Abstract
To construct a versatile model host for heterologous expression of genes encoding secondary metabolite biosynthesis, the genome of the industrial microorganism Streptomyces avermitilis was systematically deleted to remove nonessential genes. A region of more than 1.4 Mb was deleted stepwise from the 9.02-Mb S. avermitilis linear chromosome to generate a series of defined deletion mutants, corresponding to 83.12–81.46% of the wild-type chromosome, that did not produce any of the major endogenous secondary metabolites found in the parent strain. The suitability of the mutants as hosts for efficient production of foreign metabolites was shown by heterologous expression of three different exogenous biosynthetic gene clusters encoding the biosynthesis of streptomycin (from S. griseus Institute for Fermentation, Osaka [IFO] 13350), cephamycin C (from S. clavuligerus American type culture collection (ATCC) 27064), and pladienolide (from S. platensis Mer-11107). Both streptomycin and cephamycin C were efficiently produced by individual transformants at levels higher than those of the native-producing species. Although pladienolide was not produced by a deletion mutant transformed with the corresponding intact biosynthetic gene cluster, production of the macrolide was enabled by introduction of an extra copy of the regulatory gene pldR expressed under control of an alternative promoter. Another mutant optimized for terpenoid production efficiently produced the plant terpenoid intermediate, amorpha-4,11-diene, by introduction of a synthetic gene optimized for Streptomyces codon usage. These findings highlight the strength and flexibility of engineered S. avermitilis as a model host for heterologous gene expression, resulting in the production of exogenous natural and unnatural metabolites.
Keywords: genome engineering, host development, natural products
A prominent property of members of the genus Streptomyces is the ability to produce numerous secondary metabolites, including antibiotics and other biologically active compounds of proven value in human and veterinary medicine and agriculture; they are also useful as biochemical tools. These structurally diverse metabolites collectively express not only antibacterial, antifungal, antiviral, and antitumor activities but also antihypertensive and immunosuppressant properties. Streptomyces have been a rich source of pharmaceutical compounds in which common cellular intermediates, including amino acids, sugars, fatty acids, and terpenes, are combined to give more complex structures by defined biochemical pathways. Genomic analysis of three species of Streptomyces, S. avermitilis (1, 2), S. coelicolor A3(2) (3), and S. griseus (4), has revealed that these microorganisms each have large linear chromosomes that harbor over 20 secondary metabolic gene clusters encoding the biosynthesis of polyketides by polyketide synthases (PKSs), peptides by nonribosomal peptide synthetases (NRPSs), bacteriocins, terpenoids, shikimate-derived metabolites, aminoglycosides, and other natural products (5).
The identification and characterization of biosynthetic gene clusters have proved to be invaluable tools for the elucidation of the biosynthesis of secondary metabolites as well as a potentially rich source of information on cryptic metabolites encoded by silent biosynthetic pathways. Controlled genetic engineering of these biosynthetic gene clusters will allow the production of analogs by combinatorial biosynthesis. A critical requirement for such applications is the availability of the relevant biosynthetic gene clusters controlling the production of a secondary metabolite of interest as well as appropriate genetic systems for the in vivo manipulation of the corresponding genes in heterologous hosts. Furthermore, successful production of the desired products requires an optimum relationship of timing and flux between primary and secondary cellular metabolism, because all secondary metabolites are ultimately derived from primary metabolic building blocks and require an adequate source of energy as well as reducing equivalents derived from primary metabolism such as ATP and NAD(P)H.
There has been considerable recent interest in the development of engineered bacterial strains for the efficient heterologous production of secondary metabolites (6 –8). S. avermitilis, which is used for the industrial production of the important anthelmintic agent avermectin, has already proven to be a highly efficient producer of secondary metabolites. Because this strain is already optimized for the efficient supply of primary metabolic precursors and biochemical energy to support multistep biosynthesis, it is, therefore, an attractive host for the heterologous production of secondary metabolites. We now report on the construction of a versatile host for the efficient production of natural products by the controlled minimization of the genome of the S. avermitilis.
Results
Construction of Large-Deletion Mutants.
Comparative analysis of three taxonomically distinct Streptomyces genomes, S. avermitilis, S. coelicolor A3(2), and S. griseus, revealed a conserved core region of ~6.28–6.50 Mb in which the majority of the genes, including genes essential for growth, are conserved with a high degree of synteny (4). Among these three species, only the genome of S. avermitilis was asymmetric in structure. Although both S. coelicolor A3(2) and S. griseus have ~1 Mb subtelomeric regions at each end of their linear chromosomes located on genomes, the subtelomeric regions of S. avermitilis are of two different sizes; 2 Mb and 0.5 Mb are at the left and right chromosomal ends, respectively (2). These subtelomeric regions contain strain-specific genes as well as genes encoding secondary metabolite biosynthesis, and no essential genes were in these two regions. Deletion of the large left subtelomeric region of S. avermitilis would, therefore, not be expected to affect either growth or the primary metabolism that is essential for the supply of precursors for secondary metabolism.
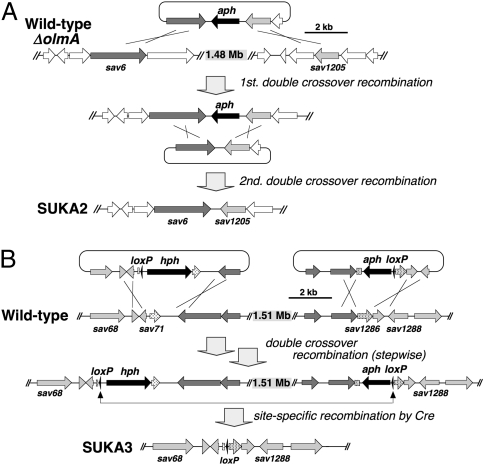
We used two complementary strategies to delete a >1.4-Mb segment from the left subtelomeric region of the S. avermitilis genome (Fig. 1). The first approach used general homologous recombination involving two homologous segments located near the left end of the genome and the 1.4-Mb region of the chromosome using the S. avermitilis ΔolmA mutant (Fig. 1A). The desired mutants should have a deletion of 1,487,159 bp spanning the region from sav6 to sav1205. This large deletion event turned out to have taken place at extremely low frequency, and almost all of the progeny that were generated by this homologous recombination strategy also contained irregular deletions. Only two correct deletion mutants, designated SUKA2 (Δ7,734–1,494,898 nt), were isolated from three trial experiments and were confirmed by clamped homogeneous electrical field (CHEF) electrophoresis and Southern hybridization analysis using sav7 and sav1204 genes as probes.
Fig. 1.
The strategy for construction of large-deletion mutants SUKA2 and SUKA3 of S. avermitilis. Detailed procedures are described in SI Materials and Methods.
The second, more successful approach involved site-specific recombination using Cre-loxP (Fig. 1B). Two loxP sequences were first introduced in the same orientation into the S. avermitilis wild-type strain at 79,454 nt and 1,595,564 nt, respectively, by stepwise homologous recombination. The desired deletion mutants were efficiently generated after induction of cre expression, and all 24 of the resultant progeny that were tested harbored the identical 1,516,020-bp deletion between sav70 and sav1287, which was confirmed by PCR (using primer pair, forward for 79,156–79,174 nt in sav70 and reverse for 1,595,769–1,595,787 nt in sav1287) and CHEF electrophoresis. These desirable large deletion mutants were designated as SUKA3 (Δ79,455–1,595,563 nt). All deletion mutants could grow on the minimum medium without any supplements, suggesting that no essential genes were in the large left subtelomeric region.
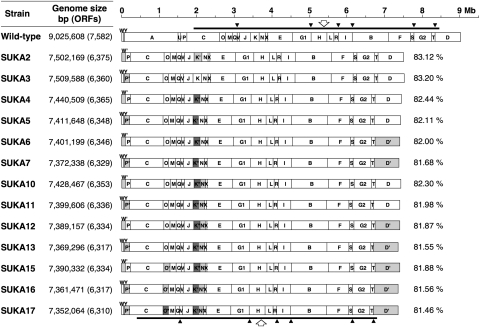
Avermectins and the related polyketides, the oligomycins and filipins, are normally major endogenous secondary metabolites produced by wild-type S. avermitilis. The gene clusters encoding both avermectin and filipin biosynthesis are located in the regions that have been removed in both large-deletion mutants, SUKA2 and SUKA3. In addition, the entire set of genes involved in oligomycin biosynthesis were deleted by site-specific recombination using Cre/loxP, giving rise two olm− derivatives, SUKA4 and SUKA5, derived from SUKA2 and SUKA3, respectively. The two prototype mutants, SUKA4 and SUKA5, could each be further modified by the deletion of specific regions of the genome or by the addition of useful marker genes (Fig. 2). HPLC-MS analysis of whole-broth EtOAc extracts of these mutants (Fig. S1) confirmed the absence of endogenous metabolites. The derived SUKA16 and SUKA17 mutants, from which the genes encoding the biosynthesis of the terpene compounds geosmin, neopentalenolactone, and carotenoid had been deleted, did not produce any of these endogenous terpene metabolites (Fig. S2).
Fig. 2.
AseI physical maps of S. avermitilis wild type and its large-deletion mutants. The genotype of large-deletion mutants were as follows: SUKA2, Δ(7,7341–1,494,898 nt) ΔolmA (Δ3,557,725–3,594,005 nt); SUKA3, Δ(79,455–1,595,563 nt)::loxP; SUKA4, SUKA2 Δ(olmA4-olmC)::mut-loxP (Δ3,536,700–3,634,730 nt); SUKA5, SUKA3 Δ(olmA4-olmC)::mut-loxP (Δ3,536,700–3,634,730 nt); SUKA6, SUKA2 Δ(8,886,025–8,925,414 nt)::loxP (containing cyp28 and fdxH); SUKA7, SUKA5 Δ(8,886,025–8,925,414 nt)::loxP; SUKA10, SUKA4 Δ(gap1-ptlL)::ermE (Δ3,745,502–3,758,936 nt); SUKA11, SUKA5 Δ(gap1-ptlL)::ermE (Δ3,745,502–3,758,936 nt); SUKA12, SUKA10 Δ(8,886,025–8,925,414 nt)::loxP; SUKA13, SUKA11 Δ(8,886,025–8,925,414 nt)::loxP; SUKA15, SUKA12 ΔgeoA::aadA; SUKA16, SUKA13 ΔgeoA::aadA; and SUKA17, SUKA13 Δ(2,633,682–2,641,994 nt)::mut-loxP. The right column indicates the percentage of the genome size compared with that of the wild type. Shaded boxes [D’; Δ(8,886,025–8,925,414 nt]::loxP, K’; ΔolmA, K”; Δ(olmA4-olmC)::mut-loxP, N’; Δ(gap1-ptlL)::ermE, O’; ΔgeoA::aadA, O” Δ(2,633,682–2,641,994 nt)::mut-loxP, P’; Δ(79,455–1,595,563 nt)::loxP and W’; Δ(7,7341–1,494,898 nt)] on the physical maps indicate the introduction of deletion(s). Thick bars at the top and bottom of the physical maps correspond to the central core region. Open arrows and filled triangles indicate the replication origin and 16S-23S-5S rRNA operon, respectively.
Transposons and insertion sequences (IS) that often promote deletions and rearrangements in the genome are associated with genetic instability. We identified 111 transposase genes including 46 IS-like structures in the S. avermitilis genome, of which 87 transposase genes were located in the left subtelomeric region and 13 were in the right subtelomeric region (Fig. S3). Most of these transposase genes were removed by the large-scale deletions. Thus, SUKA2 and SUKA3 had each lost 86 and 84 predicted transposase genes, respectively, corresponding to 78% of the total transposase genes in the wild-type genome.
Heterologeous Expression of Exogenous Gene Clusters for Secondary Metabolism.
Expression of the gene cluster from S. griseus Institute for Fermentation, Osaka (IFO) 13350 encoding biosynthesis of the aminoglycoside antibiotic streptomycin.
Analysis of the S. avermitilis genome revealed that although the microorganism harbored several biosynthetic gene clusters, no gene clusters for aminoglycoside antibiotic biosynthesis could be located. The recently completed analysis of the genome of the streptomycin producer S. griseus IFO13350 (4) indicated that at least 27 genes (sgr5914-sgr5940) are concerned with the regulation, self-resistance, and streptomycin biosynthesis. To move these genes to S. avermitilis, an ~41.2-kb fragment containing an entire set of genes for streptomycin biosynthesis was inserted into the integrating cosmid vector pKU465cos (SI Materials and Methods) to generate pSM1, which was used to transform both wild-type S. avermitilis and the large-deletion mutants, SUKA4 and SUKA5.
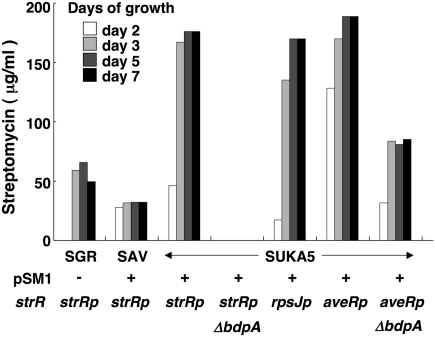
In contrast to wild-type S. avermitilis, which was sensitive to streptomycin (0.1 μg/mL), the transformants carrying pSM1 were resistant to more than 10 μg/mL of streptomycin (Fig. S4A). These transformants also produced streptomycin (Fig. S4B), suggesting that both the regulatory gene strR (sgr5931), whose gene product acts as positive regulator for the expression of the self-resistance gene (sgr5932), and the suite of genes for streptomycin biosynthesis were expressed in transformed strains of S. avermitilis and its derivatives, SUKA4 and SUKA5. The identity of the antibiotic produced by the tranformants was confirmed by direct comparison with authentic streptomycin. Some production media were examined for the optimal production of streptomycin in S. avermitilis/pSM1. Examination of a variety of production media revealed that maximum production of streptomycin by the pSM1 was obtained by culturing in an avermectin production medium rather than the usual streptomycin production medium, which is preferred for the production of streptomycin in S. griseus. The streptomycin productivity of SUKA5(pSM1) was higher than that of the S. avermitilis wild-type strain carrying pSM1 (Fig. 3). Because the deletion mutants lack the biosynthetic gene clusters for the principal endogenous natural products of S. avermitilis, the natural precursors and biochemical energy of the host are apparently efficiently used in the biosynthesis of streptomycin. Interestingly, the productivity of streptomycin in these S. avermitilis large-deletion mutants carrying pSM1 was higher than that of the wild-type strain of S. griseus under optimum production condition (Fig. 3).
Fig. 3.
Production of streptomycin in S. avermitilis wild type carrying pSM1, its large-deletion mutants SUKA5(pSM1), and the original producer S. griseus. All strains were grown in production medium at 28 °C. Quantitative analysis of streptomycin in the culture broth was carried out by the agar-diffusion method using B. subtilis as an indicator microorganism. The strRp, rpsJp, and aveRp indicate that strR was expressed by its own promoter or the promoters rpsJ and aveR in S. avermitilis, respectively. ΔbdpA indicates disruption mutants of bdpA (sav5261). Construction of pSM1 containing the entire streptomycin biosynthetic gene cluster of S. griseus is described in SI Materials and Methods. SGR, S. griseus IFO13350; SAV, S. avermitilis wild type.
The regulatory network for streptomycin biosynthesis in S. griseus has been elucidated in detail (9 –11). The expression of the positive regulatory gene, strR, in streptomycin biosynthesis is controlled by AraC-family regulatory protein AdpA, for which gene (sgr4742) expression is also regulated by A–factor receptor protein ArpA (SGR3731). Because no A-factor or its related compounds have been detected in S. avermitlis, the regulation of streptomycin biosynthesis in transformed S. avermitilis is of particular interest. Among 26 putative AraC family transcriptional regulatory genes in S. avermitilis, the predicted amino acid sequence of the gene product of sav5261 (bdpA) was highly similar to AdpA with 87% identity and 89% positive matches, suggesting that BdpA is most likely a homolog of AdpA. To examine the role of BdpA, a bdpA-disrupted mutant of SUKA5(pSM1) was constructed by homologous recombination. The resultant bdpA disruptant completely failed to produce streptomycin (Fig. 3), whereas streptomycin production could be restored to the bdpA disruptant by introduction of a copy of bdpA under control of its own promoter (Fig. S5), thereby suggesting that BdpA activates the transcription of strR. Interestingly, streptomycin formation was also restored to the bdpA disruptant by complementation with S. griseus adpA (Fig. S5). In like fashion, streptomycin production in the adpA disruptant of S. griseus was similarly restored by introduction of bdpA from S. avermitilis (Fig. S5), although it is not clear whether or not ArpA controls gene expression of the exogenous bdpA in S. griseus.
We also examined whether or not it was possible to control strR expression using heterologous promoter(s) in S. avermitilis. An strR expression cassette that used the rpsJ (sav4925) or aveR (sav935) (12) promoter was introduced using a second actinophage-based (φK38-1; AB251919) integrating vector pKU493hph, becauase the streptomycin biosynthetic gene cluster has been integrated using a φC31-based integrating vector. Although the productivity in SUKA5(pSM1) was improved slightly using the aveR promoter for the expression of strR, the rpsJ promoter had no effect on streptomycin productivity. However, introduction of an extra copy of strR under control of the aveR promoter restored streptomycin production in SUKA5 ΔbdpA(pSM1) (Fig. 3).
Expression in S. avermitilis of the S. clavuligerus American type culture collection (ATCC) 27064 gene cluster-encoding β-lactam antibiotic cephamycin C biosynthesis.
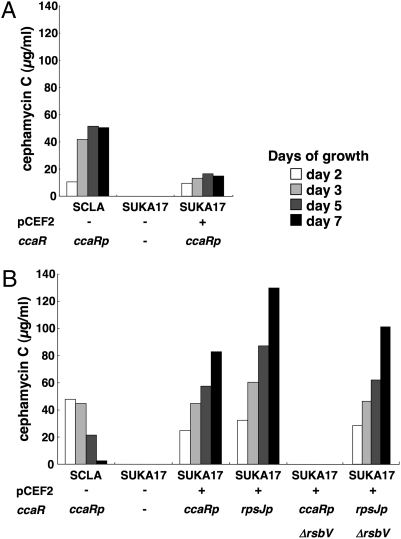
β-lactam antibiotics, which include penicillins and cephalosporins, are derived from tripeptides synthesized by nonribosomal peptide synthetases (NRPSs). At least eight different NRPS biosynthetic gene clusters are present in the S. avermitlis genome (2, 5), but none of the corresponding peptide-derived metabolites have been detected in fermentation cultures. It has not yet been confirmed whether or not any of the presumptive holo-NRPS enzymes can actually catalyze peptide-chain elongation or if the apo-NRPS proteins are converted to the corresponding active holo-NRPS enzymes by the endogenous 4′-phosphopantetheinyl transferase (PptA) in S. avermitilis. In the biosynthesis of cephamycin C, the tripeptide L-δ-(ε-aminoadipoyl)-L-cysteinyl-D-valine (ACV) is synthesized under control of a giant polypeptide NRPS (ACV synthetase) by condensation of three amino acids, L-α-aminoadipate, L-cysteine, and L-valine (13 –15). We, therefore, chose to examine heterologous expression of the gene cluster for cephamycin C biosynthesis in S. avermitilis as a model for NRPS function in this host. The full set of genes encoding cephamycin C biosynthesis has been located within a 35-kb continuous region in the S. clavuligerus chromosome (15 –18). We isolated two cosmid clones containing the cephamycin C biosynthetic gene cluster from an S. clavuligerus genomic library. The transformants, SUKA17(pCEF2), were examined to find the appropriate culture conditions to support cephamycin C production. Cephamycin C production by SUKA17(pCEF2) was first examined with the same starch asparagine-based medium (SA) used for cephamycin C production in the original producer S. clavuligerus. Although SUKA17(pCEF2) cultured in SA medium produced mainly cephamycin C, as determined by HPLC-MS analysis, the productivity was lower than that of the original producer, S. clavuligerus (Fig. 4). Interestingly, cephamycin C productivity in SUKA17(pCEF2) was improved by cultivation in the same avermectin production medium that had also been used for streptomycin production of SUKA5(pSM1), giving levels higher than that observed in the native producer S. clavuligerus. Furthermore, the production of cephamycin C in S. clavuligerus declined after 3 days, whereas the production in SUKA17(pCEF2) continued during at least 7 days of cultivation (Fig. 4).
Fig. 4.
Production of cephamycin C in S. avermitilis SUKA17(pCEF2) and in the original producer S. clavuligerus. Microorganisms were grown in the original cephamycin C production medium (A) and avermectin production medium (B) at 28 °C. The titer of cephamycin C was measured by the agar-diffusion method using C. terrigena as an indicator microorganism. SCLA indicates the original producer S. clavuligerus. The ccaRp indicates that the transcriptional regulatory gene ccaR was expressed by its own promoter, and rpsJp is as defined in Fig. 3. ΔrsbV indicates a rsbV-null mutant. Construction of pCEF2 containing the entire cephamycin C biosynthetic gene cluster of S. clavuligerus is described in SI Materials and Methods.
The ccaR gene product from the cephamycin C biosynthetic gene cluster, which activates the expression of genes involved in cephamycin C biosynthesis (16, 19), is similar to strR in streptomycin biosynthesis. We, therefore, examined cephamycin C productivity in S. avermitilis by introducing conjugation of an extra copy of ccaR expressed from the alternative rpsJ promoter. The resulting exoconjugants of SUKA17(pCEF2) produced more cephamycin C than the parent strain SUKA17(pCEF2) (Fig. 4), thereby confirming the role of CcaR in the regulation of cephamycin C biosynthesis in recombinant S. avermitilis. It has also been reported that the expression of ccaR in S. clavuligerus was dependent on the anti-sigma factor antagonist encoded by bldG (20). It was, therefore, of some interest whether or not the corresponding bldG ortholog in S. avermitilis, rsbV (sav4614), could control the expression of ccaR. In fact, the requisite rsbV-null mutant of S. avermitilis SUKA17(pCEF2), prepared by homologous recombination, failed to produce cephamycin C (Fig. 4) and express ccaR when monitored by real-time PCR analysis; this suggests that the S. avermitilis RsbV protein activates the expression of ccaR harbored in the cephamycin C biosynthetic gene cluster. Cephamycin C production was restored to the rsbV-null mutant by introduction of extra copy of ccaR expressed under control of the rpsJ promoter (Fig. 4).
Expression in S. avermitilis of the S. platensis Mer-11107 biosynthetic gene cluster for the pladienolide antitumor macrocyclic lactones.
The major endogenous secondary metabolites of wild-type S. avermitilis are the polyketide compounds (the avermectins, filipins, and oligomycin), each of which is synthesized by a modular PKS. This microorganism has the ability to synthesize polyketide compounds at the industrial production level, making S. avermtilis a particularly attractive host for the heterologous expression of modular PKS genes. Pladienolides, metabolites of S. platensis Mer-11107 (21, 22), are antitumor macrocyclic polyketides with an unusual mode of action (23). The gene cluster for pladienolide biosynthesis has been cloned and characterized (24). We used a recombinant BAC clone (pPLD30) carrying the 75-kb entire gene cluster for pladienolide biosynthesis to introduce the pathway into both wild-type S. avermtilis and the SUKA5 by conjugation. Unfortunately, none of the exoconjugants produced pladienolides in either pladienolide or avermectin production media (Fig. S6). Real-time PCR analysis revealed that pldR, the transcriptional activator for the pladienolide biosynthetic genes, was not expressed, suggesting that the appropriate regulator proteins to activate the pldR expression might not be present in the heterologous S. avermitilis host. Indeed, when we introduced an extra copy of pldR under control of the ermE promoter, we could observe production of pladienolide B and 18,19Δ-pladienolide B along with other, currently unidentified, pladienolide components. Pladienolide production in SUKA5 carrying both pPLD30 and ermEp::pldR was remarkably improved to levels more than 20-fold higher than that of the wild-type S. avermitilis host carrying only pPLD30 (Fig. S6). These observed differences might be explained by the likely competition between pladienolide biosynthesis and avermectin biosynthesis for common acyl-CoA precursors that serve as building-block units for their respective polyketide backbones. We have previously reported similar observations for the balance of avermectin and oligomycin production in wild-type S. avermitilis. Thus, a biosynthetically blocked mutant of aveA1 encoding the avermectin PKS in which avermectin production was completely abolished displayed a greater than 10-fold increase in oligomycin production (25).
Expression of gene-encoding plant sesquiterpene synthase.
We have recently examined heterologous expression of actinomycete monoterpene synthase genes in S. avermitilis (26). We have now extended these observations to the expression plant terpene synthase genes in S. avermitilis. A synthetic gene encoding amorpha-4,11-diene synthase (ads) of Artemisia annua (27, 28), in which the codon usage was optimized for expression in S. avermitilis, was expressed in S. avermitilis under control of the rpsJ promoter. The requisite farnesyl diphosphate precursor was provided by introducing a copy of the native of S. avermitilis ptlB gene (sav2997) encoding farnesyl diphosphate synthase downstream of the ads gene. The synthetic operon was introduced into SUKA17 by transformation. A single major product with mass [M+] m/z 204 corresponding to C15H24 was detected in the n-hexane extract of transformants carrying ads, but SUKA17 carrying only vector without ads and ptlB did not produce any terpenoid compounds (Fig. S7). Both the fragmentation pattern and proton-NMR spectrum of the product that eluted at 10.25 min in GC-MS were identical to those of authentic amorpha-4,11-diene (29). The productivity was estimated to 10–30 μg/mL based on the observed GC-MS TIC (total ion chromatogram) value. Thus, S. avermitilis could efficiently produce a plant terpenoid using a synthetic gene modified for S. avermitilis-specific codon usage.
Discussion
The terminally inverted repeats found at the end of the linear chromosome ends are 21,653 bp in S. coelicolor A3(2) and 132,913 bp in S. griseus. Large DNA rearrangements involving gene duplication, elimination, and acquisition were frequently observed in or near the long-terminal inverted repeats (30, 31). The chromosomal instabilities may be important driving forces in Streptomyces evolution. However, because the terminal inverted repeats of S. avermitilis are a mere 49 bp in size and the imperfect inverted repeats of 167 bp, recombination between these very short-terminal inverted repeats may rarely occur. In fact, under stressed conditions such as high temperature or hypertonic condition, the growth characteristics of S. avermitilis are more stable than those of both S. coelicolor A3(2) and S. griseus (evaluated by the appearance of bald mutants), suggesting that having the shortest terminal inverted repeats might allow for the maintenance of a stable phenotype in S. avermitilis. Because the large-scale deletions that we introduced into S. avermitilis also eliminated 78% of the putative transposase genes, this would be expected to further improve genetic stability in these mutants. Enhancement of the already intrinsic genetically stable characteristics of S. avermitilis, therefore, make it especially suitable as a source of endogenous natural products and as a host for the heterolgous production of secondary metabolites derived from exogenous biosynthetic genes.
We have shown the feasibility of using engineered S. avermitilis as a heterologous host by the efficient expression of the intact gene clusters for both streptomycin and cephamycin C biosynthesis at levels that even exceed those of each metabolite in the original native-producing strains. Moreover, because the S. avermitilis deletion mutants no longer produced the major endogenous secondary metabolites, primary metabolism seemed to be efficiently exploited to generate precursors for streptomycin or cephamycin C biosynthesis in the deletion mutants carrying pSM1 or pCEF2, respectively.
Although an understanding of the higher level regulatory system that controls the expression of pathway-specific regulatory genes for individual biosynthetic gene clusters should be essential to control and improve the secondary metabolite production, only a few cases have been studied to date (9, 10, 20). The results with the bdpA disruptants of S. avermitilis suggest that the BdpA protein, which is an AdpA homolog, has the ability to activate the expression of strR. In S. griseus, the expression of adpA is also known to be regulated by A–factor, the prototype of the important family of γ-butyrolactone compounds (10). The role of butyrolactones has not been examined yet in S. avermitilis, and it is not known if such autoregulators are involved in bdpA expression in S. avermitilis. From our results, it seems that the higher level regulatory systems for streptomycin in S. griseus and cephamycin C biosynthesis in S. clavuligerus are compatible with antibiotic expression in S. avermitilis. However, the pladienolide polyketides, which are synthesized by modular PKSs, could not be produced by S. avermitilis carrying an intact gene cluster for pladienolide biosynthesis. The specific regulator underlying the expression of pldR in S. platensis, however, remains unknown, and S. avermitilis does not seem to possess the requisite orthologs. Similar observations have been made with the low-level production of iso-migrastatin using SUKA4 and SUKA5 as heterologous hosts (7).
Active NRPS and PKS holo-enzymes must be generated from the nascent translated apo-polypeptides by modification of the constituent peptidyl carrier protein (PCP) or acyl carrier protein (ACP) domains mediated by a suitable PptA. There are no pptA genes located in either the cephamycin C or pladienolide biosynthetic gene clusters. The required posttranslational modifications might be carried out by one or more of the endogenous PptAs (SAV1748, SAV2905, SAV3193, and SAV3637) of S. avermitilis. Moreover, because the reactions of cytochrome P450, such as the hydroxylation at C6 catalyzed by PldB in pladienolide biosynthesis (24), require the electron-transport proteins, ferredoxin and ferredoxin reductase, these redox partners that support pladienolide biosynthesis in S. avermitilis are apparently supplied by the endogenous gene product of fdxB-G and fprB-F.
The large-deletion mutants of S. avermtilis have been shown to be versatile and effective hosts for the expression of heterologous gene clusters governing the production of a variety of secondary metabolites, including aminoglycosides, nonribosomal peptides, polyketides, and terpenes. Although some Streptomyces strains have previously been examined for use as model heterologous hosts, they have not proven to be sufficiently flexible or vigorous producers of foreign secondary metabolites (6 –8). S. avermitilis is also attractive as a host for heterologous expression, because the availability of the complete genome sequence makes possible analysis of global transcription using DNA microarrays. The large-deletion mutants of S. avermitilis constructed here would be applicable to use as heterologous hosts not only for the production of exogenous secondary metabolites derived from cultivable and uncultivable microorganisms but also for the production of unnatural metabolites by combinatorial biosynthesis using two or more metabolic pathways.
Materials and Methods
Bacterial Strains and Plasmid Vectors.
S. griseus IFO 13350, S. clavuligerus ATCC 27064, Bacillus subtilis ATCC 6633, and Comamonas terrigena ATCC 8461 were obtained from the culture collection of the RIKEN Bioresource Center. Integrating vectors, pKU465cos, pKU503, pKU493hph, and pKU493aad (Fig. S8), were used for cloning gene clusters for secondary metabolite biosynthesis and expression of specific transcriptional regulator genes. A small vector pRED (cassette vector for λ red-mediated recombination) (Fig. S8) was used for the construction of the gene-disruption cassette. pKU250, pKU257, and pGM160Δaac(3)I::oriT, for which replication in S. avermitilis was unstable (Fig. S8), were for gene disruption of the Streptomyces genome by allelic replacement. Cloning, transformation, conjugation, and Escherichia coli strains used were described previously (26, 32). The mutant loxP sequence (mut-loxP) (33) was used for the deletion cassettes. S. avermitilis cosmid clones used are listed at http://avermitilis.ls.kitasato-u.ac.jp.
Cultivation for Secondary Metabolite Production.
Conditions for seed culture and avermectin production medium were described previously (34). Streptomycin, cephamycin C, and pladienolide production media of the original strains are described in refs. 35, 11, and 24, respectively.
Supplementary Material
Acknowledgments
We thank H. Takano and K. Ueda of the College of Bioresource Sciences, Nihon University for providing cosmid clones SGR4C6 and SGR1A7. We also thank K. Machida, A. Arisawa, and T. Tsuchida of Mercian Corporation for provision of a pladienolide-producing S. platensis Mer-11107. We are grateful to I. Saito of The Institute for Medical Sciences, University of Tokyo, for providing pxCANCre (cre resource) and pULwL (loxP resource) and E. coli Genetic Stock Center at Yale University for E. coli BW25141, pKD46, pKD78, and pKD119. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (H.I.). This work was also supported by the Scientific Research from the Japan Society for the Promotion of Science Grants-in-Aid 20710154 (to M.K.) and 20310122 (to H.I.) and by the National Institutes of Health Grant GM30301 (to D.E.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914833107/DCSupplemental.
References
- 1.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis . Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 3.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi Y, et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez A, et al. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol. 2004;70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Z, et al. Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorg Med Chem. 2009;17:2147–2153. doi: 10.1016/j.bmc.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeifer B, Hu Z, Licari P, Khosla C. Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl Environ Microbiol. 2002;68:3287–3292. doi: 10.1128/AEM.68.7.3287-3292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. AdpA, a central transcriptional regulator in the A–factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus . Biosci Biotechnol Biochem. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 10.Tomono A, Tsai Y, Yamazaki H, Ohnishi Y, Horinouchi S. Transcriptional control by A–factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus . J Bacteriol. 2005;187:5595–5604. doi: 10.1128/JB.187.16.5595-5604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A–factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: Identification of a target gene of the A–factor receptor. Mol Microbiol. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 12.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis . Appl Microbiol Biotechnol. 2009;82:1089–1096. doi: 10.1007/s00253-008-1850-2. [DOI] [PubMed] [Google Scholar]
- 13.Coque JJR, de la Fuente JL, Liras P, Martín JF. Overexpression of the Nocardia lactamdurans α-aminoadipyl-cysteinyl-valine synthetase in Streptomyces lividans. The purified multienzyme uses cystathionine and 6-oxopiperidine 2-carboxylate as substrates for synthesis of the tripeptide. Eur J Biochem. 1996;242:264–270. doi: 10.1111/j.1432-1033.1996.0264r.x. [DOI] [PubMed] [Google Scholar]
- 14.Coque JJR, Liras P, Láiz L, Martín JF. A gene encoding lysine 6-aminotransferase, which forms the β-lactam precursor α-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans . J Bacteriol. 1991;173:6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobin MB, et al. Localization of the lysine ε-aminotransferase (lat) and δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine synthetase (pcbAB) genes from Streptomyces clavuligerus and production of lysine ε-aminotransferase activity in Escherichia coli . J Bacteriol. 1991;173:6223–6229. doi: 10.1128/jb.173.19.6223-6229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander DC, Jensen SE. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J Bacteriol. 1998;180:4068–4079. doi: 10.1128/jb.180.16.4068-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander DC, Brumlik MJ, Lee L, Jensen SE. Early cephamycin biosynthetic genes are expressed from a polycistronic transcript in Streptomyces clavuligerus . J Bacteriol. 2000;182:348–356. doi: 10.1128/jb.182.2.348-356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Llarena FJ, Rodríguez-García A, Enguita FJ, Martín JF, Liras P. The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of α-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus . J Bacteriol. 1998;180:4753–4756. doi: 10.1128/jb.180.17.4753-4756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Tahlan K, Kaziuk TL, Alexander DC, Jensen SE. Transcriptional and translational analysis of the ccaR gene from Streptomyces clavuligerus . Microbiology. 2004;150:4137–4145. doi: 10.1099/mic.0.27245-0. [DOI] [PubMed] [Google Scholar]
- 20.Bignell DRD, Tahlan K, Colvin KR, Jensen SE, Leskiw BK. Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob Agents Chemother. 2005;49:1529–1541. doi: 10.1128/AAC.49.4.1529-1541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai T, et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. I. Taxonomy, fermentation, isolation and screening. J Antibiot (Tokyo) 2004;57:173–179. doi: 10.7164/antibiotics.57.173. [DOI] [PubMed] [Google Scholar]
- 22.Mizui Y, et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 23.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 24.Machida K, et al. Organization of the biosynthetic gene cluster for the polyketide antitumor macrolide, pladienolide, in Streptomyces platensis Mer-11107. Biosci Biotechnol Biochem. 2008;72:2946–2952. doi: 10.1271/bbb.80425. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka Y, et al. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol. 2009;75:4919–4922. doi: 10.1128/AEM.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang YJ, Song SH, Park SH, Kim SU. Amorpha-4,11-diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in artemisinin biosynthesis. Arch Biochem Biophys. 2000;383:178–184. doi: 10.1006/abbi.2000.2061. [DOI] [PubMed] [Google Scholar]
- 28.Wallaart TE, Bouwmeester HJ, Hille J, Poppinga L, Maijers NC. Amorpha-4,11-diene synthase: Cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin. Planta. 2001;212:460–465. doi: 10.1007/s004250000428. [DOI] [PubMed] [Google Scholar]
- 29.Picaud S, et al. Amorpha-4,11-diene synthase: Mechanism and stereochemistry of the enzymatic cyclization of farnesyl diphosphate. Arch Biochem Biophys. 2006;448:150–155. doi: 10.1016/j.abb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Chen CW, Huang CH, Lee HH, Tsai HH, Kirby R. Once the circle has been broken: Dynamics and evolution of Streptomyces chromosomes. Trends Genet. 2002;18:522–529. doi: 10.1016/s0168-9525(02)02752-x. [DOI] [PubMed] [Google Scholar]
- 31.Choulet F, et al. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol Biol Evol. 2006;23:2361–2369. doi: 10.1093/molbev/msl108. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda H, Kotaki H, Omura S. Genetic studies of avermectin biosynthesis in Streptomyces avermitilis . J Bacteriol. 1987;169:5615–5621. doi: 10.1128/jb.169.12.5615-5621.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 34.Cane DE, He X, Kobayashi S, Omura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J Antibiot (Tokyo) 2006;59:471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- 35.Paradkar AS, Jensen SE. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus . J Bacteriol. 1995;177:1307–1314. doi: 10.1128/jb.177.5.1307-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.