SLR/IAA14-Dependent Cell Cycle Regulation.
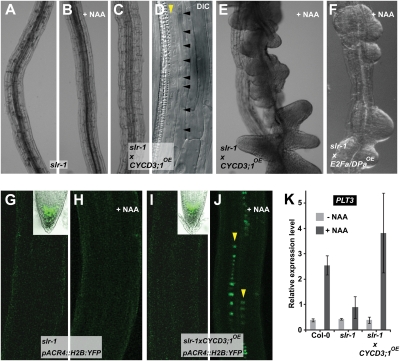
In the gain-of-function mutant slr-1, lateral root formation cannot be induced by auxin treatment or by cell cycle stimulation, although the latter results in stretches of small cells (6, 7) (Fig. 2 A–D). The absence of lateral roots is supported by the absent expression of ARABIDOPSIS CRINKLY4 (ACR4) (a receptor-like kinase marking the first divisions of the lateral root initiation site) (2) and the lack of increased expression of PLETHORA3 (PLT3) (a highly auxin-responsive AP2-domain transcription factor involved in primary root meristem formation) (7, 14) (Fig. 2 G–I and K). However, when slr-1xCYCD3;1OE seedlings were treated with auxin, ACR4 was expressed in the stretches of dividing pericycle cells (Fig. 2J), and PLT3 expression was induced (Fig. 2K). This reactivation of markers augured the formation of lateral root organs in auxin-treated slr-1xCYCD3;1OE (Fig. 2E) and slr-1xE2Fa/DPaOE (Fig. 2F). These observations imply that in the slr-1 mutants as well, genetic stimulation of the basic cell cycle machinery enhances the capacity of pericycle cells to divide and to form new lateral roots in response to auxin. But endogenous auxin is not sufficient to induce lateral roots in slr-1xCYCD3;1OE and slr-1xE2Fa/DPaOE, most likely because in slr-1, PIN-dependent auxin redistribution—which is required for lateral root development—is affected as well (7, 15), and this is largely overcome by exogenous auxin application. Very little is known about how differential cell cycle activity and auxin regulate specific division patterns during lateral root initiation. Our findings suggest that along with the SLR/IAA14-dependent cell cycle regulation, other auxin response modules might work to coordinate organogenesis and to prevent proliferative division.
Fig. 2.
Auxin response module after SLR-dependent cell cycle activation. Absence of lateral roots (A–C), lack of expression of the marker ProACR4:H2B:YFP (G–I), and lack of increased expression of PLT3 (K) in slr-1 (A, G, and K), slr-1 grown on NAA (B, H, and K), and slr-1 with increased cell cycle activity (slr-1xCYCD3;1OE) (C, I, and K), but presence of stretches of short cells in slr-1xCYCD3;1OE (D). Induction of lateral roots (E and F) and/or turning on of ProACR4:H2B:YFP (J) and PLT3 (K) in slr-1xCYCD3;1OE (E, J, and K) and slr-1xE2Fa/DPaOE (F) by the combination of NAA and increased cell cycle activity. Insets (G, and I) show the presence of ProACR4:H2B:YFP in the root tip. Seedlings 5 days after germination were transferred to 10 μM NAA for 96 h (B, E, and F) and 30 h (H and J). Arrowheads indicate the pericycle (yellow) (D and J), which consists of one layer (D) or of one layer starting to undergo periclinal divisions (J) and pericycle cell boundaries (black) (D). DIC, differential interference contrast (Nomarski) imaging. (K) Quantitative RT-PCR expression analysis of PLT3. Seedlings 5 days after germination were treated with 10 μM NAA for 24 h.
BDL-MP–Dependent Auxin Response Module Acts in Lateral Root Initiation.
To identify additional auxin response modules, we analyzed the expression profiles of other AUX/IAA and ARF genes in a highly specific transcript data set from pericycle cells undergoing lateral root initiation (2). Of the retrieved AUX/IAA and ARF genes, which showed significant transcriptional induction during lateral root initiation and for which no role in lateral root development has been documented until now (3, 16), the most intriguing were BDL/IAA12, its paralog IAA13, and its interaction partner MP/ARF5. Because the extensively studied role of the BDL/IAA12–MP/ARF5 pair in embryogenesis implies a function in cell fate determination and asymmetric cell division (5, 17 –21), we investigated the functional involvement of these two proteins in postembryonic lateral root development.
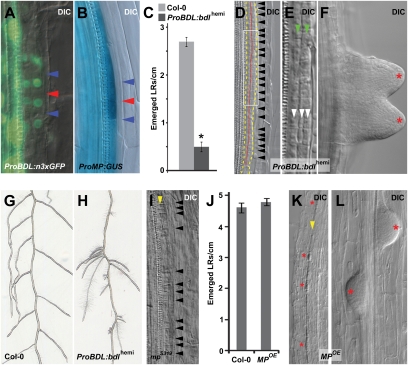
Using promoter fusions with GFP and β-glucuronidase (GUS), ProBDL:n3xGFP and ProMP:GUS, we found that BDL (Fig. 3A) and MP (Fig. 3B) were indeed expressed in the lateral root initiation site. Because homozygous gain-of-function bdl mutants lack a main root (18), we analyzed the lateral root phenotype of hemizygous ProBDL:bdl:GUS plants (ProBDL:bdlhemi) (19). At 11 days after germination, the density of emerged lateral roots was significantly lower in the mutants compared with WT seedlings (Fig. 3C). A detailed microscopic analysis revealed fused primordia (Fig. 3F) and stretches with a two- or three-layer pericycle (Fig. 3 D and E) that were not present in WT roots, which had single-layer pericycle (Fig. 1B), and clearly differed from normal lateral root initiation sites (Fig. 1A), where only a limited number of cells divide periclinally. Emerging ProBDL:bdlhemi lateral roots did not display the regular longitudinal positioning seen in the WT root primordia (Fig. 3G) but rather were grouped in clusters (Fig. 3H). Similarly, the weak loss-of-function mpS319 mutant (22) exhibited zones of ectopic pericycle cell division, although they were less pronounced (Fig. 3I). Although the 35S-driven overexpression of MP (MPOE) did not result in a significant increase in the number of emerged lateral roots (Fig. 3J), closely positioned lateral root initiation sites and aberrantly spaced lateral root primordia were occasionally seen in the MPOE roots (Fig. 3 K and L). Together, these findings demonstrate that BDL and MP are involved in lateral root organogenesis.
Fig. 3.
Role of BDL and MP in lateral root development. (A and B) Expression of ProBDL:n3xGFP (A) and ProMP:GUS (B) in the lateral root initiation site. Arrowheads indicate adjacent (red) and short cell boundaries (blue). (C–L) bdl gain of function [hemizygous ProBDL:bdl:GUS (ProBDL:bdlhemi)] (C–F and H), mp partial loss of function (mpS319) (I), and MP overexpression (MPOE) (J–L) leading to defects in pericycle cell division, lateral root initiation, and positioning compared with WT (Col-0) (C, G, and J). (C and J) Emerged lateral root density at 10 (J) and 11 (C) days after germination. (G and H) Lateral root positioning in Col-0 WT (G) and ProBDL:bdlhemi (H) at 15 days after germination. The red dotted line indicates separation between two pericycle cell layers, and the yellow dashed line indicates outer boundaries of the two- or three-layer pericycle (D). Arrowheads indicate two- (green) and three-layer pericycle (white) (E), small pericycle cell boundaries (black) (D and I), and pericycle (yellow), which is undergoing periclinal division (I) or consists of one layer (K). Red asterisks indicate lateral root primordia (F and L) and sites that strongly resemble a lateral root initiation site (K). Data are mean ± SEM. *Statistically significant differences for values compared with WT as determined by Student's t test (P < 0.01).
The BDL-MP–Dependent Auxin Response Module Follows the SLR-ARF7-ARF19–Dependent Auxin Response Module.
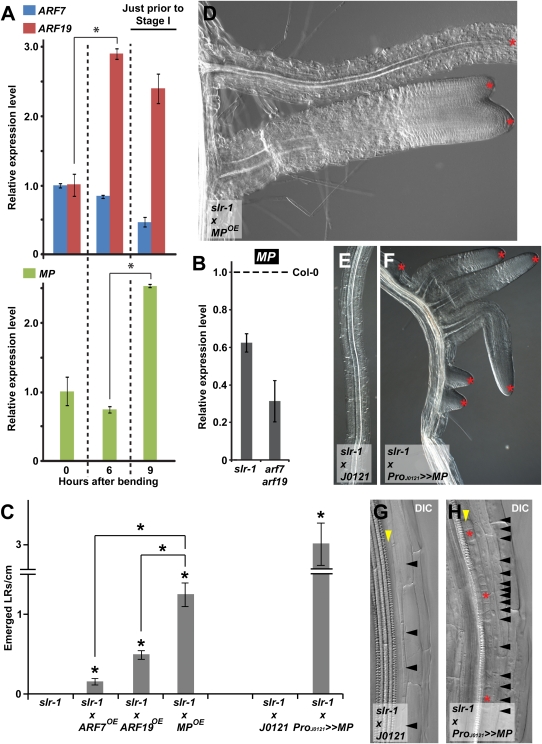
Both gain-of-function bdl and loss-of-function mp mutants displayed different defects arising at later stages than those of the lateral rootless phenotypes of slr-1 and arf7arf19 (3 –7, 23, 24). Using a system of synchronized lateral root induction after gravistimulation (25), we found that an increase in expression of at least ARF19, which together with ARF7 has been proposed to interact with SLR (26), precedes an increase in MP expression (Fig. 4A). In addition, we found a decrease in MP expression in the absence of lateral root initiation in slr-1 and arf7arf19 roots (Fig. 4B). Therefore, it is reasonable to postulate that BDL and MP act later than SLR, ARF7, and ARF19 during lateral root organogenesis.
Fig. 4.
BDL-MP–dependent auxin response module after SLR/IAA14-ARF7-ARF19 action. (A and B) Quantitative RT-PCR expression analysis of ARF7, ARF19, and MP during synchronized lateral root initiation (A) and of MP in slr-1 (5 days after germination) and arf7arf19 roots (10 days after germination) compared with WT expression level (dashed line) (B), indicating two successive modules. Just prior to stage I in (A) indicates at which time point CYCB1;1 expression is induced, which coincides with the onset of lateral root initiation. (C–H) Induction of lateral roots in slr-1 by expression in whole roots MPOE (C and D) and pericycle-specific expression (ProJ0121>>MP) of MP (C and E–H). (C) Emerged lateral root density at 12 days (slr-1, slr-1xMPOE, slr-1xARF7OE, and slr-1xARF19OE) and 15 days (slr-1xJ0121 and slr-1xProJ0121>>MP) after germination. Data are mean ± SEM. *Statistically significant differences for values compared with control or as indicated with a black line as determined by Student’s t test (P < 0.01). (D–H) Red asterisks indicate emerged lateral roots (D and F) and sites that strongly resemble a lateral root initiation site (H) in slr-1xMPOE (D) and in slr-1xProJ0121>>MP (F and H) compared with the control slr-1xJ0121 (E and G). Arrowheads indicate pericycle cell boundaries (black) (G and H) and a pericycle consisting of a single layer (yellow) (G and H). DIC, differential interference contrast (Nomarski) imaging.
To test this hypothesis, we used 35S-driven overexpression of MP in the slr-1 mutant. The number of emerged lateral roots in slr-1xMPOE increased significantly (Fig. 4C), and occasionally those lateral roots were fused or irregularly spaced, or both (Fig. 4D), in contrast to the complete absence of lateral roots in the slr-1 mutant (Figs. 2A and 4C). Such a strong rescue of the slr-1 lateral rootless phenotype could not be obtained even through overexpression of ARF19 (slr-1xARF19OE) and ARF7 (slr-1xARF7OE) (Fig. 4C); however, in slr-1xARF19OE and slr-1xARF7OE, the ARF19 and ARF7 overexpression levels, respectively, were not as high as the MP overexpression level in slr-1xMPOE. In addition, we specifically overexpressed MP in the xylem pole pericycle of slr-1 plants with a GAL4-based transactivation expression approach using the J0121 driver line (ProJ0121>>MP) (27). Although the slr-1xJ0121 control did not display any emerged lateral roots, the slr-1xProJ0121>>MP lines exhibited numerous emerged lateral roots (Fig. 4 C, E, and F). This increase in emerged lateral roots was also reflected in a significant increase in xylem pole pericycle cell divisions, resembling lateral root initiation sites that were absent in the slr-1xJ0121 control (Fig. 4 G and H). These lateral root initiation sites gave rise to closely spaced emerged lateral roots (Fig. 4F).
Although the possibility that the difference in rescue between slr-1xMPOE and slr-1xARF19OE or slr-1xARF7OE is due to unequal expression levels cannot be ruled out, we know of no evidence indicating that MP also activates direct targets of ARF7 and ARF19 in the root. For example, LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16) and LBD29 (28) showed no clear difference in expression in slr-1xProJ0121>>MP and slr-1xMPOE compared with control (Fig. S1).
These data strongly suggest the existence of a second auxin response module that is sufficient to activate lateral root initiation, that is controlled by BDL/IAA12-MP/ARF5, and that acts after the well-known SLR/IAA14-ARF7-ARF19–dependent control. However, based on the gain-of-function bdl and loss-of-function mp mutants, it appears that along with its role in activating lateral root initiation, the BDL/IAA12-MP/ARF5 module also might play a role in inhibiting lateral root formation, similar to what was observed for the receptor-like kinase ACR4 (2).
Conclusions
In multicellular organisms, growth and development, including proper pattern formation and organogenesis, must be tightly regulated. In plants, the phytohormone auxin plays a prominent role in controlling nearly every step in growth and development (1). Just how one generic signal, such as auxin, can be translated into so many diverse developmental responses is unclear, however. Our results demonstrate that the control exerted by auxin on lateral root initiation is at least bimodal and consists of the crucial early SLR/IAA14-ARF7-ARF19–dependent auxin response module (3 –7, 23, 24), followed by a second BDL/IAA12-MP/ARF5-dependent module. Most likely, more modules are involved, because other ARFs have been implicated in adventitious and lateral root development (3, 29 –34). We propose that discrete auxin response modules successively coordinate distinct developmental processes, most probably through the regulation of unique targets, comparable to the spatially distinct, bipartite auxin response during hypophysis specification (19). Thus, a genetic framework for many diverse functions is provided for a single molecule, such as auxin. We hypothesize that such bimodular or multimodular response mechanisms might represent a general principle for auxin signaling in plants.
Materials and Methods
Plant Material and Growth Conditions.
We analyzed A. thaliana (L.) Heynh. ecotypes Columbia (Col-0) and Landsberg erecta (Ler), the mutants slr-1 (6) and mpS319 (22), and the transgenic lines CYCD3;1OE (13), slr-1xCYCD3;1OE (7), slr-1xE2Fa/DPaOE (7), E2Fa/DPaOE (35), slr-1xProACR4:H2B:YFP (2), slr-1xCYCD3;1OExProACR4:H2B:YFP (2), ProMP:GUS, ProBDL:bdl:GUS (19), UAS::MP:HA (19), MPOE, arf7arf19 (24, 36), ARF7OE (37), ARF19OE (23), slr-1xJ0121 (7), and J0121 (27). For slr-1xMPOE, slr-1xARF7OE slr-1xARF19OE, and slr-1xProJ0121>>MPOE, the F1 seedling roots were analyzed. For analysis of expression patterns and lateral root densities, seedlings were grown on normal or half-strength Murashige and Skoog (MS) medium as described previously (38). For the auxin treatment, 5-day-old seedlings were transferred for 5 days from MS medium to MS medium supplemented with various concentrations of 1-naphthalene acetic acid (NAA) for analysis of lateral root densities as described previously (11), or, alternatively, 4- to 5-day-old seedlings were incubated for 24 h in liquid half-strength MS medium with or without 1% sucrose supplemented with NAA for quantitative RT-PCR analyses as described previously (15). Seedlings were incubated in a growth chamber under continuous light at 22 °C or under 16-h light/8-h dark conditions at 24 °C.
Histochemical, Histological, and Microscopic Analyses.
The GUS assays were conducted as described previously (39). Microscopic analyses we performed as described previously (2).
Quantitative RT-PCR.
RNA extraction and quantification by qRT-PCR was done in triplicate as described previously (15). The following primers were used to quantify the gene expression levels: PLT3, 5′-CGGCGAATGCAGCTTCTGACTC-3′/5′-GGTGCCATAAGTCCCATTGTTCCC-3′; ARF7, 5′-GCTCATATGCATGCTCCACA-3′/5′-GCAATGCATCTCTGTCATATTTG-3′; ARF19, 5′-CACCGATCACGAAAACGATA-3′/5′-TGTTCTGCACGCAGTTCAC-3′; and MP, 5′-CGAGCTTTGTGGGTGAGTTAGTAG-3′/5′-ACAAGCTTTAAAGACTACGAGGAGCTA-3′.
New Constructs and Transgenic Lines.
ProMP:GUS was generated as follows. The primers 5′-GGGTTCTAGATTGGAGATCCTTTGATTCAAATAT-3′/5′-CACCTCTAGAGAAGTAATACACTAAGCTCCCAA-3′ were used to amplify a 2.5-kb genomic fragment consisting of ≈2 kb of MP promoter region and further extending into exon 2 of the MP-coding region, which was inserted via XbaI into pPZP-GUS (40). pGreenIIKanProBDL:NLS:3xEGFP::nost (ProBDL:n3xGFP) was constructed by inserting ProBDL into pGreenIIKanNLS:3xEGFP::nost (41) via the EcoRI and BamHI restriction sites. ProBDL was amplified as two fragments which were fused via the PstI restriction site; those fragments were amplified with primers 5′-CCGAATTCCATGTGGTAGTGTCGAG-3′ / 5′-GGCTGCAGACAACAAGAAGAGAAAGAG-3′ and 5′-GGCTGCAGCTCCATTCTCTCTGTG-3′ / 5′-CCGGATCCGTCAATAACAAAACCC-3′, respectively. 35S::MP (MPOE) was generated by fusing the MP cDNA and a sequence encoding an HA epitope-tag (36) to the double-enhanced CaMV 35S promoter. Constructs were used for floral dip transformation of Col-0 plants (42). Transformants were selected on kanamycin and subsequently confirmed for a 3:1 segregation. For MPOE, lines were selected that had elevated MP mRNA levels in the qRT-PCR, namely an increase of ≥ 40-fold compared with WT.
Supplementary Material
Acknowledgments
We thank J. Murray, H. Fukaki, T. Berleth, and A. Theologis for sharing materials; M. Cook (Comprehensive Cancer Facility, Duke University) for providing expert assistance with cell sorting; and D. Slane, M. Bayer, and M. De Cock for critically reading the manuscript. This work was funded by grants from the Interuniversity Attraction Poles Programme (P6/33 and P5/13), initiated by the Belgian State Science Policy Office (BELSPO); the National Science Foundation AT2010 program (to P.N.B.); the Deutsche Forschungsgemeinschaft (SFB 446, to G.J.); the Austrian Science Fund (to C.L. and R.B.), Research Foundation-Flanders (travel grant, to I.D.S.), and the Netherlands Organization for Scientific Research (ALW-VIDI 864-06.012, to D.W.). Financial support was provided by the University of Nottingham , Biotechnology and Biological Sciences Research Council (BBSRC), and Engineering Physics Scientific Research Council (EPSRC) award to the Centre for Plant Integrative Biology (to U.V. and M.J.B.), a fellowship for non–European Union researchers from BELSPO (to V.V), fellowships from the Institute for the Promotion of Innovation by Science and Technology in Flanders (to I.D.S. and S.V.), the Bijzondere Onderzoeksfonds of Ghent University (B.D.R. and W.G.), the European Molecular Biology Organization (ALTF 108-2006, to I.D.S.; ALTF 142-2007, to S.V.), and the Marie Curie Intra-European Fellowship scheme (FP6 MEIF-CT-2007–041375, to I.D.S).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915001107/DCSupplemental.
References
- 1.Vanneste S, Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 2.De Smet I, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- 3.Péret B, et al. Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Mol Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- 5.Lau S, Jürgens G, De Smet I. The evolving complexity of the auxin pathway. Plant Cell. 2008;20:1738–1746. doi: 10.1105/tpc.108.060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis . Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 7.Vanneste S, et al. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana . Plant Cell. 2005;17:3035–3050. doi: 10.1105/tpc.105.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 9.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis . Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 10.Dubrovsky JG, et al. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA. 2008;105:8790–8794. doi: 10.1073/pnas.0712307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himanen K, et al. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanchenko MG, Coffeen WC, Lomax TL, Dubrovsky JG. Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J. 2006;46:436–447. doi: 10.1111/j.1365-313X.2006.02702.x. [DOI] [PubMed] [Google Scholar]
- 13.Dewitte W, et al. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinha C, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 15.Vieten A, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 16.De Smet I, Vanneste S, Inzé D, Beeckman T. Lateral root initiation or the birth of a new meristem. Plant Mol Biol. 2006;60:871–887. doi: 10.1007/s11103-005-4547-2. [DOI] [PubMed] [Google Scholar]
- 17.Hamann T, Benková E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamann T, Mayer U, Jürgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- 19.Weijers D, et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berleth T, Jürgens G. The role of the monopteros gene in organising basal development in the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- 22.Cole M, et al. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136:1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- 23.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19 . Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilmoth JC, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 25.Lucas M, Godin C, Jay-Allemand C, Laplaze L. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot. 2008;59:55–66. doi: 10.1093/jxb/erm171. [DOI] [PubMed] [Google Scholar]
- 26.Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis . Plant J. 2005;44:382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- 27.Laplaze L, et al. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana . J Exp Bot. 2005;56:2433–2442. doi: 10.1093/jxb/eri236. [DOI] [PubMed] [Google Scholar]
- 28.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian CE, et al. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- 30.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JW, et al. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis . Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez L, et al. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell. 2009;21:3119–3132. doi: 10.1105/tpc.108.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon EK, et al. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1128. 10.1093/nar/gkp1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Veylder L, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weijers D, et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardtke CS, et al. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4 . Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 38.Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beeckman T, Engler G. An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep. 1994;12:37–42. [Google Scholar]
- 40.Diener AC, et al. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell. 2000;12:853–870. doi: 10.1105/tpc.12.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada S, Jürgens G. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development. 2007;134:1141–1150. doi: 10.1242/dev.02803. [DOI] [PubMed] [Google Scholar]
- 42.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.